Abstract
In Gram-positive bacteria, the secretion of proteins requires translocation of polypeptides across the bacterial membrane into the highly charged environment of the membrane-cell wall interface. Here, proteins must be folded and often further delivered across the matrix of the cell wall. While many aspects of protein secretion have been well studied in Gram-negative bacteria which possess both an inner and outer membrane, generally less attention has been given to the mechanics of protein secretion across the single cell membrane of Gram-positive bacteria. In this review, we focus on the role of a post-translocation secretion chaperone in Listeria monocytogenes known as PrsA2, and compare what is known regarding PrsA2 with PrsA homologs in other Gram-positive bacteria. PrsA2 is a member of a family of membrane-associated lipoproteins that contribute to the folding and stability of secreted proteins as they cross the bacterial membrane. PrsA2 contributes to the integrity of the L. monocytogenes cell wall as well as swimming motility and bacterial resistance to osmotic stress; however its most critical role may be its requirement for L. monocytogenes virulence and viability within host cells. A better understanding of the role of PrsA2 and PrsA-like homologs will provide insight into the dynamics of protein folding and stability in Gram-positive bacteria and may result in new strategies for optimizing protein secretion as well as inhibiting the production of virulence factors.
Keywords: PrsA, PrsA2, bacterial pathogenesis, PrfA, PPIase, foldase, LLO, cell wall
Introduction
Bacteria are generally highly adaptable creatures that must interface with their varied environments to acquire nutrients, mitigate stress conditions, establish a replication niche, and avoid or eliminate undesirables, such as immune effector cells bent on bacterial destruction. Bacterial secreted proteins provide both structural and enzymatic functions that facilitate bacterial adaptation to environmental changes; following synthesis, these proteins must be translocated across the bacterial cell membrane and properly folded so as to carry out their functional roles. While considerable attention has been given to the process of protein secretion across the inner and outer membranes of Gram-negative bacteria, less attention has been generally focused on protein translocation across the single cell membrane of Gram-positive bacteria, which also possess a formidable cell wall. Gram-positive bacteria appear to have evolved protein secretion strategies that are both similar and distinct from those of their Gram-negative cousins.
Listeria monocytogenes is a Gram-positive facultative intracellular bacterial pathogen that has served as a model organism for studies focused on areas that include immunology, host-pathogen interactions, cell biology, and mechanisms by which environmental microbes become human pathogens (Cossart, 2011; Witte et al., 2012; Xayarath and Freitag, 2012). It has recently become apparent that L. monocytogenes is also an excellent model system for the investigation of Gram-positive protein secretion and the associated role of a critical post-translocation secretion chaperone known as PrsA2 (Alonzo et al., 2009, 2011; Zemansky et al., 2009; Alonzo and Freitag, 2010; Forster et al., 2011). PrsA2 is a member of a family of membrane-associated lipoproteins that contribute to the folding and stability of secreted proteins as they cross the bacterial membrane and enter the highly charged environment of the membrane-cell wall interface (Forster and Marquis, 2012). In this review, we describe the role of PrsA2 in L. monocytogenes protein secretion and compare what is known about L. monocytogenes PrsA2 with homologs in other Gram-positive bacterial species. Evidence indicates that when newly synthesized proteins foray into the challenging social scene of the membrane-cell wall interface, it is clearly advantageous to be properly chaperoned.
Identification of PrsA2 (and PrsA1) as post-translocation secretion chaperones in L. monocytogenes
L. monocytogenes is considered an environmental pathogen as it is capable of life as a saprophyte in the outside environment while also maintaining the ability to invade and replicate within mammalian cells (Xayarath and Freitag, 2012). As part of the transition between life in the outside environment to life within the cytosol, L. monocytogenes increases the expression of secreted virulence factors that facilitate intracellular survival by promoting cell entry, bacterial escape from host vacuoles, replication within the cytosol, and spread to adjacent cells (Shetron-Rama et al., 2003; Mueller and Freitag, 2005; Port and Freitag, 2007; Alonzo and Freitag, 2010; de Las Heras et al., 2011; Mostowy and Cossart, 2012). The expression of many of the secreted virulence gene products required for bacterial survival within the host is regulated by the transcriptional activator PrfA (positive regulatory factor A) (Leimeister-Wachter et al., 1990; Mengaud et al., 1991; Freitag et al., 1992). Experimental evidence suggests that PrfA becomes highly activated upon bacterial entry into host cells, thereby leading to large increases in the synthesis and secretion of proteins that promote L. monocytogenes intracellular survival and replication. This substantial increase in virulence factor translocation across the bacterial membrane is likely to correspondingly require an increase in the activity of proteins directing protein secretion and folding.
L. monocytogenes prsA2, encoding a post-translocation molecular chaperone, was first identified via transcriptome analysis as a gene whose expression increased as a result of PrfA activation (Chatterjee et al., 2006) and which contained a putative PrfA binding site within its promoter region located 206 base pairs upstream from its translation start codon (Milohanic et al., 2003). Subsequent proteomic analysis of L. monocytogenes secreted proteins revealed that PrsA2 levels were increased in mutant strains containing constitutively activated PrfA (PrfA*) (Port and Freitag, 2007). Later, strains containing transposon insertions within prsA2 were identified in a genetic screen designed to identify L. monocytogenes mutants with reduced hemolytic activity (Zemansky et al., 2009). While the presence of a PrfA binding site may enhance prsA2 expression upon PrfA activation, this PrfA binding site is not essential for pathogenesis (Zemansky et al., 2009). Since its initial identification, L. monocytogenes PrsA2 and its homolog PrsA1 have been characterized for their roles in protein secretion and pathogenesis (Alonzo et al., 2009, 2011; Zemansky et al., 2009; Alonzo and Freitag, 2010; Forster et al., 2011).
L. monocytogenes PrsA2 contributes to multiple facets of bacterial pathogenesis and is essential for virulence (Alonzo et al., 2009; Zemansky et al., 2009), reflecting the apparent requirement of PrsA2 for the folding and secretion of a number of proteins (Alonzo and Freitag, 2010). PrsA1, in contrast, while sharing 75% amino acid similarity and 58% identity with PrsA2, makes no detectable contribution to L. monocytogenes pathogenesis (Alonzo et al., 2009). L. monocytogenes mutants lacking PrsA2 exhibit decreased hemolytic and phospholipase activity, and are defective for cell-to-cell spread (Alonzo et al., 2009; Zemansky et al., 2009; Forster et al., 2011). In mouse models of infection, L. monocytogenes strains lacking PrsA2 have at least 100,000-fold fewer colony forming units recovered from the livers and spleens of infected animals in comparison to animals infected with the wild-type strain (Alonzo et al., 2009; Zemansky et al., 2009). These data indicate that L. monocytogenes PrsA2 has been adapted for aspects of protein secretion that are critical for L. monocytogenes pathogenesis, whereas the function of PrsA1 remains to be identified. Neither prsA1 nor prsA2 is required for bacterial growth in broth culture as both genes can be deleted without significant effects on growth (Alonzo and Freitag, 2010).
A family resemblance: comparison of L. monocytogenes PrsA2 with homologs in other gram-positive bacteria
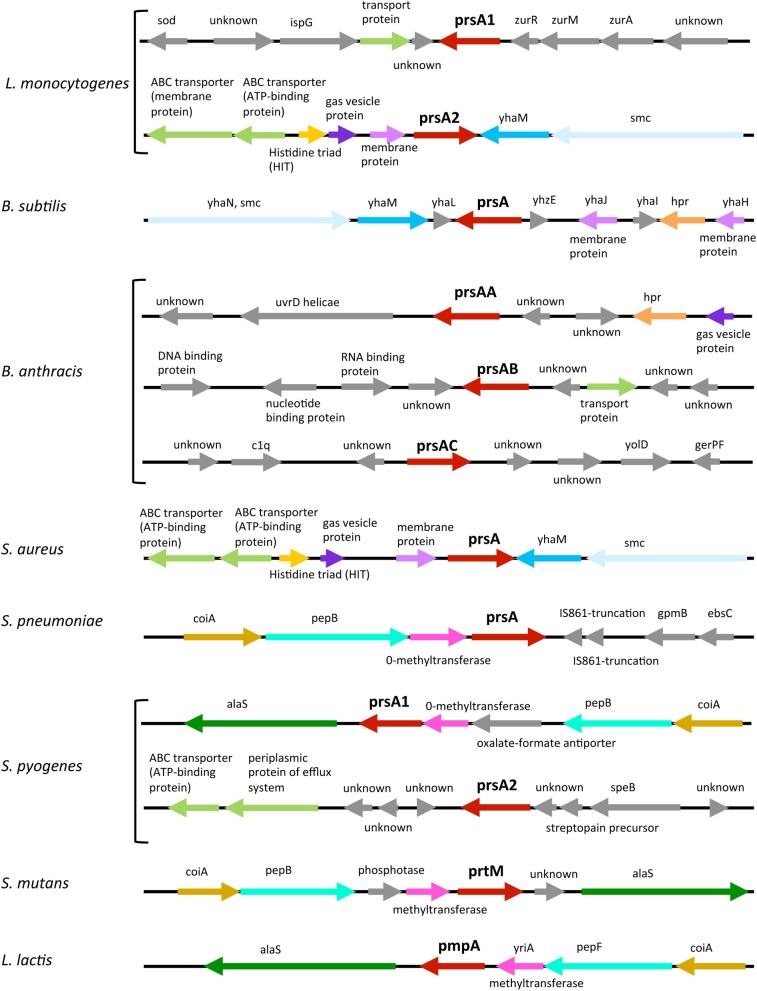
Given the critical requirement of PrsA2 for L. monocytogenes pathogenesis, how does it functionally compare with homologs expressed in other Gram-positive bacteria? prsA-like genes can be identified based on encoded amino acid sequence homology in many Gram-positive bacterial species (Figure 1). Thus far, the proteins have been best characterized in L. monocytogenes and Bacillus subtilis, although relatively little is still known about their molecular function. Interestingly, not all Gram-positive bacteria have the same number of PrsA-like proteins (Figure 2). L. monocytogenes has two, PrsA1 and PrsA2. As mentioned above, the function of L. monocytogenes PrsA1 has yet to be determined as deletion of the gene has yielded no discernable phenotype (Alonzo et al., 2009, 2011; Alonzo and Freitag, 2010). The expression of prsA1 from the prsA2 promoter does not complement a prsA2 deletion, indicating that the lack of functional complementation observed was not due to differences in the timing of gene expression (Alonzo and Freitag, 2010). Streptococcus pyogenes, similar to L. monocytogenes, has two prsA alleles that also appear to encode proteins with non-redundant functions (Ma et al., 2006), whereas the related Streptococcal species Streptococcus pneumonia and Streptococcus mutans have only one prsA allele (Drouault et al., 2002; Guo et al., 2013). B. subtilis contains one prsA gene that was at first considered essential for viability but has recently been shown to be non-essential in the presence of high concentrations of magnesium (Hyyrylainen et al., 2010). In contrast, Bacillus anthracis has three prsA alleles that can complement B. subtilis PrsA, however whether one or more of these alleles are essential for B. anthracis viability is unknown (Williams et al., 2003). The single prsA alleles of S. mutans, Staphylococcus aureus, and Lactococcus lactis have been shown to be non-essential (Drouault et al., 2002; Jousselin et al., 2012; Guo et al., 2013).
Figure 1.
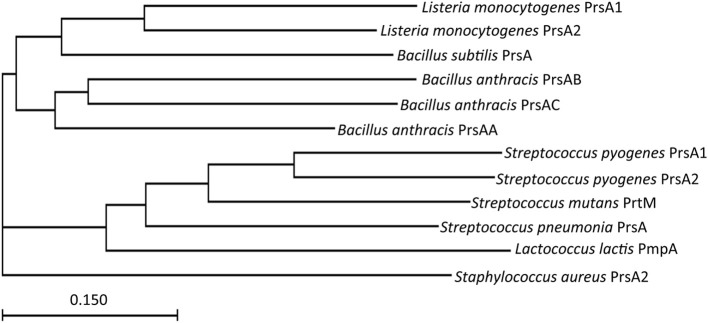
The evolutionary relationship of PrsA homologs from Gram positive bacteria. Shown is a phylogenetic tree of PrsA homologs from Listeria monocytogenes and other Gram positive bacteria created by CLUSTAL analysis. The scale bar indicates 1 substitution for every 10 nucleic acid residues.
Figure 2.
Comparative organization of prsA-like loci. The prsA-like loci from eight Gram-positive bacteria are shown where arrows represent open- reading frames. Genes with similar functions have the same color while genes that have unrelated functions are in gray. The annotation of genes mentioned in the text: yhaN encodes a putative ATPase involved in DNA metabolism containing a chromosome partition domain (smc), yhaM encodes a 3'-5' exoribonuclease, coiA encodes a competence transcription factor, pep is a peptidase, htr is a transcriptional regulator, and alaS is a alanyl-tRNA synthetase.
Interestingly, there are some common themes with respect to the genomic organization of prsA alleles. L. monocytogenes prsA2 and S. aureus prsA are most similar and are each adjacent to the 3′-5′ exoribonuclease YhaM coding sequence, as well as a gene encoding a putative ATPase involved in DNA metabolism (smc), and near an enzyme that acts as a nucleotidylyl hydrolase or transferase and ABC transporter genes (Figure 2). B. subtilis prsA is also located near yhaM (Figure 2). An alternative type of gene clustering is observed for bacteria for which the prsA allele is adjacent to methyltransferase and peptidase genes, such as the clustering found for S. pneumoniae prsA with the addition of an alanyl-tRNA synthetase gene for S. pyogenes prsA1, S. mutans prtM, and L. lactis pmpA (Figure 2). Organization of the L. monocytogenes prsA1 locus is distinct from the loci mentioned above and does not appear to share many commonalities with the other prsA allele loci with the exception of being adjacent to a gene encoding a predicted transport protein (Figure 2).
Shared family traits: PrsA2 functional domain organization
L. monocytogenes PrsA2 is a lipoprotein and a member of a more extended and diverse class of chaperones known as parvulin peptidyl-prolyl isomerases (PPIases) that catalyze the cis-trans isomerization of peptide bonds N-terminal to proline residues in polypeptide chains (Alonzo et al., 2011). L. monocytogenes PrsA2 possesses a signal peptide that directs protein translocation across the bacterial cytoplasmic membrane via the Sec secretion pathway, an N-terminus and C-terminus that comprise the chaperone-like regions, and the central PPIase domain (Figure 3A). The L. monocytogenes PPIase domain contains a signature motif shared amongst PPIase containing proteins (Sigrist et al., 2010) (Figure 3B); however, not all PrsA-like proteins contain this PPIase motif. L. lactis pmpA, S. pyogenes prsA1 and prsA2, and S. mutans prtM encoded proteins all lack the signature PPIase motif (Drouault et al., 2002; Guo et al., 2013) while L. monocytogenes prsA1 and prsA2, B. subtilis prsA, B. anthracis prsAA, prsAB, and prsAC and S. aureus prsA, and S. pneumoniae prsA encode proteins with the centrally located motif (Williams et al., 2003; Vitikainen et al., 2004; Heikkinen et al., 2009; Alonzo et al., 2011) (Figure 3B). A functional PPIase domain has been shown using in vitro assays for L. monocytogenes PrsA2 and PrsA1 as well as for B. subtilis PrsA (Vitikainen et al., 2004; Tossavainen et al., 2006; Alonzo et al., 2011). The structural differences between the PrsA-like proteins suggests they may have two distinct and separable functions, N- and C-terminal foldase activity that is conserved among family members and PPIase activity that appears to be less highly conserved.
Figure 3.

L. monocytogenes PrsA2 domain organization and the comparative alignment of PPIase domains of Gram-positive bacteria. (A) Shown are the N-terminal signal peptide, N-terminus, PPIase domain, and C-terminus where numbers designate amino acids. (B) An alignment of the PPIase domain of 12 PrsA-like homologs is shown. The sequence logo is also indicated where identical amino acids are highlighted in gray. Homologs having a PPIase or no PPIase motif are designated.
In Gram-positive bacteria, PrsA-like proteins have an N-terminal cysteine that upon signal sequence cleavage is modified with a diacylglycerol residue by the enzyme Lgt to promote membrane association (Leskela et al., 1999; Stoll et al., 2005; Baumgartner et al., 2007). In L. monocytogenes, B. subtilis, and S. aureus, disruption of lgt results in the increased release of PrsA2/PrsA and other lipoproteins into culture supernatants (Leskela et al., 1999; Stoll et al., 2005; Baumgartner et al., 2007). However, while L. monocytogenes PrsA2 is lipid modified and is membrane-associated, a significant amount of the protein is also secreted into culture supernatants (Alonzo et al., 2009). It remains to be determined whether both forms of L. monocytogenes PrsA2 (membrane-associated and secreted) are important for activity, and whether they fulfill distinct functions.
L. monocytogenes provides several advantages for studies focused on the molecular mechanisms underlying PrsA2 activity and its functional domains. For example, the bacterium is very amenable to genetic manipulation, thereby facilitating the demonstration that neither PrsA2 nor PrsA1 is essential given the fact that both genes can be deleted without deleterious effects to bacterial growth in broth culture (Alonzo and Freitag, 2010). Multiple assays can be used to assess the activity of PrsA2-dependent virulence factors in vitro, with the added dimension of tissue and animal culture models of infection. N- and C-terminal domain swap experiments between L. monocytogenes PrsA1 and PrsA2 in conjunction with site-directed mutagenesis experiments have indicated that PrsA2 tolerates substantial modifications without loss of activity. Data suggest that PrsA2 is likely to make multiple contacts with substrate proteins such that minor amino acid substitutions do not generally compromise protein function or substrate specificity (Alonzo et al., 2011). In addition, L. monocytogenes ΔprsA2 mutants complemented with a construct encoding an N-and C-terminal PrsA2 fusion protein that completely lacks the central PPIase domain exhibited wild type levels of secreted hemolysin and phospholipase activities and formed normal plaques in fibroblast cell monolayers (Alonzo et al., 2011). These studies indicate that the PPIase domain is dispensable for some PrsA2-associated activities, however the complemented mutant had altered patterns of protein secretion, exhibited increased sensitivity to penicillin, and was defective in mouse models of infection (Alonzo et al., 2011).
Signaling for a chaperone: regulation of PrsA expression
prsA2 was initially identified as a result of its increased transcription following PrfA activation (Chatterjee et al., 2006). Subsequent studies confirmed that increased amounts of PrsA2 are synthesized and secreted in constitutively activated PrfA* mutants (Port and Freitag, 2007), which led to the speculation that increased virulence factor secretion occurring as a result of PrfA activation requires the presence of additional amounts of chaperone to promote folding and activity and avoid membrane stress (Alonzo et al., 2009). In the absence of PrfA activation, prsA2 reporter gene fusions indicate that prsA2 expression increases during exponential growth and decreases upon entry into stationary phase (Forster et al., 2011). Whether other signals contribute to the regulation of prsA2 expression is not known, and still less is known regarding the regulation of expression or function of PrsA1 in L. monocytogenes. Similarly, limited information is currently available regarding the regulation of PrsA-like homologs in other bacteria. In S. pyogenes the expression of prsA2 appears to be negatively regulated either directly or indirectly by MtsR (metal transporter of streptococcus regulator) (Olsen et al., 2010). Disruption of mtsR increased the expression of prsA2 in addition to altering transcript levels of multiple operons (Olsen et al., 2010). In S. aureus, prsA expression is directly regulated by the VraRS two-component sentinel system of cell wall stress (Jousselin et al., 2012). A VraR binding site was identified 84 base pairs upstream of the S. aureus prsA and deletion of vraRS caused a significant decrease in prsA basal mRNA levels (Jousselin et al., 2012). In addition, the VraRS two component regulator was triggered by cell wall active antibiotics vancomycin and teicoplanin which increased the expression of prsA (Jousselin et al., 2012). Lastly, in L. lactis some evidence suggests that nitrogen source may regulate pmpA expression but a regulator has not been identified (Drouault et al., 2002). prsA expression in Gram-positive bacteria thus appears to be regulated in response to different external stimuli. In the case of L. monocytogenes and S. pyogenes, each of which has two prsA alleles, only the regulation of prsA2 has been observed.
Deciding who needs a chaperone: identifying PrsA2 substrates
Studies in B. subtilis first identified a role for PrsA as a post-translocation secretion chaperone located at the membrane-cell wall interface (Kontinen and Sarvas, 1993). Because prsA initially appeared essential for B. subtilis viability, the influence of PrsA on patterns of protein secretion and folding were assessed using IPTG-inducible prsA constructs and monitoring the membrane and extracellular proteome of PrsA depleted cells compared to PrsA replete cells (Kontinen and Sarvas, 1993; Hyyrylainen et al., 2000; Wahlstrom et al., 2003; Vitikainen et al., 2004). Depletion of B. subtilis PrsA results in the accumulation of misfolded proteins at the cell envelope and triggers a membrane stress response (Hyyrylainen et al., 2001). The identification of non-essential PrsA2 in L. monocytogenes and the regulation of prsA2 expression following PrfA activation enabled comparison of secreted protein profiles in ΔprsA2, wild type, and constitutively activated prfA* strains (Alonzo and Freitag, 2010). Currently, many direct and/or indirect PrsA-like substrates have been identified in L. monocytogenes and B. subtilis through proteomic analyses (Milohanic et al., 2003; Alonzo and Freitag, 2010; Hyyrylainen et al., 2010). These studies suggest that both L. monocytogenes and B. subtilis share many direct and/or indirect PrsA2/PrsA substrates with roles in cell wall metabolism (penicillin binding proteins), swimming motility (flagellin) and chemotaxis, oligopeptide transport (OppA), as well as a quinol oxidase associated with membrane bioenergetics, surface-localized enolase, and secreted superoxide dismutase (Milohanic et al., 2003; Alonzo and Freitag, 2010; Hyyrylainen et al., 2010) (Table 1). The fact that L. monocytogenes PrsA2 and B. subtilis PrsA appear to share common substrates suggests that these two PrsA-like proteins have conserved foldase and PPIase functions. However, not all potential substrates are shared between the two organisms. B. subtilis PrsA has been associated with the folding and activity of alpha-amylase, an enzyme which hydrolyses starch to generate glucose, and subtilisin, an alkaline protease (Jacobs et al., 1993); L. monocytogenes does not secrete these enzymes. In contrast, L. monocytogenes PrsA2 has been linked to the folding and activity of several L. monocytogenes secreted virulence factors not expressed by B. subtilis (Alonzo and Freitag, 2010). Evidence suggests, for example, that loss of PrsA2 leads to improper folding and degradation of secreted pore forming toxin listeriolysin O (LLO) as indicated by Western blot analyses (Alonzo et al., 2009; Zemansky et al., 2009).
Table 1.
Potential PrsA2/PrsA substrates relevant to bacterial cell physiology.
| Bacteria | PrsA-like cell effector | Gene function | References |
|---|---|---|---|
| Listeria monocy- | Lmo2522 | LysM domain penicillin binding protein | Alonzo and Freitag, 2010 |
| togenes | PbpA | Penicillin binding protein 2a transpeptidase | Alonzo and Freitag, 2010 |
| PbpB | Penicillin binding protein 2b transpeptidase | Alonzo and Freitag, 2010 | |
| Lmo0540 | Penicillin binding protein, putative | Alonzo and Freitag, 2010 | |
| OppA | Oligopeptide ABC-transporter, oligopeptide-binding protein | Alonzo and Freitag, 2010 | |
| FlaA | Flagellin | Alonzo and Freitag, 2010 | |
| QoxA | Quinol oxidase subunit II | Alonzo and Freitag, 2010 | |
| Eno | Enolase | Alonzo and Freitag, 2010 | |
| Sod | Superoxide dismutase | Alonzo and Freitag, 2010 | |
| Bacillus subtilis | AmyQ | α-amylase-hydrolyses starch to generate glucose | Kontinen and Sarvas, 1993 |
| AprE | Subtilisin- alkaline protease | Kontinen and Sarvas, 1993 | |
| PbpA | Penicillin binding protein 2a transpeptidase | Hyyrylainen et al., 2010 | |
| PbpB | Penicillin binding protein 2b transpeptidase | Hyyrylainen et al., 2010 | |
| PbpC | Penicillin binding protein 3 transpeptidase | Hyyrylainen et al., 2010 | |
| PbpD | Penicillin binding protein 4 transpeptidase | Hyyrylainen et al., 2010 | |
| YvrA | ABC transporter ATP-binding protein | Hyyrylainen et al., 2010 | |
| OppA | Oligopeptide-binding protein oppA precursor | Hyyrylainen et al., 2010 | |
| FliF | Flagellar M-ring protein | Hyyrylainen et al., 2010 | |
| FlhA | Flagellar biosynthesis protein flhA | Hyyrylainen et al., 2010 | |
| YlxF | FlaA locus 22.9 kDa protein | Hyyrylainen et al., 2010 | |
| QoxB | Quinol oxidase subunit I | Hyyrylainen et al., 2010 | |
| Eno | Enolase | Vitikainen et al., 2004 | |
| SodA | Superoxide dismutase | Vitikainen et al., 2004 |
Activation of the central virulence regulator PrfA results in increased expression of a number of secreted proteins that play central roles in L. monocytogenes pathogenesis (Leimeister-Wachter et al., 1990; Mengaud et al., 1991; Port and Freitag, 2007; Scortti et al., 2007; de Las Heras et al., 2011). These include the internalins InlA and InlB, associated with bacterial entry into host cells (Gaillard et al., 1991; Lingnau et al., 1995), LLO (Gaillard et al., 1987; Kuhn et al., 1988; Bielecki et al., 1990), the phospholipases PlcA and PlcB which mediate vacuole membrane lysis (Marquis et al., 1995; Smith et al., 1995; Grundling et al., 2003), the surface protein ActA which mediates actin polymerization (Domann et al., 1992; Vazquez-Boland et al., 1992; Brundage et al., 1993; Sokolovic et al., 1993), InlC which reduces cortical tension to facilitate bacterial cell-to-cell spread with in the host (Engelbrecht et al., 1996; Rajabian et al., 2009), and PrsA2 (Port and Freitag, 2007; Alonzo et al., 2009). Comparisons of the secreted protein profiles between L. monocytogenes wild type and strains lacking PrsA2 or containing constitutively activated PrfA suggests that PrsA2 contributes to the folding and activity of a number of PrfA-regulated gene products, including LLO, ActA, PlcB, InlC, and the metalloprotease that processes pro-PlcB to its activated form, Mpl (Alonzo and Freitag, 2010; Forster et al., 2011). Additional gene products that contribute to L. monocytogenes pathogenesis are associated with PrsA2 activity; these gene products do not appear to be directly regulated by PrfA but their secretion increases as a result of PrfA activation (Alonzo and Freitag, 2010). These include CtaP, a cysteine-transport associated lipoprotein associated with host cell adhesion, acid resistance, and bacterial membrane integrity (Xayarath et al., 2009), and ChiA, a chitinase that inhibits the expression of host inducible nitric oxide synthase (iNOS) (Chaudhuri et al., 2013). Both of these proteins have been identified as potential substrates of PrsA2 via proteomic analysis (Alonzo and Freitag, 2010). In the absence of prsA2, PrfA activation reduces L. monocytogenes fitness and compromises bacterial membrane integrity (Alonzo and Freitag, 2010). Taken together, these findings suggest that PrsA2 has evolved to promote the folding and activity of virulence factors whose expression is increased as a result of PrfA activation within mammalian host cells.
PrsA-like proteins have been associated with virulence factor secretion and activity in other Gram-positive bacterial pathogens. S. pyogenes PrsA2 is required for production of the fully mature, enzymatically active streptococcal cysteine protease SpeB (Ma et al., 2006) and loss of PrsA2 and thus SpeB activity has been associated with a reduction in the ability of S. pyogenes to cause necrotizing fasciitis (Olsen et al., 2010). In B. anthracis, production of protective antigen PA, a component of lethal and edema toxins, is dependent on PrsA (Williams et al., 2003). As we gain a better understanding of the activities of PrsA chaperones and how substrate recognition occurs, it is likely that the list of secreted proteins that depend upon PrsA-like molecules for folding and activity will increase in number.
Building a better bacterium: the role of PrsA-like homologs in cell wall biosynthesis and cell physiology
While evidence indicates that PrsA2 plays a critical role in the folding and stabilization of L. monocytogenes secreted virulence factors, PrsA2 has additional roles through its contributions to cell wall biosynthesis, resistance to osmotic stress, and swimming motility. L. monocytogenes strains lacking prsA2 exhibit increased sensitivity to penicillin, which targets peptidoglycan transpeptidation (Alonzo et al., 2011), and are also more sensitive to incubation in the presence of lysozyme (Forster et al., 2011). Strains that lack prsA2 have reduced levels of several penicillin binding proteins (PBPs) as well as cell wall modifying enzymes (Alonzo and Freitag, 2010), suggesting that selected aspects of L. monocytogenes cell wall biosynthesis may be altered in ΔprsA2 strains. Interestingly, PrsA2 mutants that lack the central PPIase domain but retain the N- and C-terminal foldase domains exhibit normal patterns of secretion and activity for a number of potential substrate proteins, including LLO and PlcB, but remain sensitive to penicillin (Alonzo et al., 2011). This finding suggests that PrsA2 substrates may be of two classes: those that require foldase function without PPIase activity, and those dependent on PPIase. Several PrsA2-associated PBPs have significant numbers of proline residues (ranging from 2 to 4% of the total amino acids), and thus may require PPIase activity for proper folding.
Evidence for PrsA-like protein contributions to cell wall biosynthesis and integrity in other bacteria is accumulating. In B. subtilis, PrsA depleted cells show a decrease in the degree of peptidoglycan cross-linkages (Hyyrylainen et al., 2010). In S. mutans, physical cell wall integrity of the prsA mutant was tested with sonication and revealed a drastic reduction in viability when compared to the wild-type strain (Guo et al., 2013). Disruption of prsA in S. aureus was found to cause bacterial sensitivity to both glycopeptides and oxacillin (Jousselin et al., 2012). In L. lactis, the inactivation of the prsA-like gene, pmpA caused increased sensitivity to ionic and non-ionic osmotic shock (Drouault et al., 2002). In addition in L. lactis, inactivation of pmpA caused a 3-fold decrease in growth rate when exposed to ionic (0.25 M NaCl) or non-ionic (0.6 M sorbitol) shock (Drouault et al., 2002). These prsA-associated phenotypes may be a direct result of a compromised cell wall and/or result from defects in the folding and activity of membrane and cell wall proteins associated with resistance to osmotic stress.
Another potential PrsA2/PrsA substrate identified in L. monocytogenes and B. subtilis was flagellin, required for the assembly of flagella and bacterial swimming motility (Alonzo and Freitag, 2010; Hyyrylainen et al., 2010). It has not yet been determined whether the FlaA subunit itself or alternatively some component(s) required for the assembly of the flagellar apparatus may require PrsA2 for folding. L. monocytogenes strains are flagellated and motile at low temperatures but have reduced motility at higher temperatures (37°C) associated with mammalian hosts (Peel et al., 1988). L. monocytogenes strains lacking prsA2 exhibit significantly reduced swimming motility at both low and high temperatures (Zemansky et al., 2009). Thus, while PrsA2 is required for L. monocytogenes pathogenesis and full bacterial viability during bacterial replication within the cytosol, it additionally contributes to several important aspects of L. monocytogenes physiology (osmotic resistance and bacterial motility) during bacterial growth outside of the host.
Putting it all together: modeling the functional contributions of PrsA2 to L. monocytogenes protein secretion both inside and outside of host cells
In Gram-positive bacteria, most secreted proteins are thought to cross the single bacterial cell membrane in an unfolded state to enter the space that exists between the cell membrane and the cell wall (Matias and Beveridge, 2005, 2006). This inner wall space characteristically contains high concentrations of cations bound to teichoic acid, a high density of negative charge, and a low pH which presents a challenging environment for protein folding and function (Sarvas et al., 2004). The proposed function of L. monocytogenes PrsA2 and related PrsA-like proteins in other Gram-positive bacteria is to promote post-membrane translocation protein folding and maintain optimal secretion homeostasis. Although L. monocytogenes PrsA2 contributes to several aspects of L. monocytogenes physiology during bacterial growth in broth culture or on solid media, its most striking contributions are detected during bacterial infection of mammalian cells where it is essential for the folding and activity of secreted proteins required for bacterial virulence and intracellular bacterial replication (Alonzo et al., 2009, 2011; Zemansky et al., 2009; Alonzo and Freitag, 2010) (Figure 4).
Figure 4.
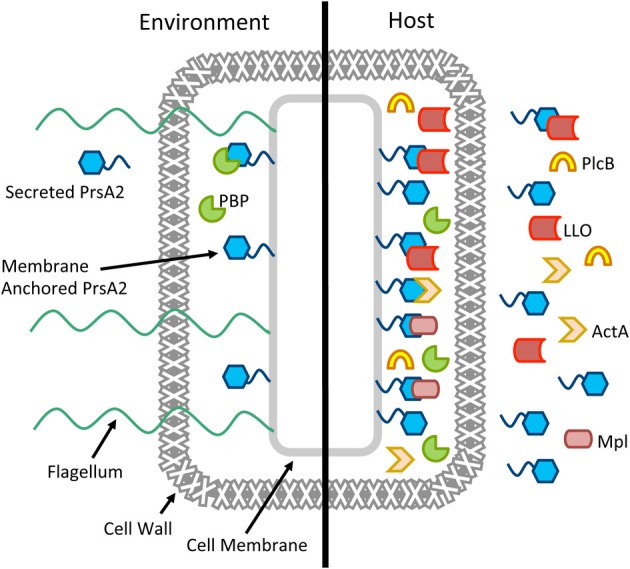
Working model for the multiple roles of PrsA2 in L. monocytogenes. A cartoon of a single L. monocytogenes cell is shown in the environment and in the host, with PrsA2 both tethered to the cell membrane and secreted. When L. monocytogenes is in the outside environment, PrsA2 directly or indirectly is required for functional penicillin binding proteins (PBP), flagellin, and other factors that contribute to cell wall integrity, swimming motility, and resistance to osmotic stress. When L. monocytogenes is in the host, activation of the central virulence regulatory protein PrfA leads to increased expression of several secreted virulence factors as well as PrsA2, which is required for the folding and stability of listeriolysin (LLO), the Mpl protease that activates the broad-range phospholipase (PlcB), ActA, and other factors.
During bacterial growth in rich media, prsA2 is expressed at a basal level with maximal expression occurring during exponential growth; expression rapidly decreases upon entry into stationary phase (Forster et al., 2011). PrsA2 associates with the bacterial membrane presumably as a result of the diacylglycerol modification of its N-terminal cysteine by the enzyme Lgt (Baumgartner et al., 2007). A significant fraction of PrsA2 can also be detected in bacterial supernatants; interestingly, secreted PrsA2 still appears to retain the N-terminal lipid modification (Baumgartner et al., 2007). PrsA2 assists indirectly or directly in the proper folding of substrates required for swimming motility, peptidoglycan synthesis, and osmotic stability (Zemansky et al., 2009; Alonzo and Freitag, 2010; Forster et al., 2011). Secretion of a second PrsA-like protein, PrsA1, has been detected for L. monocytogenes grown in broth culture and experimental evidence suggests that the protein is functional based on its PPIase activity (Alonzo et al., 2011), but the substrates for this chaperone have not been identified and its functional significance is not yet known.
Following L. monocytogenes entry into host cells, the virulence regulator PrfA becomes activated and increases gene expression by binding to target promoters that contain a PrfA palindromic DNA binding site (or PrfA box) (Xayarath and Freitag, 2012). PrfA activation leads to increased expression and secretion of a variety of protein products associated with bacterial virulence, including PrsA2 (Port and Freitag, 2007). The PrfA-dependent increase in secreted PrsA2 appears necessary for: (1) the proper folding and stability of several secreted virulence factors, including LLO, Mpl, and PlcB; (2) the maintenance of bacterial membrane integrity; and (3) the proper biosynthesis and modulation of the bacterial cell wall (Figure 4). L. monocytogenes strains lacking PrsA2 exhibit reduced viability under conditions of PrfA activation and are severely attenuated for virulence in mice (Alonzo et al., 2009, 2011; Alonzo and Freitag, 2010).
What are the critical aspects of PrsA2 function that promote L. monocytogenes survival and replication within the infected host? Mutants lacking PrsA2 and the secreted chaperone-protease HtrA rapidly lose viability under conditions of PrfA activation, suggesting that the loss of foldase activity and presumably HtrA protease degradation of misfolded proteins leads to a membrane stress response that reduces bacterial fitness within the cytosol (Alonzo and Freitag, 2010). It is possible that it is the reduction in bacterial fitness resulting from misfolded proteins and membrane stress, rather than a reduction in the secretion of a particular set of virulence factors, that leads to the dramatic 100,000-fold level of attenuation observed for ΔprsA2 mutants in mice (Alonzo et al., 2009). Interestingly, evidence suggests that both the foldase activity and the PPIase activity are required for full virulence, as PrsA2 mutants that lack PPIase activity but retain foldase remain attenuated in mouse models of infection, albeit to a lesser degree than full deletion mutants (Alonzo et al., 2011).
There are many functional aspects of L. monocytogenes PrsA2 and PrsA-like proteins in other Gram-positive bacteria that remain unknown. As yet, no structural information is available for this class of chaperones other than the reported tendency of L. monocytogenes PrsA2 and B. subtilis PrsA to form dimers (Hyyrylainen et al., 2010; Alonzo et al., 2011). How PrsA2 recognizes and interacts with its target substrates remains to be determined, as does the active form(s) of the protein (membrane associated vs. secreted). It is possible that the membrane-associated and secreted forms of PrsA2 serve distinct functions, such that the membrane-associated form is required for the correct folding of proteins following membrane translocation, while the secreted form may remain bound to the folded substrate to prevent aggregation or degradation until substrate activity is required. A more detailed understanding of PrsA2 and related homologs will not only increase current understanding of the process of protein secretion in Gram-positive bacteria, but may yield new strategies for the optimization of active protein secretion for purification or foreign antigen secretion in Gram-positive bacteria-based vaccine vectors. PrsA-like proteins may also be effective targets for small molecule inhibitors that could potentially reduce the production of secreted virulence factors during host infection. Given these possibilities, PrsA2 and PrsA-like proteins are chaperones whose molecular résumés are definitely worth a detailed look.
Authors and contributions
Laty A. Cahoon and Nancy E. Freitag contributed to the conception and writing of this review.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Freitag laboratory and the UIC Positive Thinking Group for helpful discussions. This work was supported by Public Health Service grants AI083241 (Nancy E. Freitag) from NIAID. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources.
References
- Alonzo F., 3rd., Freitag N. E. (2010). Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect. Immun. 78, 4944–4957 10.1128/IAI.00532-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F., 3rd., Port G. C., Cao M., Freitag N. E. (2009). The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 77, 2612–2623 10.1128/IAI.00280-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo F., 3rd., Xayarath B., Whisstock J. C., Freitag N. E. (2011). Functional analysis of the Listeria monocytogenes secretion chaperone PrsA2 and its multiple contributions to bacterial virulence. Mol. Microbiol. 80, 1530–1548 10.1111/j.1365-2958.2011.07665.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner M., Karst U., Gerstel B., Loessner M., Wehland J., Jansch L. (2007). Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189, 313–324 10.1128/JB.00976-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecki J., Youngman P., Connelly P., Portnoy D. A. (1990). Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature 345, 175–176 10.1038/345175a0 [DOI] [PubMed] [Google Scholar]
- Brundage R. A., Smith G. A., Camilli A., Theriot J. A., Portnoy D. A. (1993). Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 90, 11890–11894 10.1073/pnas.90.24.11890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. S., Hossain H., Otten S., Kuenne C., Kuchmina K., Machata S., et al. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338 10.1128/IAI.74.2.1323-1338.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Gantner B. N., Ye R. D., Cianciotto N. P., Freitag N. E. (2013). The Listeria monocytogenes ChiA chitinase enhances virulence through suppression of host innate immunity. MBio 4, e00617–e00612 10.1128/mBio.00617-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 108, 19484–19491 10.1073/pnas.1112371108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Las Heras A., Cain R. J., Bielecka M. K., Vazquez-Boland J. A. (2011). Regulation of listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127 10.1016/j.mib.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Domann E., Wehland J., Rohde M., Pistor S., Hartl M., Goebel W., et al. (1992). A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11, 1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouault S., Anba J., Bonneau S., Bolotin A., Ehrlich S. D., Renault P. (2002). The peptidyl-prolyl isomerase motif is lacking in PmpA, the PrsA-like protein involved in the secretion machinery of Lactococcus lactis. Appl. Environ. Microbiol. 68, 3932–3942 10.1128/AEM.68.8.3932-3942.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht F., Chun S. K., Ochs C., Hess J., Lottspeich F., Goebel W., et al. (1996). A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21, 823–837 10.1046/j.1365-2958.1996.541414.x [DOI] [PubMed] [Google Scholar]
- Forster B. M., Marquis H. (2012). Protein transport across the cell wall of monoderm Gram-positive bacteria. Mol. Microbiol. 84, 405–413 10.1111/j.1365-2958.2012.08040.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster B. M., Zemansky J., Portnoy D. A., Marquis H. (2011). Posttranslocation chaperone PrsA2 regulates the maturation and secretion of Listeria monocytogenes proprotein virulence factors. J. Bacteriol. 193, 5961–5970 10.1128/JB.05307-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag N. E., Youngman P., Portnoy D. A. (1992). Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J. Bacteriol. 174, 1293–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J. L., Berche P., Frehel C., Gouin E., Cossart P. (1991). Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65, 1127–1141 10.1016/0092-8674(91)90009-N [DOI] [PubMed] [Google Scholar]
- Gaillard J. L., Berche P., Mounier J., Richard S., Sansonetti P. (1987). In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55, 2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A., Gonzalez M. D., Higgins D. E. (2003). Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J. Bacteriol. 185, 6295–6307 10.1128/JB.185.21.6295-6307.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wu T., Hu W., He X., Sharma S., Webster P., et al. (2013). Phenotypic characterization of the foldase homologue PrsA in Streptococcus mutans. Mol. Oral Microbiol. 28, 154–165 10.1111/omi.12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen O., Seppala R., Tossavainen H., Heikkinen S., Koskela H., Permi P., et al. (2009). Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA–implications for the catalytic mechanism of parvulins. BMC Struct. Biol. 9:17 10.1186/1472-6807-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyyrylainen H. L., Bolhuis A., Darmon E., Muukkonen L., Koski P., Vitikainen M., et al. (2001). A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41, 1159–1172 10.1046/j.1365-2958.2001.02576.x [DOI] [PubMed] [Google Scholar]
- Hyyrylainen H. L., Marciniak B. C., Dahncke K., Pietiainen M., Courtin P., Vitikainen M., et al. (2010). Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol. Microbiol. 77, 108–127 10.1111/j.1365-2958.2010.07188.x [DOI] [PubMed] [Google Scholar]
- Hyyrylainen H. L., Vitikainen M., Thwaite J., Wu H., Sarvas M., Harwood C. R., et al. (2000). D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J. Biol. Chem. 275, 26696–26703 10.1074/jbc.M003804200 [DOI] [PubMed] [Google Scholar]
- Jacobs M., Andersen J. B., Kontinen V., Sarvas M. (1993). Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol. Microbiol. 8, 957–966 10.1111/j.1365-2958.1993.tb01640.x [DOI] [PubMed] [Google Scholar]
- Jousselin A., Renzoni A., Andrey D. O., Monod A., Lew D. P., Kelley W. L. (2012). The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 3629–3640 10.1128/AAC.06264-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontinen V. P., Sarvas M. (1993). The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8, 727–737 10.1111/j.1365-2958.1993.tb01616.x [DOI] [PubMed] [Google Scholar]
- Kuhn M., Kathariou S., Goebel W. (1988). Hemolysin supports survival but not entry of the intracellular bacterium Listeria monocytogenes. Infect. Immun. 56, 79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister-Wachter M., Haffner C., Domann E., Goebel W., Chakraborty T. (1990). Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 87, 8336–8340 10.1073/pnas.87.21.8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskela S., Wahlstrom E., Kontinen V. P., Sarvas M. (1999). Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene. Mol. Microbiol. 31, 1075–1085 10.1046/j.1365-2958.1999.01247.x [DOI] [PubMed] [Google Scholar]
- Lingnau A., Domann E., Hudel M., Bock M., Nichterlein T., Wehland J., et al. (1995). Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63, 3896–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Bryant A. E., Salmi D. B., Hayes-Schroer S. M., McIndoo E., Aldape M. J., et al. (2006). Identification and characterization of bicistronic speB and prsA gene expression in the group A Streptococcus. J. Bacteriol. 188, 7626–7634 10.1128/JB.01059-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis H., Doshi V., Portnoy D. A. (1995). The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63, 4531–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias V. R., Beveridge T. J. (2005). Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56, 240–251 10.1111/j.1365-2958.2005.04535.x [DOI] [PubMed] [Google Scholar]
- Matias V. R., Beveridge T. J. (2006). Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 188, 1011–1021 10.1128/JB.188.3.1011-1021.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Dramsi S., Gouin E., Vazquez-Boland J. A., Milon G., Cossart P. (1991). Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5, 2273–2283 10.1111/j.1365-2958.1991.tb02158.x [DOI] [PubMed] [Google Scholar]
- Milohanic E., Glaser P., Coppee J. Y., Frangeul L., Vega Y., Vazquez-Boland J. A., et al. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47, 1613–1625 10.1046/j.1365-2958.2003.03413.x [DOI] [PubMed] [Google Scholar]
- Mostowy S., Cossart P. (2012). Virulence factors that modulate the cell biology of Listeria infection and the host response. Adv. Immunol. 113, 19–32 10.1016/B978-0-12-394590-7.00007-5 [DOI] [PubMed] [Google Scholar]
- Mueller K. J., Freitag N. E. (2005). Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect. Immun. 73, 1917–1926 10.1128/IAI.73.4.1917-1926.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. J., Sitkiewicz I., Ayeras A. A., Gonulal V. E., Cantu C., Beres S. B., et al. (2010). Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc. Natl. Acad. Sci. U.S.A. 107, 888–893 10.1073/pnas.0911811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel M., Donachie W., Shaw A. (1988). Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and western blotting. J. Gen. Microbiol. 134, 2171–2178 [DOI] [PubMed] [Google Scholar]
- Port G. C., Freitag N. E. (2007). Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 75, 5886–5897 10.1128/IAI.00845-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabian T., Gavicherla B., Heisig M., Muller-Altrock S., Goebel W., Gray-Owen S. D., et al. (2009). The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 11, 1212–1218 10.1038/ncb1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvas M., Harwood C. R., Bron S., Van Dijl J. M. (2004). Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694, 311–327 10.1016/j.bbamcr.2004.04.009 [DOI] [PubMed] [Google Scholar]
- Scortti M., Monzo H. J., Lacharme-Lora L., Lewis D. A., Vazquez-Boland J. A. (2007). The PrfA virulence regulon. Microbes Infect. 9, 1196–1207 10.1016/j.micinf.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Shetron-Rama L. M., Mueller K., Bravo J. M., Bouwer H. G., Way S. S., Freitag N. E. (2003). Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48, 1537–1551 10.1046/j.1365-2958.2003.03534.x [DOI] [PubMed] [Google Scholar]
- Sigrist C. J., Cerutti L., De Castro E., Langendijk-Genevaux P. S., Bulliard V., Bairoch A., et al. (2010). PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166 10.1093/nar/gkp885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. A., Marquis H., Jones S., Johnston N. C., Portnoy D. A., Goldfine H. (1995). The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63, 4231–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolovic Z., Riedel J., Wuenscher M., Goebel W. (1993). Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol. Microbiol. 8, 219–227 10.1111/j.1365-2958.1993.tb01566.x [DOI] [PubMed] [Google Scholar]
- Stoll H., Dengjel J., Nerz C., Gotz F. (2005). Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73, 2411–2423 10.1128/IAI.73.4.2411-2423.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossavainen H., Permi P., Purhonen S. L., Sarvas M., Kilpelainen I., Seppala R. (2006). NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis. FEBS Lett. 580, 1822–1826 10.1016/j.febslet.2006.02.042 [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland J. A., Kocks C., Dramsi S., Ohayon H., Geoffroy C., Mengaud J., et al. (1992). Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitikainen M., Lappalainen I., Seppala R., Antelmann H., Boer H., Taira S., et al. (2004). Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J. Biol. Chem. 279, 19302–19314 10.1074/jbc.M400861200 [DOI] [PubMed] [Google Scholar]
- Wahlstrom E., Vitikainen M., Kontinen V. P., Sarvas M. (2003). The extracytoplasmic folding factor PrsA is required for protein secretion only in the presence of the cell wall in Bacillus subtilis. Microbiology 149, 569–577 10.1099/mic.0.25511-0 [DOI] [PubMed] [Google Scholar]
- Williams R. C., Rees M. L., Jacobs M. F., Pragai Z., Thwaite J. E., Baillie L. W., et al. (2003). Production of Bacillus anthracis protective antigen is dependent on the extracellular chaperone, PrsA. J. Biol. Chem. 278, 18056–18062 10.1074/jbc.M301244200 [DOI] [PubMed] [Google Scholar]
- Witte C. E., Archer K. A., Rae C. S., Sauer J. D., Woodward J. J., Portnoy D. A. (2012). Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv. Immunol. 113, 135–156 10.1016/B978-0-12-394590-7.00002-6 [DOI] [PubMed] [Google Scholar]
- Xayarath B., Freitag N. E. (2012). Optimizing the balance between host and environmental survival skills: lessons learned from Listeria monocytogenes. Future Microbiol. 7, 839–852 10.2217/fmb.12.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xayarath B., Marquis H., Port G. C., Freitag N. E. (2009). Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol. Microbiol. 74, 956–973 10.1111/j.1365-2958.2009.06910.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemansky J., Kline B. C., Woodward J. J., Leber J. H., Marquis H., Portnoy D. A. (2009). Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191, 3950–3964 10.1128/JB.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]