Abstract
In recent years, the need to identify molecular markers characterized by high sensitivity and specificity in detecting and monitoring early and colorectal cancer lesions has increased. Up to now, none of the markers or panels of markers analyzed have met the rigorous standards required of a screening program. The important discovery of circulating nucleic acids in biological fluids has aroused intense scientific interest because of their usefulness in malignant and non malignant diseases. Over time, their yield and stability have been identified and compared with other “standard” biomarkers. The analysis of circulating DNA from blood and stool is a relatively simple and non-invasive procedure, representing a very attractive marker to detect genetic and epigenetic mutations and to monitor disease progression. A correlation between blood and stool biomarkers could also help to enhance currently available diagnostic approaches. However, various processing and analytic problems need to be resolved before such an approach can be applied in clinical practice.
Keywords: Colorectal cancer, Circulating DNA, Circulating RNA, Early diagnosis, DNA integrity
Core tip: Although the importance of circulating free DNA is widely recognized, numerous studies evaluating its presence in blood and stool samples have reported analytic variability and non conforming approaches. Nonetheless, circulating free DNA has shown high potential as a biomarker for the early non-invasive detection of cancer and for monitoring disease progression. Population studies are now needed to confirm its usefulness for colorectal cancer diagnosis.
HISTORY OF CIRCULATING NUCLEIC ACIDS
Circulating nucleic acids in the plasma of healthy and diseased individuals were identified[1] a few years before the discovery of the double helical structure of DNA[2]. Almost 20 years later, circulating DNA was identified in the serum and plasma of subjects with systemic lupus erythematosus[3]. Around the same time, circulating DNA was identified in other diseases characterized by tissue destruction such as hepatitis, metastatic carcinoma and miliary tuberculosis, suggesting that serum DNA might originate from endogenous tissue breakdown[4]. In 1977, levels of circulating DNA in the serum of individuals with different types of cancer were found to be related to response to radiotherapy treatment. In particular, for the first time, circulating DNA was more accurately quantified using a sensitive radioimmunoassay based on anti-DNA antibodies obtained from lupus erythematosus patients[5]. This quantification revealed that increased or high DNA circulating levels were mainly present in patients characterised by a lack of response to treatment.
The presence of extractable amounts of DNA in the plasma of cancer patients was also identified, suggesting that circulating DNA may be shed from tumours[6]. Other researchers reported findings of KRAS and NRAS gene mutations from the primary tumor in the plasma and serum of individuals with cancer, providing clear evidence of the origin of circulating DNA from tumors[7-14]. Extracellular nucleic acids, present in different body fluids such as plasma, serum, bronchial lavage, urine and faecal fluids, have aroused the interest of the scientific community in recent years[15,16] representing a valid biomarker for the early, non-invasive detection of cancer or for the monitoring of disease progression. Early diagnosis is fundamental to reduce morbidity and mortality, especially as patients diagnosed at early stages show long-term survival[17].
Unfortunately, the quantity of circulating free DNA in these body fluids is usually low and its isolation remains a challenge. However, rapid technological advances have led to an improved sensitivity and specificity for the detection of cell-free nucleic acids, opening up new possibilities for the non-invasive detection and monitoring of various malignant diseases[15].
ORIGIN OF CIRCULATING FREE DNA
Circulating free DNA is a double-stranded molecule of low molecular weight which, although mainly fragmented in 70-200 base pairs (bp), also has sections up to 21 kilobases in length[18]. In healthy individuals, apoptosis and necrosis of lymphocytes and other nucleated cells are mainly involved in the release of circulating nucleic acids into the blood. Apoptosis leads to DNA degradation in which chromosomal DNA is first cleaved into large fragments (50-300 kb) and then into multiples of nucleosomal units (180-200 bp)[19]. The contents of apoptotic cells are rapidly ingested by phagocytes or neighbouring cells[20] and the DNA is consequently completely digested by DNase II in lysosomes[19]. Thus, DNA fragments released by apoptosis may be removed before entering the circulation[19,20]. However, apoptotic DNA is probably the primary source of circulating nucleic acids, especially if we take into account the fact that normal plasma DNA on electrophoresis exhibits band sizes equivalent to whole-number multiples of nucleosomal DNA (185-200 bp)[21]. In cancer patients, the origin of circulating nucleic acids remained unknown for many years. Although increased circulating free DNA levels cannot be regarded as specific to cancer, different size distributions have been observed in cancer patients[22,23]. Currently, the hypothesis on the endogenous origin of circulating DNA proposed by Tan et al[3] is widely accepted[4]. Initially, circulating DNA was thought to be a derivative of increased and abnormal apoptotic pathways in cancerous lesions[24,25] because of its ladder pattern revealed by gel electrophoresis similar to the one shown by apoptotic cells[26,27]. However, it must be remembered that apoptosis is a mechanism apparently lost by proliferating cancer cells and that its restoral is highly problematic[9,24,27]. Another hypothesis is that circulating DNA derives from “micrometastatic” tumor cells shed in the circulation. However, some authors reported that the amount of DNA isolated from the plasma of cancer patients was very high and did not correspond to the number of cancer cells present in the circulation[28,29]. Tumor necrosis is thought to be related to high amounts of DNA fragments found in the plasma of patients with large or advanced/metastatic tumors, suggesting that this mechanism may be related to circulating DNA[5,30,31]. However, other pathways could also be involved[4], and probably abnormal DNA degradation or secretion mechanisms may lead to increased DNA levels and differing DNA fragmentation, contributing to the presence of high levels of circulating free DNA[24,32] (Figure 1).
Figure 1.
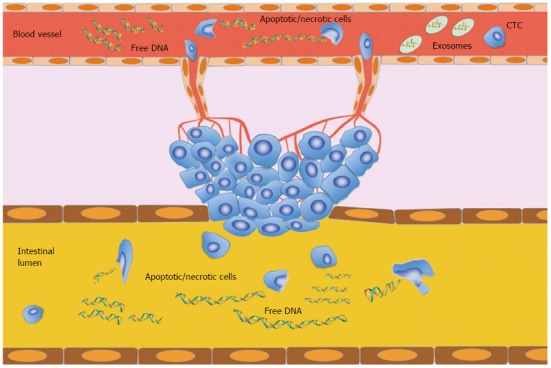
Hypothesis for circulating free DNA development. The primary tumor releases cells into the bloodstream or intestinal lumen. In healthy individuals, apoptosis and necrosis are the main pathways linked to cell degradation and, consequentially, to DNA fragmentation. In cancer patients, in addition to the aforementioned necrosis and apoptosis, there would seem to be abnormal mechanisms of DNA degradation or secretion that increase levels and fragmentation of DNA. CTC: Circulating tumor cells.
ORIGIN OF CIRCULATING FREE RNA
Less is known on the origin of circulating free RNA[33]. More than 25 years ago, RNA in proteolipid complexes were first identified in the serum of cancer patients[34]. Initially, circulating RNA was found in the serum of healthy individuals and patients with melanoma, breast cancer and hepatocellular carcinoma[35-37]. Numerous studies have reported that specific RNA is present in the plasma of patients with a variety of cancers and that these molecules are more stable than expected[38], suggesting that free circulating RNA is probably protected by vesicles or vesicle-like structures. Apoptosis would also appear to be involved in the release of circulating free RNA, and the binding of proteins or phospholipids may explain the resistance to RNase degradation in the bloodstream[39,40]. Moreover, mRNA and miRNA are found in particles such as exosomes released into the bloodstream, which may help to preserve these nucleic acids in the blood and increase the amount in circulation. In fact, it has been seen that the higher levels of mRNA identified in cancer patients than in healthy individuals are mainly associated with exosome fraction[37].
EARLY DIAGNOSIS OF COLORECTAL CANCER
Colorectal cancer (CRC) incidence and mortality rates vary markedly around the world. However, rates are substantially higher in males than in females[41], representing the third most commonly diagnosed cancer in males and the second in females[42]. CRC is caused by a molecular alteration in the epithelial cells of the colon, specifically, in proto-oncogenes and tumor suppressor genes such as APC, KRAS, SMAD 2/4 and p53. Epigenetic alterations have gained recognition as a key mechanism in colorectal carcinogenesis. In particular, hypermethylation of CpG islands present in gene promoter sequences leads to the inactivation of tumor suppressor genes. The vast majority of tumors (about 50%-80%) present chromosomal instability, while a smaller fraction (10%-15%) is characterized by microsatellite instability (MSI). CRCs with hypermethylation changes in numerous different CpG-rich DNA regions are defined as showing CpG island methylation phenotype (CIMP). CIMP-positive cancer also seems to be associated with MSI and BRAF mutations[43,44]. Conversely, hypomethylation of specific sequences may decrease the fidelity of chromosomal segregation[45], suggesting that it could be involved in the chromosomal instability phenotype[46]. DNA methylation changes probably cause adenomatous precursor lesions to progress into malignant tumors.
The importance of screening tools to identify early stage CRC is acknowledged worldwide. Colonoscopy is currently considered to be the “gold standard” for CRC screening. However, it is estimated that less than 60% of eligible individuals over 50 years of age have undergone this test for various reasons, the main one being the invasiveness of the procedure[47]. The immunochemical fecal occult blood test (iFOBT), a non invasive screening CRC approach that uses antibodies against human globulin, has reduced CRC mortality by 15%-33%[48,49]. However, this test is characterized by frequent false-negative and false-positive results, and its sensitivity in detecting precursor lesion such as adenomas is very low (10%-20%)[50].
NUCLEIC ACIDS IN SERUM AND PLASMA
Numerous studies have been published on circulating free DNA in both plasma and serum of different tumor types including colorectal[51-60]. A summary of the most important studies on colorectal cancer patients is shown in Table 1.
Table 1.
Summary of studies that have tested DNA integrity and genetic alteration markers in blood samples in colorectal cancer
| Study | Biomarker(s) | Methods | Case analyzed | Main results | |
| Flamini et al[17], 2006 | Serum cfDNACEAcfDNA and CEA | qRT PCR | CRC: 75HD: 75 | Sensitivity | Specificity |
| 81% | 73% | ||||
| 39% | 97% | ||||
| 88% | 71% | ||||
| Frattini et al[51], 2006 | Plasma cfDNA | DNA Dipstick Kit | CRC: 70HD: 20 | CRC (T0) mean value DNA: 495.7 ng/mL | |
| CRC (FU at 4 mo) mean value DNA: 170.6 ng/mL | |||||
| CRC (FU at 10 mo) mean value DNA: 240.9 ng/mL | |||||
| CRC (FU at 10 mo)(DFP) mean value DNA: 136.2 ng/mL | |||||
| CRC (FU at 10 mo)(RP) mean value DNA: 694.4 ng/mL | |||||
| HD: mean value DNA 10.3 ng/mL | |||||
| Altered in only 37% of CRC patients | |||||
| Umetani et al[61], 2006 | Plasma cfDNASerum cfDNA | ALU qRT PCR | Breast, Colorectal, Thyroid Cancer: 22Thyroid A: 2 | Mean ± SD value plasma cfDNA: 180 ± 150 pg/μL | |
| Mean ± SD value serum cfDNA: 970 ± 730 pg/μL | |||||
| Umetani et al[64], 2006 | Absolute serum DNAIntegrity serum DNA Alu repeats (2 sites) | ALU qRT PCR | CRC: 32HD: 51 | CRC (I-II) mean absolute value DNA: 1.63 ng/μL | |
| CRC (I-II) mean integrity DNA: 0.22 ng/μL | |||||
| CRC (III-IV) mean absolute DNA: 1.73 ng/μL | |||||
| CRC (III-IV) mean integrity DNA: 0.22 ng/μL | |||||
| Boni et al[52], 2007 | Plasma cfDNACEA | qRT PCR | CRC: 67HD: 67 | CRC: Mean value DNA 57.93 ng/mL | |
| HD: Mean value DNA 0.85 ng/mL | |||||
| Evaluated in only 47% of patients | |||||
| Danese et al[65], 2010 | Serum cfDNACEA | RT-PCR | CRC: 118P: 49HD: 26 | Sensitivity | Specificity |
| 83% | 92% | ||||
| 36% | 100% | ||||
| Mead et al[31], 2011 | Plasma cfDNA (4 DNA markers) CEA | PCRELISA | CRC: 24P: 26HD: 35 | AUC: cfDNA: 0.81 | |
| AUC (CEA and cfDNA): 0.855 | |||||
| P: Sensitivity: 83%; Specificity 72% | |||||
| Czeiger et al[66], 2011 | Serum cfDNA | Fluorometric Assay | CRC: 38HD: 34 | Sensitivity | Specificity |
| 42% | 94% | ||||
| da Silva Filho et al[67], 2013 | Serum cfDNAAlu repeats (2 sites) | qPCR | CRC not operated on: 27CRC operated on: 33HD: 30 | CRC not operated on: 0.08-62.10 pg/μL | |
| CRC operated on: 0.01-186.7 pg/μL | |||||
| HD: 0.01-26.11 pg/μL | |||||
| Mean value range (according to different Alu analysis) | |||||
cfDNA: Cell-free circulating DNA; HD: Healthy donors; A: Adenomas; P: Polyps; T0: Time 0; FU: Follow up; DFP: Disease free patients; RP: Recurrence patients; AUC: Area under the receiver-operating characteristic (ROC) curve; qRT PCR: Quantitative real time polymerase chain reaction; ELISA: Enzyme-linked immunosorbent assay.
It is known that serum contains a higher amount of free circulating DNA than plasma. Different hypotheses for this have been put forward, e.g., an unequal distribution of DNA during separation from whole blood[61]. However, differing levels of circulating free DNA have been observed in experiments using serum and plasma, and the optimal material to process remains open to debate[31].
The most important serum markers for CRC detection are carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9) and tissue inhibitor of matrix metalloproteinases (TIMP)-1. However, CEA is currently the only marker used for prognosis, follow-up and monitoring of disease status. Although high levels of serum CEA are often associated with an increased risk of recurrence and poor prognosis. It is not uncommon to detect normal levels in patients with advanced CRC or early stage patients who subsequently develop recurrence or distant metastases[62]. Thus, the use of CEA assessment for the early diagnosis and monitoring of CRC is limited by relatively poor sensitivity and specificity[63]. In 2006, Frattini et al[51] quantified circulating DNA plasma levels of 70 patients submitted to surgery for primary CRC at the time of surgery and during follow-up and comparing this marker with CEA. It was found that circulating DNA levels in all patients at the time of surgery were about 25-fold higher those of 20 healthy donors. Of note, only 37% of patients had altered CEA levels[51]. Moreover, Boni et al[52] observed a statistically significant difference in plasma circulating DNA levels between healthy donors and CRC patients. A significant difference was also observed between circulating DNA and CEA values in CRC cases, calculated in only 47% of cases[52].
Sensitivity and specificity of serum free circulating DNA and CEA values were also examined in other studies, confirming high levels of circulating free DNA in CRC patients with respect to healthy individuals. Conversely, sensitivity values of CEA were around 35%, even when specificity was higher[17,65]. Sensitivity and specificity values increased to 88% and 71%, respectively, considering the two markers in combination[17]. Moreover, Frattini et al[51] observed that circulating free DNA levels decreased progressively during follow-up in disease-free patients but increased in those who relapsed. This finding is in line with the first observation on circulating free DNA made by Leon et al[5] who suggested that the decrease in free DNA in cancer patients before and after treatment may be due to the therapy’s inhibitory effect on the proliferation of cancer cells. Conversely, in the early stages of cancer, when little cell death occurs, circulating DNA may already be present in higher than normal concentrations.
Analysing two sites of Alu repeats by a quantitative PCR approach, Umetani et al[64] found that serum DNA integrity values were higher in 32 CRC patients than in controls. Interestingly, the serum free circulating DNA concentration was 4- to 6-fold higher in patients than in healthy individuals. This ALU sequencing approach would also appear to be able to discriminate between healthy individuals, CRC patients who have undergone surgery and those who have not been submitted to surgical treatment[67].
These studies revealed a higher sensitivity of circulating DNA then CEA quantitation in CRC patients. Serum CA19-9, CA72-4 and TIMP-3 are currently being evaluated for screening purposes[68-70], while serum methylation markers such as TAC1, SEPT9 and EYA4 are under investigation as biomarkers for adenoma and early CRC detection[71]. In particular, SEPT9 has been investigated with the aim of improving detection rates of malignant lesion precursors[72], but sensitivity values do not appear to be better than those obtained with stool[73]. Ahlquist et al[73] reported higher SEPT9 values than those of the stool DNA multimarker test only for stage IV CRC. Available data do not, therefore, justify the application of SEPT9 as a single biomarker for the detection of premalignant and early malignant lesions in a screening population. The analysis of paired samples, i.e., stool DNA multimarker test and plasma SEPT 9, showed a sensitivity of 82% and 14% for adenoma detection, respectively, and 87% and 60% for CRC, respectively[73].
Interestingly, a recent meta-analysis by Yang et al[74] concluded that stool DNA was not suitable for CRC screening in average- rather than high-risk individuals, yielding a low detection value for precancerous lesions. Amir et al[75] demonstrated a correlation between SEPT9_V1 overexpression and drug resistance in various cancer cell lines. The potential of plasma biomarkers to identify individuals at risk of developing drug resistance was also demonstrated by Misale et al[76] in a metastatic colorectal cancer population.
Cell free mRNA has also been evaluated in plasma, in particular by the quantification of mRNA levels of hTERT, a ribonucleoprotein involved in the maintenance of correct length of telomeric chromosome ends, overexpressed in a wide number of tumors, including colon and rectal cancers[77,78]. In particular, a correlation has been found between plasma mRNA levels and tumor stage in CRC patients, suggesting that plasma RNA quantification could be useful for early diagnosis and follow up[77,78].
NUCLEIC ACIDS IN STOOL
Non-invasive, no bowel preparation and the sampling design are some of the main advantages of molecular stool analysis. A greater understanding of the molecular pathogenesis and the natural history of CRC has helped researchers to improve methods for molecular stool screening[79]. In 1952, Bader et al[80] reported that cancer of the rectum, sigmoid and descending colon could be detected by the cytological analysis of colonocytes. DNA from colonocytes shed in stool can be used to characterize the colonic epithelium involved in carcinogenesis. Colonocytes are, in fact, exfoliated continuously into the fecal stream[81], and their concentration in the intestinal lumen of CRC patients can increase of 4.5-fold with respect to healthy individuals[82]. It has been seen that another important stool element are mucus and its cellular cargo, found on the surface of stool after defecation[83,84]. Colonic mucus would seem to have protective properties that create a niche in which colonocytes are preserved in relative abundance[81,84]. For these reasons, although the mucus present on stool surface after defecation may have been picked up in the distal large bowel, it may also contain cells derived from the entire colorectal mucosa[85]. This would explain why malignant cells from the cecum and other right-sided CRCs can be isolated from stool after defecation[86] despite the luminal contents generally being liquid in the most proximal part of the large bowel. Stool tests based on isolated colonocytes rather than blood markers may offer better results due to the higher rate of neoplastic cell exfoliation, especially when used alone or in combination with current routine diagnostic tests, such as iFOBT[70]. However, as it is extremely difficult to discriminate between normal and malignant cells using standard morphological criteria, considerable interest has arisen in identifying biomarkers secreted by CRC cells rather than normal colonocytes[79]. Adenomas and CRC characteristically exfoliate non-apoptotic colonocytes, unlike normal colonic mucosa, which typically sheds apoptotic colonocytes[87]. The carcinogenetic process can lead to genetic mutations and/or epigenetic alterations that prevent normal colonocytes apoptosis[50]. Non-apoptotic colonocytes shed from diseased mucosa and isolated in the stool can release segments of 200 bp or more in length of intact DNA (L-DNA), making the latter a potentially effective stool biomarker.
A number of authors have proposed different methods to develop valid DNA integrity analysis (DIA) assays to improve sensibility and specificity in the detection of pre-malignant and malignant lesions. Results from studies carried out on stool biomarkers since 2000 are summarized in Table 2.
Table 2.
Summary of studies that have tested DNA integrity and genetic alteration markers in stool samples
| Study | Biomarker(s) | Methods | Assay for Long DNA | Case analyzed | Sensitivity | Specificity |
| Ahlquist et al[88], 2000 | Stool DNA integrity | L-DNA (4 sites) | Hybrid | CRC: 22 | 91% | 93% |
| KRAS, TP53, APC | Mutation analysis | Capture | A ≥ 1 cm: 11 | 82% | NA | |
| BAT 26 | MSI | PCR | HD: 28 | |||
| Tagore et al[89], 2003 | Stool DNA integrity | L-DNA (6 sites) | Hybrid | CRC: 52 | 64% | 96% |
| KRAS, TP53, APC | Mutation analysis | Capture | AA: 28 | |||
| BAT 26 | MSI | PCR | HD: 212 | 57% | NA | |
| Calistri et al[90], 2003 | Stool DNA integrity | L-DNA (8 sites) | PCR | CRC: 56 | 62% | 97% |
| KRAS, TP53, APC | Mutation analysis | HD: 38 | ||||
| D2S123, D5S346, D17S250 | ||||||
| BAT 25, BAT 26 | MSI | |||||
| Boyton et al[91], 2003 | Stool DNA integrity | L-DNA (6 sites) | Hybrid | CRC: 27 | 56% | 97% |
| Capture | HD: 77 | |||||
| PCR | ||||||
| Calistri et al[92], 2004 | Stool DNA integrity | FL-DNA (8 sites) | Capillary | CRC: 85 | 76% | 93% |
| Electrophoresis | HD: 59 | |||||
| Whitney et al[93], 2004 | Stool DNA integrity | L-DNA (4 sites) | Magnetic | CRC: 86 | 70% | 96% |
| KRAS, TP53, APC | Mutation analysis | Bead-Based | HD: 100 | |||
| BAT 26 | MSI | Sequence-Specific Purification | ||||
| Capture | ||||||
| Imperiale et al[94], 2004 | Stool DNA integrity | L-DNA (4 sites) | Hybrid Capture | CRC: 31 | 52% | 95% |
| KRAS, TP53, APC | Mutation analysis | PCR | AA: 407 | 15% | NA | |
| BAT 26 | MSI | P: 648 | 8% | NA | ||
| HD: 1423 | ||||||
| Kutzner et al[95], 2005 | Stool DNA integrity | L-DNA (4 sites) | Hybrid | CRC: 57 | 65% | 91% |
| APC | Mutation analysis | Capture | HD: 44 | |||
| BAT 26 | MSI | PCR | ||||
| Zou et al[96], 2006 | Stool DNA integrity | L-DNA: Alu-assay (2 sites) | RT PCR | CRC: 18 | 44% | 100% |
| HD: 20 | ||||||
| Itzkowitz et al[97], 2007 | Stool DNA integrity | L-DNA (4 sites Locus D, Locus Y) | RT PCR | CRC: 40 | 88% | 82% |
| Vimentin | Methylation analysis | HD: 122 | ||||
| Abbaszadegan et al[98], 2007 | Stool DNA integrity | L-DNA: (1476 bp fragments) | PCR | CRC: 25 | 64% | 95% |
| p16 | Methylation analysis | HD: 20 | ||||
| BAT 26 | MSI | |||||
| Ahlquist et al[99], 2008 | Stool DNA integrity | L-DNA (4 sites) | PCR | CRC: 12 | 25% | 96% |
| KRAS, APC, TP53 | Mutations Analysis | A ≥ 1 cm: 135 | 17% | NA | ||
| BAT 26 | MSI | A < 1 cm: 469 | ||||
| Stool DNA test 1 | P: 341 | 4% | NA | |||
| HD: 1473 | 5% | NA | ||||
| Itzkowitz et al[47], 2008 | Stool DNA integrity | L-DNA ( 4 sites, Locus D, Locus Y) | RT PCR | CRC: 82 | 83% | 82% |
| Vimentin | Methylation Analysis | HD: 363 | ||||
| Calistri et al[100], 2009 | Stool DNA integrity | FL-DNA (8 sites) | Capillary | CRC: 100 | 79% | 89% |
| Electrophoresis | HD: 100 | |||||
| Calistri et al[101], 2010 | Stool DNA integrity | FL-DNA (8 sites) | Capillary | CRC: 26 | Cancer risk prediction with markers combination | |
| iFOBT | iFOBT | Electrophoresis | A HR: 264 | |||
| A LR: 54 | ||||||
| HD: 216 | ||||||
| Kalimutho et al[102], 2011 | Stool DNA integrity | L-DNA (4 sites) | QdHPLC | CRC: 28 | 86% | 81% |
| Calprotectin | ELISA | A: 69 | 17% | NA | ||
| HD: 95 | 72% | 75% | ||||
| Ahlquist et al[103], 2012 | Stool DNA integrity | L-DNA | QuARTS | CRC: 252 | 85% | 89% |
| KRAS | Mutations Analysis | A ≥ 1 cm: 133 | 63% | NA | ||
| NDRG4, BMP3, vimentin, | Methylation Analysis | A < 1 cm: 94 | 54% | NA | ||
| TFP12/α-actine | HD: 293 | |||||
L-DNA: Long-DNA; FL-DNA: Fluorescence long DNA; CRC: Colorectal cancer; A: Adenoma; AA: Advanced Adenoma; A HR: Adenoma high risk; A LR: Adenoma low risk; HD: Healthy donors; HP: Hyperplasic polyps; NA: Not available; RT-PCR: Real time polymerase chain reaction; QdHPLC: Quantitative denaturing high performance liquid chromatografy; QuARTS: Quantitative allele specific real-time target and signal amplification; MSI: Microsatellite instability.
In one of the first studies to focus on this topic, Boynton et al[91] evaluated the length and integrity of L-DNA fragments using oligonucleotide-based hybrid captures with specific target sequences of 200 bp, 400 bp, 800 bp, 1.3 kb, 1.8 kb and 24 kb in PCR reactions; 56% sensitivity and 97% specificity were obtained. The authors concluded that CRC was related to the presence of high molecular weight bands[91]. Another approach to quantify stool L-DNA was carried out using fluorescence primers, capillary electrophoresis and standard curves (fluorescence long DNA, FL-DNA)[92]. In this pilot study on 56 patients and 38 healthy volunteers, FL-DNA evaluation using a cut-off of 25 ng showed a sensitivity of about 76% and a specificity of 93% compared to a specificity of 97% and a sensitivity of only 50% when a non quantitative DNA amplification method was utilized[92].
Long DNA has been also evaluated by analysing human Alu repeats using Real Time PCR assay showing a specificity of 100% but a sensitivity of 44%[96]. To improve accuracy in detecting neoplastic and pre-neoplastic lesions, different combinations of DNA integrity and genetic alteration analyses have been proposed in the last few years. Ahlquist et al[88] explored the feasibility of stool assays evaluating panels of selected DNA alterations to discriminate between subjects with colorectal cancer and healthy individuals. The pilot study, using an assay based on L-DNA analysis, APC, TP53 and KRAS gene determination and BAT26 microsatellite instability evaluation, reported 91% sensitivity and 93% specificity in detecting CRC[88]. This high accuracy was not confirmed in a large multicenter study of more than 4000 subjects conducted by Imperiale, who reported obtaining 52% sensitivity in detecting CRC[94]. However, results from Imperiale’s study confirmed that molecular analysis identified CRC and adenomas more accurately than the standard FOBT[94]. Another large study performed by Ahlquist some years later using a similar approach based on L-DNA evaluation, an assessment of 21 tumor-specific point mutations and BAT 26 microsatellite analysis reported a sensitivity of only 25% in detecting cancer[99]. However, the author also showed that sensitivity could be increased to 58% by using a different molecular approach based on 3 broadly informative markers (KRAS mutation, APC mutator cluster region and vimentin gene methylation)[99]. Around the same time, Itzkowitz et al[47] used an approach based on the analysis of vimentin and different-sized DNA fragments of different loci, obtaining a sensitivity of 83% and a specificity of 82% in detecting CRC. Other combinations of genetic and epigenetic, MSI and DNA integrity markers has been tested over the past ten years in an attempt to define an effective assay for CRC detection. Kalimutho et al[102] used an approach based on quantitative-denaturing high performance liquid chromatography detection of APC, BRAF, KRAS and p53 genes to quantify fecal DNA integrity status. Results showed that the four-gene amplification analysis increased sensitivity with respect to single gene amplification, calprotectin evaluation or iFOBT, the latter two assessed using commercial kits[102]. More recently, a next generation stool DNA test based on a quantitative allele-specific real-time target and signal amplification assay was developed with the aim of detecting early-stage CRC, despite the laboriousness and cost of this approach, a sensitivity of 85% for CRC and a specificity of 89% were obtained[103]. Long DNA values have also been evaluated in combination with iFOBT, showing that a combined approach could better predict the presence of tumor or high risk adenoma lesions in the colon[101].
An interesting study by Kanaoka et al[104] comparing stool cyclooxygenase-2 (COX-2) and CEA mRNA levels revealed that faecal CEA mRNA specificity was lower than that of COX-2. Furthermore, no significant differences in median faecal CEA mRNA values between CRC patients and control subjects were detected by Koga et al[105]. Using the RNA extraction method published by Kanaoka et al[104], Hamaya et al[106], confirmed significantly higher faecal COX-2 mRNA expression levels in CRC patients than in controls. Moreover, faecal mRNA levels of CEA, E-cad and CD45 showed significantly higher values in CRC patients than in controls.
CONCLUSION
Circulating cell-free nucleic acids are potentially excellent marker for early diagnosis, disease monitoring and more accurate tumour staging.
Molecular markers that could be used to monitor or predict a relapse in a presymptomatic phase of follow-up could have a great impact on the management and, potentially the survival of CRC patients. Several studies have proposed the use of the circulating free DNA quantification as a screening method for CRC diagnosis. Extraction of circulating DNA and RNA from biological fluids e.g., blood and stool, is a simple, relatively noninvasive and low cost procedure, thus representing a very attractive tool to detect genetic and epigenetic mutations, whereas analysis of gene alterations are generally expensive and time consuming. Furthermore, the evaluation of circulating free DNA exclusively derived from tumor cells represents a useful strategy to monitor disease progression. In fact, molecular alterations present in metastatic and primary tumors from the same patient can vary, determining different aggressiveness and/or responsiveness to treatments. Thus, the possibility of monitoring molecular alterations using simple tests based on nucleic acids obtained from blood samples could permit a more efficient assessment of disease status and response to treatments.
Unfortunately, the results from studies carried out in this area also highlight great variability in terms of DNA and RNA concentration, yield, and sensitivity and specificity, indicating the presence of various pre-analytic (serum preparation with or without coagulation accelerator, interval between collection and centrifugation, storage and cryopreservation of samples) and analytic factors (type of extraction with organic solvents, commercial kits, use of magnetic beads) that could influence the diagnostic value of the method. In addition, human faecal RNA is an understudied type of biospecimen due to the difficulty of sample preservation[107]. In particular, sample collection, storage and handling are very important issues for DNA and RNA extracted from stool and could have a substantial impact on the performance of a specific test[107,108]. For these reasons, standardization of sample collection and analysis is fundamental to ensure good reproducibility and large, multicenter studies are needed to clarify the role of these molecular markers in a clinical setting.
ACKNOWLEDGMENTS
The authors would like to thank Ursula Elbling for editing the manuscript.
Footnotes
P- Reviewers: Aglietta M, Beaulieu JF, Pavlidis TE S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
References
- 1.Mandel P, Metais P. [Les acides nucleiques du plasma sanguin chez l’Homme] C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. A structure for deoxyribose nucleic acid. 1953. Nature. 2003;421:397–398; discussion 396. [PubMed] [Google Scholar]
- 3.Tan EM, Schur PH, Carr RI, Kunkel HG. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest. 1966;45:1732–1740. doi: 10.1172/JCI105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev. 2002;28:255–271. doi: 10.1016/s0305-7372(02)00077-4. [DOI] [PubMed] [Google Scholar]
- 5.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 6.Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 7.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67–71. [PubMed] [Google Scholar]
- 8.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86:774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 9.Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 10.Sorenson GD. Detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Clin Cancer Res. 2000;6:2129–2137. [PubMed] [Google Scholar]
- 11.Kopreski MS, Benko FA, Borys DJ, Khan A, McGarrity TJ, Gocke CD. Somatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNA. J Natl Cancer Inst. 2000;92:918–923. doi: 10.1093/jnci/92.11.918. [DOI] [PubMed] [Google Scholar]
- 12.Yamada T, Nakamori S, Ohzato H, Oshima S, Aoki T, Higaki N, Sugimoto K, Akagi K, Fujiwara Y, Nishisho I, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res. 1998;4:1527–1532. [PubMed] [Google Scholar]
- 13.Castells A, Puig P, Móra J, Boadas J, Boix L, Urgell E, Solé M, Capellà G, Lluís F, Fernández-Cruz L, et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17:578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- 14.Ryan BM, Lefort F, McManus R, Daly J, Keeling PW, Weir DG, Kelleher D. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut. 2003;52:101–108. doi: 10.1136/gut.52.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong YK, Lo YM. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta. 2006;363:187–196. doi: 10.1016/j.cccn.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 16.Kirsch C, Weickmann S, Schmidt B, Fleischhacker M. An improved method for the isolation of free-circulating plasma DNA and cell-free DNA from other body fluids. Ann N Y Acad Sci. 2008;1137:135–139. doi: 10.1196/annals.1448.035. [DOI] [PubMed] [Google Scholar]
- 17.Flamini E, Mercatali L, Nanni O, Calistri D, Nunziatini R, Zoli W, Rosetti P, Gardini N, Lattuneddu A, Verdecchia GM, et al. Free DNA and carcinoembryonic antigen serum levels: an important combination for diagnosis of colorectal cancer. Clin Cancer Res. 2006;12:6985–6988. doi: 10.1158/1078-0432.CCR-06-1931. [DOI] [PubMed] [Google Scholar]
- 18.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 19.Nagata S, Nagase H, Kawane K, Mukae N, Fukuyama H. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 2003;10:108–116. doi: 10.1038/sj.cdd.4401161. [DOI] [PubMed] [Google Scholar]
- 20.Viorritto IC, Nikolov NP, Siegel RM. Autoimmunity versus tolerance: can dying cells tip the balance? Clin Immunol. 2007;122:125–134. doi: 10.1016/j.clim.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 22.Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC, Roden R, Sokoll LJ, Chan DW, Shih IeM. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- 23.Ellinger J, Bastian PJ, Haan KI, Heukamp LC, Buettner R, Fimmers R, Mueller SC, von Ruecker A. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. Int J Cancer. 2008;122:138–143. doi: 10.1002/ijc.23057. [DOI] [PubMed] [Google Scholar]
- 24.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 25.Tuaeva NO, Abramova ZI, Sofronov VV. The origin of elevated levels of circulating DNA in blood plasma of premature neonates. Ann N Y Acad Sci. 2008;1137:27–30. doi: 10.1196/annals.1448.043. [DOI] [PubMed] [Google Scholar]
- 26.Fournié GJ, Courtin JP, Laval F, Chalé JJ, Pourrat JP, Pujazon MC, Lauque D, Carles P. Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett. 1995;91:221–227. [Google Scholar]
- 27.Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids--a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Rhodes CH, Honsinger C, Porter DM, Sorenson GD. Analysis of the allele-specific PCR method for the detection of neoplastic disease. Diagn Mol Pathol. 1997;6:49–57. doi: 10.1097/00019606-199702000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Bonnefoi H, Diebold-Berger S, Lyautey J, Lederrey C, Faltin-Traub E, Stroun M, Anker P. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clin Cancer Res. 1999;5:2297–2303. [PubMed] [Google Scholar]
- 30.Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- 31.Mead R, Duku M, Bhandari P, Cree IA. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer. 2011;105:239–245. doi: 10.1038/bjc.2011.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drew WL. Laboratory diagnosis of cytomegalovirus infection and disease in immunocompromised patients. Curr Opin Infect Dis. 2007;20:408–411. doi: 10.1097/QCO.0b013e32821f6010. [DOI] [PubMed] [Google Scholar]
- 33.González-Masiá JA, García-Olmo D, García-Olmo DC. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther. 2013;6:819–832. doi: 10.2147/OTT.S44668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieczorek AJ, Sitaramam V, Machleidt W, Rhyner K, Perruchoud AP, Block LH. Diagnostic and prognostic value of RNA-proteolipid in sera of patients with malignant disorders following therapy: first clinical evaluation of a novel tumor marker. Cancer Res. 1987;47:6407–6412. [PubMed] [Google Scholar]
- 35.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961–1965. [PubMed] [Google Scholar]
- 36.Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy HE, Meyer P, Stroun M, et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823–3826. [PubMed] [Google Scholar]
- 37.Ng EK, Tsui NB, Lam NY, Chiu RW, Yu SC, Wong SC, Lo ES, Rainer TH, Johnson PJ, Lo YM. Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clin Chem. 2002;48:1212–1217. [PubMed] [Google Scholar]
- 38.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 39.Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488–1489. [PubMed] [Google Scholar]
- 40.Kopreski MS, Gocke CD. Cellular- versus extracellular-based assays. Comparing utility in DNA and RNA molecular marker assessment. Ann N Y Acad Sci. 2000;906:124–128. doi: 10.1111/j.1749-6632.2000.tb06601.x. [DOI] [PubMed] [Google Scholar]
- 41.Ahnen DJ, Macrae FA. Colorectal cancer: Epidemiology, risk factors, and protective factors. 2013. Available from: http://www.uptodate.com/contents/colorectal-cancer-epidemiology-risk-factors-and-protective-factors.
- 42.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 43.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 44.Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M, Ehrlich M. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res. 1997;379:33–41. doi: 10.1016/s0027-5107(97)00088-2. [DOI] [PubMed] [Google Scholar]
- 46.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L, Schroy PC, Sontag S, Johnson D, Markowitz S, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 48.Brenner DE, Rennert G. Fecal DNA biomarkers for the detection of colorectal neoplasia: attractive, but is it feasible? J Natl Cancer Inst. 2005;97:1107–1109. doi: 10.1093/jnci/dji244. [DOI] [PubMed] [Google Scholar]
- 49.Lenhard K, Bommer GT, Asutay S, Schauer R, Brabletz T, Göke B, Lamerz R, Kolligs FT. Analysis of promoter methylation in stool: a novel method for the detection of colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:142–149. doi: 10.1016/s1542-3565(04)00624-x. [DOI] [PubMed] [Google Scholar]
- 50.Kanthan R, Senger JL, Kanthan SC. Fecal molecular markers for colorectal cancer screening. Gastroenterol Res Pract. 2012;2012:184343. doi: 10.1155/2012/184343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frattini M, Gallino G, Signoroni S, Balestra D, Battaglia L, Sozzi G, Leo E, Pilotti S, Pierotti MA. Quantitative analysis of plasma DNA in colorectal cancer patients: a novel prognostic tool. Ann N Y Acad Sci. 2006;1075:185–190. doi: 10.1196/annals.1368.025. [DOI] [PubMed] [Google Scholar]
- 52.Boni L, Cassinotti E, Canziani M, Dionigi G, Rovera F, Dionigi R. Free circulating DNA as possible tumour marker in colorectal cancer. Surg Oncol. 2007;16 Suppl 1:S29–S31. doi: 10.1016/j.suronc.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Tomita H, Ichikawa D, Sai S, Morimura R, Murayama Y, Komatsu S, Ikoma D, Tani N, Ikoma H, Fujiwara H, et al. [Quantification of circulating plasma DNA fragments as tumor markers in patients with esophageal and gastric cancer] Gan To Kagaku Ryoho. 2007;34:1908–1910. [PubMed] [Google Scholar]
- 54.Kolesnikova EV, Tamkovich SN, Bryzgunova OE, Shelestyuk PI, Permyakova VI, Vlassov VV, Tuzikov AS, Laktionov PP, Rykova EY. Circulating DNA in the blood of gastric cancer patients. Ann N Y Acad Sci. 2008;1137:226–231. doi: 10.1196/annals.1448.009. [DOI] [PubMed] [Google Scholar]
- 55.Catarino R, Ferreira MM, Rodrigues H, Coelho A, Nogal A, Sousa A, Medeiros R. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol. 2008;27:415–421. doi: 10.1089/dna.2008.0744. [DOI] [PubMed] [Google Scholar]
- 56.Yoon KA, Park S, Lee SH, Kim JH, Lee JS. Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls. J Mol Diagn. 2009;11:182–185. doi: 10.2353/jmoldx.2009.080098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellinger J, Müller SC, Stadler TC, Jung A, von Ruecker A, Bastian PJ. The role of cell-free circulating DNA in the diagnosis and prognosis of prostate cancer. Urol Oncol. 2011;29:124–129. doi: 10.1016/j.urolonc.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Dobrzycka B, Terlikowski SJ, Mazurek A, Kowalczuk O, Niklinska W, Chyczewski L, Kulikowski M. Circulating free DNA, p53 antibody and mutations of KRAS gene in endometrial cancer. Int J Cancer. 2010;127:612–621. doi: 10.1002/ijc.25077. [DOI] [PubMed] [Google Scholar]
- 59.Liggett T, Melnikov A, Yi QL, Replogle C, Brand R, Kaul K, Talamonti M, Abrams RA, Levenson V. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer. 2010;116:1674–1680. doi: 10.1002/cncr.24893. [DOI] [PubMed] [Google Scholar]
- 60.van der Drift MA, Hol BE, Klaassen CH, Prinsen CF, van Aarssen YA, Donders R, van der Stappen JW, Dekhuijzen PN, van der Heijden HF, Thunnissen FB. Circulating DNA is a non-invasive prognostic factor for survival in non-small cell lung cancer. Lung Cancer. 2010;68:283–287. doi: 10.1016/j.lungcan.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 61.Umetani N, Hiramatsu S, Hoon DS. Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci. 2006;1075:299–307. doi: 10.1196/annals.1368.040. [DOI] [PubMed] [Google Scholar]
- 62.Hara M, Kanemitsu Y, Hirai T, Komori K, Kato T. Negative serum carcinoembryonic antigen has insufficient accuracy for excluding recurrence from patients with Dukes C colorectal cancer: analysis with likelihood ratio and posttest probability in a follow-up study. Dis Colon Rectum. 2008;51:1675–1680. doi: 10.1007/s10350-008-9406-1. [DOI] [PubMed] [Google Scholar]
- 63.Winawer S, Fletcher R, Rex D, Bond J, Burt R, Ferrucci J, Ganiats T, Levin T, Woolf S, Johnson D, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-Update based on new evidence. Gastroenterology. 2003;124:544–560. doi: 10.1053/gast.2003.50044. [DOI] [PubMed] [Google Scholar]
- 64.Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, Bilchik AJ, Hoon DS. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin Chem. 2006;52:1062–1069. doi: 10.1373/clinchem.2006.068577. [DOI] [PubMed] [Google Scholar]
- 65.Danese E, Montagnana M, Minicozzi AM, De Matteis G, Scudo G, Salvagno GL, Cordiano C, Lippi G, Guidi GC. Real-time polymerase chain reaction quantification of free DNA in serum of patients with polyps and colorectal cancers. Clin Chem Lab Med. 2010;48:1665–1668. doi: 10.1515/CCLM.2010.301. [DOI] [PubMed] [Google Scholar]
- 66.Czeiger D, Shaked G, Eini H, Vered I, Belochitski O, Avriel A, Ariad S, Douvdevani A. Measurement of circulating cell-free DNA levels by a new simple fluorescent test in patients with primary colorectal cancer. Am J Clin Pathol. 2011;135:264–270. doi: 10.1309/AJCP4RK2IHVKTTZV. [DOI] [PubMed] [Google Scholar]
- 67.da Silva Filho BF, Gurgel AP, Neto MÁ, de Azevedo DA, de Freitas AC, Silva Neto Jda C, Silva LA. Circulating cell-free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol. 2013;66:775–778. doi: 10.1136/jclinpath-2013-201521. [DOI] [PubMed] [Google Scholar]
- 68.Holten-Andersen MN, Christensen IJ, Nielsen HJ, Stephens RW, Jensen V, Nielsen OH, Sørensen S, Overgaard J, Lilja H, Harris A, et al. Total levels of tissue inhibitor of metalloproteinases 1 in plasma yield high diagnostic sensitivity and specificity in patients with colon cancer. Clin Cancer Res. 2002;8:156–164. [PubMed] [Google Scholar]
- 69.Carpelan-Holmström M, Louhimo J, Stenman UH, Alfthan H, Haglund C. CEA, CA 19-9 and CA 72-4 improve the diagnostic accuracy in gastrointestinal cancers. Anticancer Res. 2002;22:2311–2316. [PubMed] [Google Scholar]
- 70.Carpelan-Holmström M, Louhimo J, Stenman UH, Alfthan H, Järvinen H, Haglund C. CEA, CA 242, CA 19-9, CA 72-4 and hCGbeta in the diagnosis of recurrent colorectal cancer. Tumour Biol. 2004;25:228–234. doi: 10.1159/000081385. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Tham CK, Ong SY, Ho KS, Lim JF, Chew MH, Lim CK, Zhao Y, Tang CL, Eu KW. Serum methylation levels of TAC1. SEPT9 and EYA4 as diagnostic markers for early colorectal cancers: a pilot study. Biomarkers. 2013;18:399–405. doi: 10.3109/1354750X.2013.798745. [DOI] [PubMed] [Google Scholar]
- 72.Grützmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, Miehlke S, Stolz T, Model F, Roblick UJ, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10:272–7.e1. doi: 10.1016/j.cgh.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang H, Xia BQ, Jiang B, Wang G, Yang YP, Chen H, Li BS, Xu AG, Huang YB, Wang XY. Diagnostic value of stool DNA testing for multiple markers of colorectal cancer and advanced adenoma: a meta-analysis. Can J Gastroenterol. 2013;27:467–475. doi: 10.1155/2013/258030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amir S, Mabjeesh NJ. SEPT9_V1 protein expression is associated with human cancer cell resistance to microtubule-disrupting agents. Cancer Biol Ther. 2007;6:1926–1931. doi: 10.4161/cbt.6.12.4971. [DOI] [PubMed] [Google Scholar]
- 76.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Terrin L, Rampazzo E, Pucciarelli S, Agostini M, Bertorelle R, Esposito G, DelBianco P, Nitti D, De Rossi A. Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res. 2008;14:7444–7451. doi: 10.1158/1078-0432.CCR-08-0478. [DOI] [PubMed] [Google Scholar]
- 78.Pucciarelli S, Rampazzo E, Briarava M, Maretto I, Agostini M, Digito M, Keppel S, Friso ML, Lonardi S, De Paoli A, et al. Telomere-specific reverse transcriptase (hTERT) and cell-free RNA in plasma as predictors of pathologic tumor response in rectal cancer patients receiving neoadjuvant chemoradiotherapy. Ann Surg Oncol. 2012;19:3089–3096. doi: 10.1245/s10434-012-2272-z. [DOI] [PubMed] [Google Scholar]
- 79.Davies RJ, Miller R, Coleman N. Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer. 2005;5:199–209. doi: 10.1038/nrc1569. [DOI] [PubMed] [Google Scholar]
- 80.Bader GM, Papanicolaou GN. The application of cytology in the diagnosis of cancer of the rectum, sigmoid, and descending colon. Cancer. 1952;5:307–314. doi: 10.1002/1097-0142(195203)5:2<307::aid-cncr2820050217>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 81.Ahlquist DA, Harrington JJ, Burgart LJ, Roche PC. Morphometric analysis of the “mucocellular layer” overlying colorectal cancer and normal mucosa: relevance to exfoliation and stool screening. Hum Pathol. 2000;31:51–57. doi: 10.1016/s0046-8177(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 82.Loktionov A, O’Neill IK, Silvester KR, Cummings JH, Middleton SJ, Miller R. Quantitation of DNA from exfoliated colonocytes isolated from human stool surface as a novel noninvasive screening test for colorectal cancer. Clin Cancer Res. 1998;4:337–342. [PubMed] [Google Scholar]
- 83.Baig MK, Stebbing JF, Marks CG. Anal canal metastases from left sided colorectal cancer. Colorectal Dis. 2002;4:371–372. doi: 10.1046/j.1463-1318.2002.00405.x. [DOI] [PubMed] [Google Scholar]
- 84.Loktionov A. Cell exfoliation in the human colon: myth, reality and implications for colorectal cancer screening. Int J Cancer. 2007;120:2281–2289. doi: 10.1002/ijc.22647. [DOI] [PubMed] [Google Scholar]
- 85.White V, Scarpini C, Barbosa-Morais NL, Ikelle E, Carter S, Laskey RA, Miller R, Coleman N. Isolation of stool-derived mucus provides a high yield of colonocytes suitable for early detection of colorectal carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18:2006–2013. doi: 10.1158/1055-9965.EPI-08-1145. [DOI] [PubMed] [Google Scholar]
- 86.Davies RJ, Freeman A, Morris LS, Bingham S, Dilworth S, Scott I, Laskey RA, Miller R, Coleman N. Analysis of minichromosome maintenance proteins as a novel method for detection of colorectal cancer in stool. Lancet. 2002;359:1917–1919. doi: 10.1016/S0140-6736(02)08739-1. [DOI] [PubMed] [Google Scholar]
- 87.Bedi A, Pasricha PJ, Akhtar AJ, Barber JP, Bedi GC, Giardiello FM, Zehnbauer BA, Hamilton SR, Jones RJ. Inhibition of apoptosis during development of colorectal cancer. Cancer Res. 1995;55:1811–1816. [PubMed] [Google Scholar]
- 88.Ahlquist DA, Skoletsky JE, Boynton KA, Harrington JJ, Mahoney DW, Pierceall WE, Thibodeau SN, Shuber AP. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology. 2000;119:1219–1227. doi: 10.1053/gast.2000.19580. [DOI] [PubMed] [Google Scholar]
- 89.Tagore KS, Lawson MJ, Yucaitis JA, Gage R, Orr T, Shuber AP, Ross ME. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer. 2003;3:47–53. doi: 10.3816/CCC.2003.n.011. [DOI] [PubMed] [Google Scholar]
- 90.Calistri D, Rengucci C, Bocchini R, Saragoni L, Zoli W, Amadori D. Fecal multiple molecular tests to detect colorectal cancer in stool. Clin Gastroenterol Hepatol. 2003;1:377–383. doi: 10.1053/s1542-3565(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 91.Boynton KA, Summerhayes IC, Ahlquist DA, Shuber AP. DNA integrity as a potential marker for stool-based detection of colorectal cancer. Clin Chem. 2003;49:1058–1065. doi: 10.1373/49.7.1058. [DOI] [PubMed] [Google Scholar]
- 92.Calistri D, Rengucci C, Lattuneddu A, Francioni G, Polifemo AM, Nanni O, Saragoni L, Monti F, Ravaioli A, Zoli W, et al. Detection of colorectal cancer by a quantitative fluorescence determination of DNA amplification in stool. Neoplasia. 2004;6:536–540. doi: 10.1593/neo.04190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Whitney D, Skoletsky J, Moore K, Boynton K, Kann L, Brand R, Syngal S, Lawson M, Shuber A. Enhanced retrieval of DNA from human fecal samples results in improved performance of colorectal cancer screening test. J Mol Diagn. 2004;6:386–395. doi: 10.1016/S1525-1578(10)60536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 95.Kutzner N, Hoffmann I, Linke C, Thienel T, Grzegorczyk M, Urfer W, Martin D, Winde G, Traska T, Hohlbach G, et al. Non-invasive detection of colorectal tumours by the combined application of molecular diagnosis and the faecal occult blood test. Cancer Lett. 2005;229:33–41. doi: 10.1016/j.canlet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 96.Zou H, Harrington JJ, Klatt KK, Ahlquist DA. A sensitive method to quantify human long DNA in stool: relevance to colorectal cancer screening. Cancer Epidemiol Biomarkers Prev. 2006;15:1115–1119. doi: 10.1158/1055-9965.EPI-05-0992. [DOI] [PubMed] [Google Scholar]
- 97.Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, Sontag S, Johnson D, Skoletsky J, Durkee K, Markowitz S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 98.Abbaszadegan MR, Tavasoli A, Velayati A, Sima HR, Vosooghinia H, Farzadnia M, Asadzedeh H, Gholamin M, Dadkhah E, Aarabi A. Stool-based DNA testing, a new noninvasive method for colorectal cancer screening, the first report from Iran. World J Gastroenterol. 2007;13:1528–1533. doi: 10.3748/wjg.v13.i10.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ, Knigge K, Lance MP, Burgart LJ, Hamilton SR, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149:441–50, W81. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Calistri D, Rengucci C, Molinari C, Ricci E, Cavargini E, Scarpi E, Milandri GL, Fabbri C, Ravaioli A, Russo A, et al. Quantitative fluorescence determination of long-fragment DNA in stool as a marker for the early detection of colorectal cancer. Cell Oncol. 2009;31:11–17. doi: 10.3233/CLO-2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Calistri D, Rengucci C, Casadei Gardini A, Frassineti GL, Scarpi E, Zoli W, Falcini F, Silvestrini R, Amadori D. Fecal DNA for noninvasive diagnosis of colorectal cancer in immunochemical fecal occult blood test-positive individuals. Cancer Epidemiol Biomarkers Prev. 2010;19:2647–2654. doi: 10.1158/1055-9965.EPI-10-0291. [DOI] [PubMed] [Google Scholar]
- 102.Kalimutho M, Del Vecchio Blanco G, Cretella M, Mannisi E, Sileri P, Formosa A, Pallone F, Federici G, Bernardini S. A simplified, non-invasive fecal-based DNA integrity assay and iFOBT for colorectal cancer detection. Int J Colorectal Dis. 2011;26:583–592. doi: 10.1007/s00384-010-1128-x. [DOI] [PubMed] [Google Scholar]
- 103.Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat LF, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–56; quiz e25-6. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kanaoka S, Yoshida K, Miura N, Sugimura H, Kajimura M. Potential usefulness of detecting cyclooxygenase 2 messenger RNA in feces for colorectal cancer screening. Gastroenterology. 2004;127:422–427. doi: 10.1053/j.gastro.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 105.Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, Kozu T, Baba H, Matsumura Y. Detection of colorectal cancer cells from feces using quantitative real-time RT-PCR for colorectal cancer diagnosis. Cancer Sci. 2008;99:1977–1983. doi: 10.1111/j.1349-7006.2008.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hamaya Y, Yoshida K, Takai T, Ikuma M, Hishida A, Kanaoka S. Factors that contribute to faecal cyclooxygenase-2 mRNA expression in subjects with colorectal cancer. Br J Cancer. 2010;102:916–921. doi: 10.1038/sj.bjc.6605564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu YJ, Majumdar AP, Nechvatal JM, Ram JL, Basson MD, Heilbrun LK, Kato I. Exfoliated cells in stool: a source for reverse transcription-PCR-based analysis of biomarkers of gastrointestinal cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:455–458. doi: 10.1158/1055-9965.EPI-07-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olson J, Whitney DH, Durkee K, Shuber AP. DNA stabilization is critical for maximizing performance of fecal DNA-based colorectal cancer tests. Diagn Mol Pathol. 2005;14:183–191. doi: 10.1097/01.pas.0000176768.18423.7e. [DOI] [PubMed] [Google Scholar]


