Abstract
Autophagy (macroautophagy) is a highly conserved intracellular and lysosome-dependent degradation process in which autophagic substrates are enclosed and degraded by a double-membrane vesicular structure in a continuous and dynamic vesicle transport process. The Rab protein is a small GTPase that belongs to the Ras-like GTPase superfamily and regulates the vesicle traffic process. Numerous Rab proteins have been shown to be involved in various stages of autophagy. Rab1, Rab5, Rab7, Rab9A, Rab11, Rab23, Rab32, and Rab33B participate in autophagosome formation, whereas Rab9 is required in non-canonical autophagy. Rab7, Rab8B, and Rab24 have a key role in autophagosome maturation. Rab8A and Rab25 are also involved in autophagy, but their role is unknown. Here, we summarize new findings regarding the involvement of Rabs in autophagy and provide insights regarding future research on the mechanisms of autophagy regulation.
Keywords: autophagy, autophagosome, Rab GTPase
Facts
Autophagy (macroautophagy) is a highly conserved intracellular and lysosome-dependent degradation process in eukaryotes.
In autophagy, cytoplasmic compartments are enclosed by an isolation membrane that elongates to form an autophagosome with a double membrane and eventually fuses with the lysosome to degrade the enclosed contents.
Autophagy is involved in vesicular formation, transport, tethering, and fusion.
As a vesicular trafficking protein, many Rab GTPases regulate various steps of autophagy.
Open Questions
The regulatory mechanisms of autophagy are not fully understood.
Other Rab proteins may regulate autophagy.
The detailed molecular mechanisms of autophagy regulated by Rab proteins remain unclear.
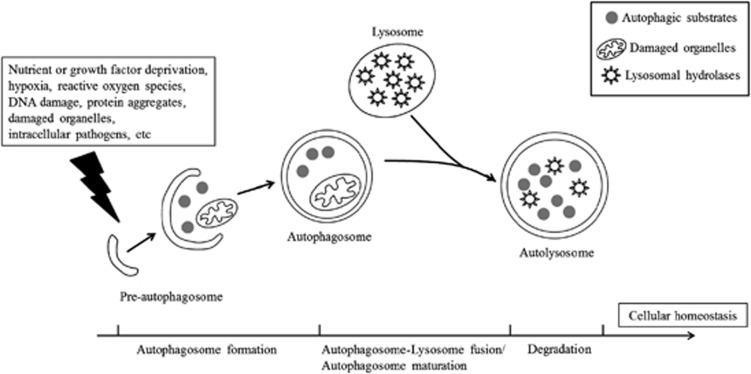
Autophagy (macroautophagy) is an ancient and highly conserved intracellular degradation process during which cytoplasmic compartments (including protein, sugars, lipids, and organelles such as mitochondria, peroxisomes, and endoplasmic reticulum (ER)) are enclosed by an isolation membrane called a pre-autophagosome or phagophore that elongates to form an autophagosome with a double membrane and eventually fuses with the lysosome or vacuole to degrade the enclosed contents.1, 2, 3, 4 Autophagy can be induced by various forms of cellular stress, including nutrient or growth factor deprivation, hypoxia, reactive oxygen species, DNA damage, protein aggregates, damaged organelles, or intracellular pathogens.5 As a cytoplasmic quality-control mechanism, autophagy is vital for cellular homeostasis, and the dysfunction of autophagy may lead to many diseases such as neurodegenerative diseases, cardiomyopathy, tumorigenesis, and pathogenic infection (Figure 1).6, 7, 8, 9, 10
Figure 1.
Schematic model of autophagy. Autophagy is induced by various cellular stressors and is related to cellular homeostasis. Following the induction of autophagy, autophagic substrates are enclosed by an isolation membrane called a pre-autophagosome or phagophore that elongates to form an autophagosome with a double membrane (autophagosome formation), and eventually fuses with the lysosome to degrade the enclosed contents (autophagosome–lysosome fusion/autophagosome maturation and degradation)
As previously described, the autophagic process can be roughly divided into three steps: autophagosome formation, autophagosome–lysosome/late endosome fusion (autophagosome maturation), and degradation.3 Although all steps of the dynamic autophagic process are known, several stages (especially in the initial steps of omegasome formation and isolation membrane initiation) remain controversial. Clues regarding the origin of the autophagosomal membrane have been provided by studies involving yeast in which autophagosome formation occurs at phagophore assembly site (PAS) where key proteins involved in autophagosome formation colocalize.11, 12 However, in mammalian cells, the existence of PAS remains unclear, as multiple autophagosomal assembly sites, including the ER, Golgi, endosomes, and mitochondria, have been detected.12, 13 The predominant view is that autophagosomes are formed from ER-associated membrane compartments enriched in phosphatidylinositol 3-phosphate (PI3P) and double FYVE domain-containing protein 1; these compartments are termed omegasomes for their omega-like shape.14
Genetic studies in Saccharomyces cerevisiae15, 16 identified ATG (Autophagy-related) genes conserved from yeast to mammals. Most ATG-encoding proteins (Atg proteins) are involved in different steps of autophagy, and the ‘core' Atg protein complexes are crucial.3, 17 The ‘core' Atg proteins are separated into five subgroups:3 the Atg1/ULK1 protein kinase complex, the Atg9·Atg2-Atg18 complex, the Vps34-Atg6/beclin1 class III PI3-kinase complex, the Atg12 conjugation system, and the Atg8/LC3 conjugation system. Each complex has a different role in autophagy; the Atg9·Atg2-Atg18 complex is required for the initial step in autophagosome formation,18, 19, 20 the Vps34-Atg6/beclin1 class III PI3-kinase complex produces PI3P that contributes to the localization of Atg proteins on the autophagic membrane,21, 22, 23 and the Atg12 and the Atg8/LC3 conjugation systems are responsible for the lipidation process of LC3-I to LC3-II that is localized to the autophagosome until its fusion with lysosomes and is thus a well-known marker of autophagosomes.24, 25, 26
It is known that Rab proteins constitute the largest family in the Ras-like GTPase superfamily and shuttle between an active state (GTP-bound) and an inactive state (GDP-bound) to have a key role in diverse vesicular trafficking events. Because GDP tightly binds Rab proteins and Rab-bound GTP is hydrolyzed very slowly, the switch is assisted by auxiliary proteins: guanine nucleotide exchange factors (GEFs) catalyze GDP dissociation to allow its replacement with GTP, whereas GTPase-activating proteins (GAPs) facilitate hydrolysis of GTP to GDP.27, 28 A common regulatory mechanism of transport vesicle traffic by GEFs, GAPs, and Rab proteins is termed as the Rab GEF/GAP cascade. For example, Rab5 is vital for the early endocytic pathway and Rab7 regulates late endosome transport. In Caenorhabditis elegans, after recruitment of Rab5 to the endosomal membrane, Rab5 is activated by its GEF, namely Rabex-5, and binds various effectors such as early endosome antigen 1 to promote endosome fusion. At the same time, SAND-1/Mon1p (monensin sensitivity 1) is recruited to the membrane by interacting with Rabex-5 and PI3P, and it recruits Rab7 through homotypic fusion and the vacuole protein sorting (HOPS) complex. Moreover, Mon1 forms a complex with calcium caffeine zinc sensitivity (Ccz1) to activate Rab7 as its GEF. When Rab7 is activated to the appropriate level, TBC-2 (Tre-2/Bub2/Cdc16), a homolog of human TBC1D2 and TBC1D2B, is localized to the endosomal membrane by Rab7, working as a GAP for Rab5 to inactivate it. Through the sequential activity of GEFs, Rabex-5, Mon1, and TBC-2, Rab5 and Rab7 can be activated/inactivated at the appropriate time and place to regulate endosome traffic.29
It is well established that autophagy is involved in vesicular formation, transport, tethering, and fusion. Thus, as molecular ‘switches' to regulate the formation, transport, tethering, and fusion of transport vesicles, Rab small GTPases have been found to participate in the autophagic process.2, 30 Recent studies have shown that some Rabs, including Rab1, Rab5, Rab7, Rab8A, Rab8B, Rab9, Rab11, Rab23, Rab24, Rab25, Rab32, and Rab33B, are essential for autophagy.
Ypt1 Module and Autophagosome Formation in Yeast
Yeast Ypt1 and its human functional homolog Rab1 are required for ER-to-Golgi transport.31, 32 De Antoni et al.33 first discovered that Ypt1 may be involved in autophagy. In their study of different Ypt1-GAP deletions, mutants that expressed the lasting active GTPase-deficient Ypt1(Q67L)p exhibited growth defects at a low or permissive temperature and exhibited various morphological alterations resembling autophagy. Subsequent studies showed that Rab1/Ypt1 has an important role in autophagy, especially in autophagosome formation.
The GEF for Ypt1 is called the Transport protein Particle (TRAPP) complex,34 and was previously identified in two forms: TRAPPI and TRAPPII.35 Previous studies showed that Trs85, a nonessential subunit of the TRAPP complexes,36 is required in yeast for both nonselective and selective macroautophagy.37, 38, 39 A recent report40 described a new form of the TRAPP complex, TRAPPIII, that contains Trs85 and acts as a GEF for Ypt1. The Trs85 subunit of the TRAPPIII complex directs this Ypt1 GEF and Ypt1 to the PAS, which is the autophagosome formation site.
Although Ypt1 and its GEF have been implicated in autophagy, the mechanism by which Ypt1 regulates autophagy is unknown. Lipatova et al.41 recently identified a Ypt1 GTPase module containing Trs85, Ypt1, and Atg11 that regulates selective autophagy in yeast. Atg11, which is a PAS organizer in selective autophagy and interacts with Atg9 to affect its cellular localization,14 was found to interact specifically with the GTP form of Ypt1 in the CC2-3 domain of Atg11, and in ypt1-1 mutant cells, that are defective in the interaction with Atg11, GFP-Atg11 appears as multiple puncta in contrast to the single-dot conformation in wild-type (wt) cells.42, 43 Furthermore, the two other PAS components, Atg1 and Atg8, show altered localization, and the mutants show defective PAS assembly. These observations support the idea that Atg11 is a downstream effector of Ypt1, and that the Ypt1-Atg11 interaction is vital for PAS assembly and function. In addition, multicolor BiFC shows that Trs85, Ypt1, and Atg11 colocalize and interact in the PAS. Importantly, overexpression of Ypt1 can suppress defects in the autophagic process caused by Trs85 deletion but not Atg11 deletion. These results indicate that Trs85, Ypt1, and Atg11 function as a GEF–GTPase–effector module in selective autophagy. A similar Ypt1 GTPase module was not discovered in nonselective autophagy, but Atg17 was suggested to be a potential effector of Ypt1. Trs85 and Ypt1 were also shown to interact on Atg9-containing membranes and were involved in Atg9 localization. Thus, Ypt1 was speculated to interact first with Trs85 on Atg9-containing membranes and then with Atg11 to promote PAS assembly through the interaction of Atg9 and Atg11. The results of another study performed in the same year supported this hypothesis.44
Rab1 and Autophagosome Formation in Mammals
Under normal conditions
It is unclear whether a module similar to the Trs85-Ypt1-Atg11 module in yeast is present in mammals. However, an earlier study showed that autophagosome formation in mammals depends on the mammalian Ypt1 ortholog Rab.45 Rab1B wt and Rab1B Q67L (GTPase deficient) were observed to colocalize with LC3, and the extent of colocalization was found to increase when cells were starved or subjected to inhibition of autophagosome maturation. Moreover, in cells overexpressing either Rab1B wt or Rab1B Q67L, the number of autophagosomes was markedly increased, and the amount of endogenous LC3-II indicated that overexpression of Rab1B or Rab1B Q67L affects autophagosome formation. In contrast to the functional enhancement of Rab1, inhibition of Rab1 function by overexpression of the nucleotide empty-form mutant Rab1B N121I or by silencing of Rab1B decreases autophagosome formation. Surprisingly, although the secretory pathway downstream of Sar1 and Rab1 was confirmed to be involved in autophagosome formation, disruption of the secretory pathway by brefeldin A, which inhibited Arf1 GEF and assembly of COPI, did not affect autophagosome formation under starvation, which suggests that the later stages of the early secretory pathway are not essential for autophagosome biogenesis.
Under pathological conditions
Parkinson's disease is characterized by the accumulation of intraneuronal α-synuclein protein, which forms protein aggregates called Lewy bodies.46 Early studies showed that the pathogenic α-synuclein mutants appeared to obstruct the chaperone-mediated autophagy pathway.47 Winslow et al.48 recently found that overexpression of wt α-synuclein in mammals not only perturbed the secretory pathway but also inhibited autophagy by impairing omegasome formation, which is mediated through inhibition of Rab1A. In addition, Rab1A knockdown or α-synuclein overexpression caused the mislocalization of Atg9. Collectively, these data support a model in which α-synuclein impairs Rab1A activity and then influences Atg9 function to decrease autophagosome formation (Figure 2). It is important to note that Winslow et al.48 found that knockdown of Rab1B increased LC3-II levels that is not consistent with the results of a study by Zoppino et al.45 Therefore, it is necessary to identify the true function of Rab1B in autophagy.
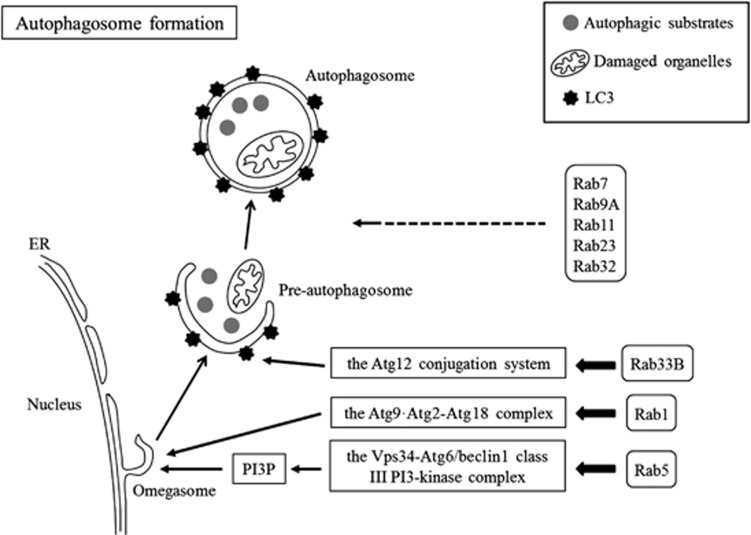
Figure 2.
Schematic diagram illustrating the regulation of autophagosome formation by Rab proteins. Following the induction of autophagy, autophagosome formation is regulated by Rab1 through the Atg9·Atg2-Atg18 complex, by Rab5 through the Vps34-Atg6/beclin1 class III PI3-kinase complex, by Rab33B through the Atg12 conjugation system, and by Rab7, Rab9A, Rab11, Rab23, and Rab32 via unknown molecular mechanisms. ER, endoplasmic reticulum
Autophagy has a key role in both innate and adaptive immunity, and is a recently identified component of the immune response to bacterial infection.49, 50 Huang et al.51 showed that the formation of bacteria-containing autophagosomes for antibacterial autophagy of Salmonella typhimurium occurs at Ptdlns(3)P-enriched domains of the ER and requires Rab1. Although Rab1 seems to be involved in protection of the cytoplasm from bacterial colonization by participating in autophagy in Salmonella typhimurium infection, it also takes part in the survival of some species of intracellular bacteria that use certain strategies to avoid the host autophagy pathway.
The GAPs for Rab GTPases contain a TBC Rab-binding domain.52 Shigella flexneri can resist autophagy-mediated defense in infected epithelial cells and translocates the effector VirA, which is vital for intracellular bacterial persistence, into host cells by a type III secretion apparatus.53 A recent study54 demonstrated that VirA possesses TBC-like GAP activity for Rab1, and also found that deletion of VirA and deficiency of GAP activity in Shigella flexneri mutants caused >20% of infected cells to form autophagosomes containing intracellular bacteria. In addition, expression of the domain-negative GDP-bound form Rab1 S25N absolutely blocked autophagosome formation around the GAP-inactive VirA-RQ mutant. These results indicate that VirA inactivation of Rab1 counteracted the autophagy-mediated host defense to keep intracellular Shigella flexneri alive.
Joshi et al.55 predicted that, in response to Legionella pneumophila infection, some effectors released by Legionella pneumophila would regulate macrophage autophagy as a barrier to cytosolic infection. In their speculative model, after macrophages engulf Legionella pneumophila within the autophagosomal membrane derived from the ER, bacterial proteins DrrA and LidA persistently activate Rab1, RalF, and Arf1 to prevent the Legionella pneumophila-containing autophagosome from fusing with lysosomes. A few hours later, Legionella pneumophila secretes LepB that inactivates Rab1. As a result, the autophagosome–lysosome fuses to the autolysosome. During the intentional pause caused by proteins released from the bacteria, the pathogen develops acid resistance and other features that exploit autolysosome as a replication niche.
Endocytic Rab Proteins and Autophagosome Formation
Rab5, Vps34, and Beclin1
The Rab5 protein is present in the plasma membrane, clathrin-coated vesicles, and early endosome that mediates fusion of endocytic vesicles to form early endosomes.56 In cell and fly models of Huntington's disease, Rab5 was shown to participate in autophagosome formation to regulate autophagy, which is used to eliminate toxic mutant huntingtin.57 Rab5 was found to activate Vps34, and inhibition of Rab5 or Vps34 activity reduced Atg5-Atg12 conjugation, which resulted in decreased autophagosome formation. In addition, Rab5 interacts with Beclin1 only when Vps34 is present. Vps34 and Beclin1 are present in a complex involved in autophagosome formation.3 Thus, these results indicate that Rab5 is a part of the macromolecular complex containing Beclin1 and Vps34 that regulates autophagosome formation (Figure 2).
Another group of PI3 kinases, the class IA PI3-kinase complex, is considered to inhibit autophagy by activating the Akt/mTOR-signaling pathway,10 but Dou et al.58 found that that the p110β subunit of the complex regulates the catalytic activity of the Vps34 complex to promote PI3P generation, which is essential for autophagosome formation. It was recently found that constitutively active Rab5 improved the autophagy deficiency caused by p110β knockdown, and p110β associated with Rab5 to promote the transition from Rab5-GDP to Rab5-GTP. In addition, only under growth factor limitation, such as in serum deprivation, the p110β-Rab5 and Rab5-Vps34 interactions were selectively enhanced, and the enhancement of the latter depended on the former that resulted in enhanced production of PI3P that was visualized as an increase in the formation of GFP-FYVE puncta.59 Taken together, these data provide a direct molecular connection mechanism where in response to growth factor limitation, the class IA PI3-kinase p110β subunit promotes Rab5 activation, followed by activation of the macromolecular complex composed of Rab5, Vps34, and Beclin1, which in turn leads to autophagosome formation (Figure 2).
In another study, Rab5 and Vps34 were shown to be important for hepatitis C virus NS4B-induced autophagy.60 NS4B is a nonstructural hepatitis C virus protein derived from the proteolytic process of a single polyprotein and it induces formation of the membranous web to provide a site for viral RNA replication.61, 62 In cells expressing NS4B, GFP-LC3 puncta formation was clearly observed and a high level of LC3-II was detected, which indicates that NS4B can induce autophagosome formation. When Rab5 was knocked down by short hairpin RNA, LC3-II formation was significantly decreased upon exogenous NS4B expression. Imaging and quantification data showed that expression of dominant-negative GFP-Rab5 not only changed the distribution pattern of mCherry-LC3 but also severely weakened NS4B-induced autophagosome formation. In addition, inhibition of Vps34 function by 3-methyladenine or RNA interference drastically diminished NS4B-induced puncta formation for GFP-LC3 and LC3-II formation. In the co-immunoprecipitation assay, NS4B, Rab5, Vps34, and beclin1 coprecipitated with each other. Thus, the previous findings collectively suggest that NS4B participates in the macromolecular complex containing Rab5, Vps34, and Beclin1 to induce autophagosome formation in hepatitis C virus infection-induced autophagy (Figure 2).
Although many studies have suggested that Rab5 is vital for autophagy, disruption of genes such as rab5 and its GEF rabx5 that regulate endocytic pathways actually activates autophagy in C. elegans.63
Rab7 and other Rab proteins in Group A Streptococcus-containing autophagosome-like vacuole formation
The Rab7 protein is another common endocytic Rab protein that coordinates traffic between the late endosome and lysosome.56 Many studies have shown that Rab7 is involved in autophagosome maturation. Rab7 also participates in the initial step of Group A Streptococcus (GAS)-containing autophagosome-like vacuole (GcAV) formation.64 GcAVs contain GAS escaping from the endolysosomal pathway and share several characteristics with autophagosomes.65 After internalization of GAS into cells, small GcAVs fuse to form a large, terminal GcAV through four morphological changes. In cells expressing dominant-negative Rab7 (T22N), the GFP-LC3 signal slightly accumulated around GAS and no GAS was enclosed in GcAV-like membrane-delimited structures. In addition, imaging data demonstrated colocalization of mRFP-Rab7 and GFP-Atg5, which is a marker of GcAV precursor structure, at DAPI-stained GAS. These findings indicate that Rab7 takes part in an early step in GcAV formation, perhaps in the interaction between the GcAV precursor structure and intracellular GAS.
In addition to Rab7, Rab9A and Rab23 have been confirmed to be involved in GcAV formation.66 Rab23 is recruited to developing GcAVs, and knockdown of Rab23 reduced GcAV formation by >50%. Furthermore, the number of Atg5-associated GAS and GcAVs decreased, which suggests that Rab23 is required for GcAV formation. Furthermore, GAS infection increased the number of LC3-positive dots that did not associate with GAS but associated with Atg5 in Rab23-knockdown cells or cells expressing Rab23 S23N, which is the GDP-bound form. Accordingly, the role of Rab23 in GcAV formation is similar to that of Rab7. In contrast to Rab7 and Rab23, Rab9A has a role in homotypic fusion between small GcAVs and lysosomal fusion. Knockdown of Rab9A or overexpression of the GDP-bound form of Rab9A S21N notably decreased the size of terminal GcAVs but increased the number of GcAVs per cell.
Rab11 and amphisome formation
Morphological, molecular, and biochemical studies have shown that autophagosomes fuse with multivesicular bodies (MVBs) or endosomes to form specific autophagosomes called amphisomes.67, 68, 69
Rab11 decorates MVBs in K562 cells, although it is usually localized to the Golgi, recycling endosome (RE), or early endosomes.70 Treatment of K562 cells with starvation or rapamycin not only induces autophagy but also promotes the fusion of Rab11-decorated MVBs with LC3-labeled autophagosomes to generate enlarged hybrid organelles, that is, amphisomes. K562 cells cotransfected with RFP-LC3 and GDP-bound GFP-Rab11S25N showed autophagy, but the colocalization between GFP-Rab11 with RFP-LC3 was decreased by starvation, which suggests that loss of Rab11 function does not affect autophagy, and that fusion of MVBs with the autophagosome seems to require GTP-Rab11 protein.69
As a TBC domain protein (i.e., a Rab-binding domain of GAPs as previously mentioned), TBC1D14 was recently found to regulate autophagosome formation through Rab11/ULK1-positive RE.71 A yeast two-hybrid assay revealed a specific interaction between TBC1D14 and Rab11 that promotes autophagosome formation, but structural analysis of the TBC1D14 TBC domain suggested that it did not have GAP function.
Transferrin (Tfn) is internalized into early endosomes and is efficiently recycled out of the cell by the endocytic recycling compartment. In TBC1D14- and Rab11-transfected cells, a small amount of Tfn was internalized and was then delivered normally to the TBC1D14-induced tubular structure containing ULK1, Rab11, TBC1D14, Atg13, FIP200, and the Tfn receptor, rather than recycling out of the cell, which indicates that RE function was affected by Rab11 and TBC1D14, most likely as a consequence of the tubulation of REs. Representative images showed that Tfn receptor was present at the edges of GFP-LC3-positive autophagosomes. In addition, both the REs and autophagosomes contained the Tfn-HRP-DAB reaction product, and the reaction product was concentrated at the edges of the autophagosomes. We therefore hypothesize that TBC1D14 binds specifically to Rab11, which induces the formation of RE tubules that contribute to autophagosome formation.
Rab32 and Autophagosome Formation
Rab32 is thought to localize to mitochondria and melanosomes to regulate the membrane trafficking of the trans-Golgi network and mitochondrial fission.30 A recent study showed that Rab32 also localizes to the ER and is required for autophagosome formation.72 In cells overexpressing Rab32WT and Rab32Q85L (a GTP-bound form), many large spherical structures appeared in the perinuclear region and were stained by an antibody against LC3, which indicated that these structures were autophagic vacuoles. However, expression of the GTP non-binding form of Rab32, Rab32T39N, and Rab32N143I caused the formation of large dot-like or chain-like ubiquitinated protein-containing aggresome-like structures. In these cells, a large amount of p62 was degraded by autophagy and accumulated in Rab32T39N- and Rab32N143I-positive aggresome-like structures, and most of the LC3 was distributed throughout the cytoplasm rather than forming punctate structures. Therefore, Rab32 not only regulates autophagosome formation by affecting the recruitment of LC3 but also impacts aggresome formation.
In their analysis of lipid storage in Drosophila, Wang et al.73 found that loss-of-function mutants of Rab32 and the Rab32 GEF claret had smaller lipid droplets than the wt, which suggests that Rab32 and its GEF claret have a role in maintaining normal lipid droplet size in Drosophila. In addition, expression of RFP-Atg8a and Rab32-EYFP in fat bodies demonstrated colocalization of Atg8a and Rab32. In the homozygous loss-of-function mutants of Rab32 and the Rab32 GEF claret, the number of autophagosomes was significantly reduced, and there was a persistent cortical signal that should have been lost with autophagic cell death. The impairment of autophagy led to small lipid droplets. Thus, we hypothesize that Rab32 and its GEF claret are involved in autophagy and may govern the lipid storage of Drosophila through autophagy.
Golgi-resident Rab33B and Autophagosome Formation
The Golgi is another supplier of the autophagosomal membrane. Golgi-resident Rab33B has recently been confirmed to interact with Atg16L and modulate autophagosome formation.74 In a GST pull-down assay, Atg16L was found to specifically and strongly interact with Rab33B. In addition, either transient expression or constitutive expression of a constitutively active mutant of Rab33B, Rab33B-QL, increased the amount of LC3-II even under full nutrient conditions. LC3-II also accumulated upon treatment with the H+/K+ ionophore nigericin, which implies that the increased LC3 lipidation resulted from increased autophagosome formation and not blockage of autophagic degradation. In contrast, knockdown of Rab33B markedly decreased autophagosome formation. Thus, these results suggest that Atg16L is an effector of Rab33B that participates in autophagosome formation (Figure 2).
Rab9 and Autophagosome Formation in Non-canonical Autophagy
Atg5 and Atg7 proteins are essential for the two ubiquitin-like conjugation systems in macroautophagy. However, a recent report described Atg5/Atg7-independent macroautophagy.75 Typical autophagic structures including double-membrane structures, autophagosomes, amphisomes, and autolysosomes were observed in etoposide-treated Atg5−/− mouse embryonic fibroblasts (MEFs) and Atg7−/− MEFs. Similar macroautophagy was also observed in staurosporine-treated Atg5−/− MEFs and Atg5−/− thymocytes, and this pathway also demonstrated proteolytic function. ULK1, FIP200, Beclin1, and Vps34 (but not Atg7, Atg12, Atg16, or Atg9) were involved in non-canonical autophagy. The colocalization of organelle markers with Lamp2-positive vacuoles, representing autolysosomes, in Atg5−/− MEFs confirmed the involvement of the trans-Golgi or late endosomes in non-canonical autophagy and also predicted the association of Rab9, which is involved in trafficking from late endosomes to the trans-Golgi.30 As expected, GFP-Rab9 colocalized with Lamp2-positive autolysosomes and the colocalization was enhanced for GFP-Rab9(Q66L), a GTP-bound form, and reduced with GFP-Rab9(S21N), a GDP-bound form, in etoposide-treated Atg5−/− MEFs. Moreover, Rab9 silencing decreased the quantity of autophagic vacuoles but increased the accumulation of isolation membranes, which represent the membrane origin of autophagic vacuoles. These findings suggest that Rab9 is involved in non-canonical autophagosome generation.
As a D2 dopamine receptor antagonist, raclopride induces autophagy in cardiac myocytes.76 In this type of autophagy, Rab9 protein expression is increased at 12 h and there is no change in the protein level of Atg7 within 24 h after raclopride treatment. Downregulation of Rab9 in raclopride-treated cells caused reduced clearance of p62, which is a substrate of autophagy, and lipidation of LC3-I to LC3-II. However, downregulation of Atg7 had no effect on LC3 lipidation or p62 clearance. These results suggest that raclopride-induced autophagy requires Rab9 but probably does not require Atg7.
Key Role of Rab7 in Autophagosome Maturation
Many studies have confirmed that Rab7 has a key role in autophagosome maturation.77, 78, 79, 80, 81 Rab7 promotes microtubule plus-end-directed transport and fusion of autophagosomes with lysosomes through a novel FYVE and coiled-coil domain-containing protein FYCO1 (Figure 3).82
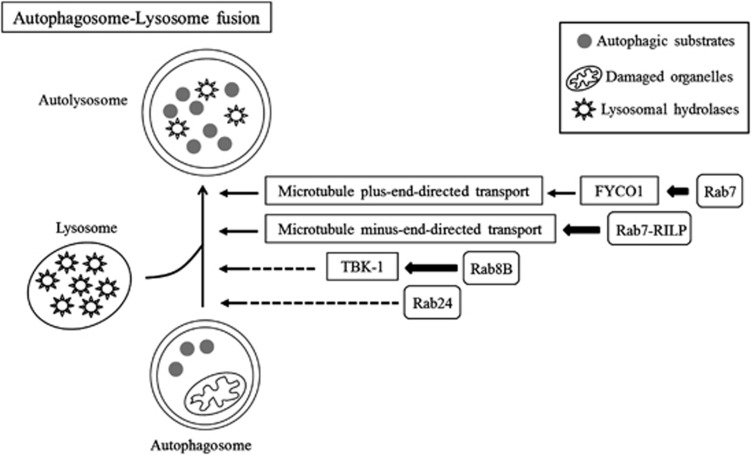
Figure 3.
Schematic diagram illustrating the regulation of autophagosome–lysosome fusion by Rab proteins. Following autophagosome formation, autophagosome–lysosome fusion is regulated by Rab7 through microtubule plus/minus-end-directed transport, by Rab8B through its downstream effector TBK-1, and by Rab24 via unknown molecular mechanisms
In trophic factor withdrawal (TFW)-induced neuronal death, insulin-like growth factor-I (IGF-I) protects Purkinje neurons from cell death by increasing the rate of autophagosome-to-lysosome fusion.83 Bains et al.84 demonstrated that Rab7 is involved in the regulation of autophagosome-to-lysosome fusion mediated by IGF-I. Under TFW conditions, Rab7 bound to Rab7-interacting lysosomal protein (RILP), which only binds to the active GTP-bound form of Rab7, at a lower level, and autophagosomes accumulated in neurons. However, IGF-I treatment during TFW restored Rab-RILP binding to normal levels and reduced the accumulation of autophagosomes. Because the Rab7-RILP complex regulates the microtubule minus-end-direct transport of late endocytic vesicles and phagosomes by recruiting the dynein–dynactin motor complex,85, 86, 87 IGF-I probably promotes Rab7-RILP binding under TFW conditions and, as a result, the Rab7-RILP complex recruits the dynein–dynactin motor complex to facilitate the transport of autophagosomes to lysosomes for fusion (Figure 3).
The COP9 signalosome (CSN) comprises eight essential subunits (CSN1–CSN8) and is responsible for the regulation of the ubiquitin proteasome system.88, 89 A recent report90 showed that cardiomyocyte-restricted Csn8 knockout (CR-Csn8KO) resulted in extensive accumulation of autophagosomes that did not contain degraded substrates. Quantification of autophagosomes, lysosomes, and autolysosomes in CR-Csn8KO hearts revealed a decreased amount of autolysosomes relative to autophagosomes and lysosomes; therefore, the accumulation of autophagosomes in CR-Csn8KO hearts was attributed to the defective fusion of autophagosomes with lysosomes. Furthermore, extensive accumulation of autophagosomes was accompanied by minimal Rab7 expression in CR-Csn8KO hearts. In cardiomyocytes, downregulation of Rab7 inhibited the fusion of autophagosomes with lysosomes and exacerbated cell necrosis or apoptosis. Collectively, these results suggest that Csn8 likely downregulates Rab7 to impair autophagosome maturation that causes cardiomyocytes to undergo necrosis or apoptosis.
Rab8B and its Downstream Effector TANK-binding kinase 1 in Autophagosome Maturation
Autophagy has been shown to confer resistance to Mycobacterium tuberculosis,91 and a recent report92 showed that Rab8B and TANK-binding kinase 1(TBK-1), which is a downstream interaction partner of Rab8B and a pivotal innate immunity regulator,93, 94 participate in this autophagy-mediated antimicrobial defense by controlling autophagosome maturation. In M. tuberculosis var. bovis BCG infection, both knockdown of Rab8B or TBK-1 reduced autophagic killing of mycobacteria, which suggests that Rab8B regulates autophagic elimination of mycobacteria possibly through TBK-1. In addition, TBK-1 knockdown increased the number of autophagosomes but decreased the number of autolysosomes with accumulation of LC3-II, which implies that TBK-1 is involved in autophagosome maturation. In addition, Rab8B interacted with TBK-1 in LC3-labeled autophagic organelles, and some autophagic components, such as LC3-II, p62, and UVRAG, were cofractionated with TBK-1 and Rab8B. These findings suggest that Rab8B facilitates autophagic elimination of mycobacteria by regulating autophagosome maturation through TBK-1 (Figure 3).
Rab24 and Autophagosome Maturation
Rab24 is localized in the ER and is thought to primarily participate in autophagosome maturation.30 Under normal conditions, Rab24 presents a reticular distribution around the nucleus. In contrast, in amino-acid starvation-induced autophagy, the distribution of Rab24 changes into punctate spots, larger dots, ring-shaped small vesicles, and some tubular-like structures, and these structures colocalize with markers for autophagic vacuoles, such as LC3 and monodansylcadaverine. The overexpression of wt Rab24 caused an increase in autophagosomes, whereas in cells overexpressing the mutant Rab24S67L, the amount of autophagosomes decreased. These results demonstrated for the first time that Rab24 is involved in autophagy.95 Egami et al.96 found that in neuronal PC12 cells treated with a proteasomal inhibitor (MG132), expression of Rab24 and LC3 markedly increased and Rab24 colocalized with LC3, which further confirms the involvement of Rab24 in autophagy.
The drs gene products suppress apoptosis-inducing tumors.97 In MEFs deficient in drs (KO MEF), the maturation process from the fusion of autophagosomes with lysosomes to form autolysosomes was inhibited, which indicates that autolysosome formation requires drs. Peptide mass fingerprinting analyses and a pull-down assay revealed that Rab24 binds to drs and that the binding requires the transmembrane domain of drs. Under low serum level conditions, drs is colocalized with Rab24 in LC3-positive vesicle-like structures. These results suggest that drs interacts with Rab24 in the autophagosome and may regulate the fusion of autophagosomes with lysosomes through Rab24.98
Rab7 and Curcumin-induced Autophagy
Curcumin, a hydrophobic polyphenol of the golden spice turmeric, influences various cellular biochemical and molecular functions through effects on multiple targets.99 It was recently reported that curcumin induces autophagy to protect vascular endothelial cells from oxidative stress damage and that Rab7 participates in this process.100 When vascular endothelial cells were subjected to H2O2-induced oxidative stress injury, curcumin treatment induced autophagy by upregulating Beclin1 and decreasing the interaction between Beclin1 and Bcl-2, thus enabling the cells to recover from oxidative stress. However, when autophagy was inhibited, curcumin treatment did not result in improved survival. These results suggest that autophagy mediates the protective effect of curcumin. Moreover, both mRNA and protein expression of Rab7 increased drastically during curcumin-induced autophagy, implying that curcumin may facilitate autophagy by increasing Rab7 expression.
Rab8A and an Autophagy-based Unconventional Secretory Pathway for IL-1β
Autophagy has long been considered to be an intracellular degradation system in mammalian cells. However, a recent study101 showed that in mammals, autophagy also contributes to the biogenesis and secretion of the proinflammatory cytokine IL-1β. Although previous studies102, 103, 104 showed that basal autophagy inhibits IL-1β secretion, the study described above found that the inflammasome and autophagy apparatus were synergized to intensify IL-1β secretion in cells stimulated to induce autophagy, and that knockdown of Rab8A or overexpression of the dominant-negative Rab8A mutant inhibited the secretion, which suggests that this autophagy-based unconventional secretory pathway for IL-1β involves Rab8A.
Rab25 and Autophagy in Ovarian Cancer
Rab25 expression only occurs in epithelial tissue and is associated with liver cancer, bladder cancer, ovarian cancer, and breast cancer.105, 106, 107, 108 A recent report109 showed that knockdown of Rab25 by siRNA in HEY and ES-2 ovarian cancer cells promoted autophagy, and that this process was related to ERK1/2 signaling. The amount of intracellular acidic vesicle organelles, which are a characteristic of autophagic cell death, and the amount of GFP-LC3 puncta in Rab25-knockdown cells was noticeably higher than the respective amounts in the control. In addition, knockdown of Rab25 promoted Beclin1 expression and LC3-I lipidation. However, all changes were reversed by treatment with U0126, an ERK1/2 signaling pathway inhibitor.
Concluding Remarks
In this review, we discussed in detail the role of Rab proteins in various stages of autophagy and the relevant molecular mechanisms (Figure 4, Table 1). At this stage, studies on the involvement of Rabs in autophagy focus more on discovering different functions of Rab proteins in autophagy and less on investigating the underlying molecular mechanisms. Such studies have only clarified some molecular events related to the involvement of Rab1, Rab5, Rab11, Rab32, and Rab33B in autophagosome formation and Rab7, Rab8B, and Rab24 in autophagosome maturation; thus, an intensive study of detailed molecular mechanisms should be conducted in the future. Many natural compounds, such as curcumin, cannabinoids, and resveratrol, exert biological or pharmacological effects through autophagy.110, 111, 112 However, more experimental results are needed to determine whether Rab proteins are involved in natural compound-related autophagy. Moreover, recent studies113, 114, 115 have shown that other vesicle-trafficking proteins, such as soluble NSF attachment protein receptors and Ras-like GTPase (Ral), participate in autophagy, which suggests that, in addition to Rab proteins, other vesicle-trafficking proteins may have a role in autophagy. Thus, the function of other vesicle-trafficking proteins in autophagy needs to be further explored. Autophagy has a key role in organism growth and development, cell differentiation, and the response to cellular stress, and also in many diseases and aging. Therefore, comprehensive understanding of the role of autophagic membrane trafficking regulated by Rab proteins will not only provide knowledge regarding the mechanisms underlying autophagy regulation but will also contribute to the search for new therapeutic targets to cure diseases through regulation of autophagy.
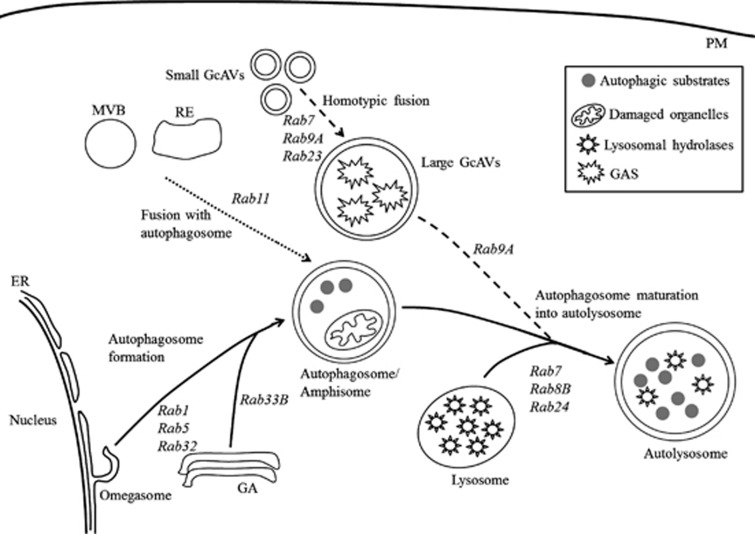
Figure 4.
Schematic diagram illustrating the regulation of autophagy by Rab proteins. The solid line shows the canonical autophagic process in which Rab1, Rab5, and Rab32 regulate autophagosome formation from the ER. Rab33B is involved in autophagosome formation from the GA, and autophagosome maturation into autolysosome is mediated by Rab7, Rab8B, and Rab24. The dotted line points to the fusion of MVB and RE with the autophagosome to form an amphisome that is regulated by Rab11. The dashed line indicates the formation and maturation of GcAVs; the homotypic fusion mediated by Rab7, Rab9A, and Rab23. Rab9A also participates in the fusion of GcAVs with lysosomes. ER, endoplasmic reticulum; GA, Golgi apparatus; GAS, Group A Streptococcus; GcAVs, GAS-containing autophagosome-like vacuoles; MVB, multivesicular bodies; PM, plasma membrane; RE, recycling endosome
Table 1. The Rab proteins: involved autophagy stage, related effector proteins in autophagy, and physiological role in membrane traffic.
| Involved autophagy stage | Rab protein | Related effectors | Membrane traffic pathway/function | References |
|---|---|---|---|---|
| Autophagosome formation | Rab1 | Atg9, Bacterial proteins (VirA, DrrA, LidA, and LepB) | ER to Golgi | 30, 45, 48, 54, 55 |
| Rab5 | p110β, Beclin1, Vps34, and HCV protein NS4B | Early endosome fusion | 30, 57, 58, 60 | |
| Rab7 | Unknown | Late endosome to lysosome | 30, 64 | |
| Rab9A | Raclopride | Endosome to TGN | 30, 66, 76 | |
| Rab11 | TBC1D14 | TGN/RE to plasma membrane | 30, 69, 71 | |
| Rab23 | Unknown | Protein recycling/transport to plasma membrane | 30, 66 | |
| Rab32 | Claret | TGN to melanosome, mitochondrial fission | 30, 72, 73 | |
| Rab33B | Atg16L | Only functions in autophagy | 30, 74 | |
| Autophagosome maturation | Rab7 | FYCO1, RILP, and Csn8 | Late endosome to lysosome | 30, 77, 78, 79, 80, 81, 82, 84, 90 |
| Rab8B | TBK-1 | Exocytosis, TGN/RE to plas- ma membrane | 30, 92 | |
| Rab24 | Drs | Chromosome segregation, and cytokinesis in normal cell division | 30, 95, 96, 98 | |
| Unknown | Rab7 | Curcumin | Late endosome to lysosome | 30, 100 |
| Rab8A | Unknown | Exocytosis, TGN/RE to plas- ma membrane | 30, 101 | |
| Rab25 | Unknown | RE (apical) to plasma membrane | 30, 109 |
Abbreviations: ER, endoplasmic reticulum; HCV, hepatitis C virus; RE, recycling endosome; RILP, Rab7-interacting lysosomal protein; TBK, TANK-binding kinase 1; TGN, trans-Golgi network
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant no. 31028007), the National Science Foundation of Chongqing (grant no. 2010bb5173), and the National College Students' Innovation and Entrepreneurship Training Program (grant nos. 201290035014 and 201390035019).
Glossary
- ATG
Autophagy-related genes
- Atg proteins
ATG-encoding proteins
- AVOs
acidic vesicle organelles
- Ccz1
calcium caffeine zinc sensitivity
- CSN
COP9 signalosome
- ER
endoplasmic reticulum
- GAPs
GTPase-activating proteins
- GcAV
Group A Streptococcus (GAS)-containing autophagosome-like vacuole
- GEF
guanine nucleotide exchange factor
- HOPS complex
homotypic fusion and vacuole protein sorting complex
- IGF-I
insulin-like growth factor-I
- MVB
multivesicular body
- PAS
phagophore assembly site
- PI3P
phosphatidylinositol 3-phosphate
- Ral
Ras-like GTPase
- RE
recycling endosome
- RILP
Rab7-interacting lysosomal protein
- SNAREs
soluble NSF attachment protein receptors
- TBC
Tre-2/Bub2/Cdc16
- TBK-1
TANK-binding kinase 1
- Tfn
transferrin
- TFW
trophic factor withdrawal
- TRAPP complex
Transport protein particle complex
The authors declare no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CE, Gan BQ, Tang BL. Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci. 2011;68:3349–3358. doi: 10.1007/s00018-011-0748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Physiological functions of autophagy. Curr Top Microbiol Immunol. 2009;335:71–84. doi: 10.1007/978-3-642-00302-8_3. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Beau I, Mehrpour M, Codogno P. Autophagosomes and human diseases. Int J Biochem Cell Biol. 2011;43:460–464. doi: 10.1016/j.biocel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung YT, Wang BJ, Hu MK, Hsu WM, Lee H, Huang WP, et al. Autophagy: a double-edged sword in Alzheimer's disease. J Biosci. 2012;37:157–165. doi: 10.1007/s12038-011-9176-0. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Tanida I. Autophagy basics. Microbiol Immunol. 2011;55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieter E, Vinke F, Bakula D, Cebollero E, Ungermann C, Proikas-Cezanne T, et al. Atg18 function in autophagy is regulated by specific sites within its beta-propeller. J Cell Sci. 2013;126:593–604. doi: 10.1242/jcs.115725. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- Homma K, Suzuki K, Sugawara H. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 2011;39:D986–D990. doi: 10.1093/nar/gkq995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman C, Ktistakis NT. Autophagosome formation in mammalian cells. Semin Immunopathol. 2010;32:397–413. doi: 10.1007/s00281-010-0222-z. [DOI] [PubMed] [Google Scholar]
- Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science. 1991;252:1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- Pind SN, Nuoffer C, McCaffery JM, Plutner H, Davidson HW, Farquhar MG, et al. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Schmitzova J, Trepte HH, Gallwitz D, Albert S. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J Biol Chem. 2002;277:41023–41031. doi: 10.1074/jbc.M205783200. [DOI] [PubMed] [Google Scholar]
- Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol. 2010;11:759–763. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- Zou S, Liu Y, Zhang XQ, Chen Y, Ye M, Zhu X, et al. Modular TRAPP complexes regulate intracellular protein trafficking through multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics. 2012;191:451–460. doi: 10.1534/genetics.112.139378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Barrowman J, Schieltz D, Yates JR, 3rd, Ferro-Novick S. Identification and characterization of five new subunits of TRAPP. Eur J Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, et al. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005;1:37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20:43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci USA. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem. 2012;287:44261–44269. doi: 10.1074/jbc.M112.411454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- Olanow CW, Brundin P. Parkinson's disease and alpha synuclein: is Parkinson's disease a prion-like disorder. Mov Disord. 2013;28:31–40. doi: 10.1002/mds.25373. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- Gong L, Devenish RJ, Prescott M. Autophagy as a macrophage response to bacterial infection. IUBMB Life. 2012;64:740–747. doi: 10.1002/iub.1070. [DOI] [PubMed] [Google Scholar]
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabernet-Castello C, O'Reilly AJ, Dacks JB, Field MC. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol Biol Cell. 2013;24:1574–1583. doi: 10.1091/mbc.E12-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiya K, Tobe T, Komatsu K, Suzuki T, Watarai M, Fukuda I, et al. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol Microbiol. 1995;17:241–250. doi: 10.1111/j.1365-2958.1995.mmi_17020241.x. [DOI] [PubMed] [Google Scholar]
- Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, Shao F. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- Joshi AD, Swanson MS. Secrets of a successful pathogen: legionella resistance to progression along the autophagic pathway. Front Microbiol. 2011;2:138. doi: 10.3389/fmicb.2011.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N, Fukuda M. Role of Rab family GTPases and their effectors in melanosomal logistics. J Biochem. 2012;151:343–351. doi: 10.1093/jb/mvs009. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, et al. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol. 2010;191:827–843. doi: 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Pan JA, Dbouk HA, Ballou LM, Deleon JL, Fan Y, et al. Class IA PI3K p110beta subunit promotes autophagy through Rab5 Small GTPase in response to growth factor limitation. Mol Cell. 2013;50:29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol. 2011;85:10561–10571. doi: 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Dwivedi M, Sung H, Shen H, Park BJ, Lee S. Disruption of endocytic pathway regulatory genes activates autophagy in C. elegans. Mol Cells. 2011;31:477–481. doi: 10.1007/s10059-011-1035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Nakagawa I, Yamamoto A, Amano A, Noda T, Yoshimori T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nozawa T, Aikawa C, Goda A, Maruyama F, Hamada S, Nakagawa I. The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection. Cell Microbiol. 2012;14:1149–1165. doi: 10.1111/j.1462-5822.2012.01792.x. [DOI] [PubMed] [Google Scholar]
- Morvan J, Kochl R, Watson R, Collinson LM, Jefferies HB, Tooze SA. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy. 2009;5:676–689. doi: 10.4161/auto.5.5.8378. [DOI] [PubMed] [Google Scholar]
- Fader CM, Sanchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Fader CM, Savina A, Sanchez D, Colombo MI. Exosome secretion and red cell maturation: Exploring molecular components involved in the docking and fusion of multivesicular bodies in K562 cells. Blood Cells Mol Dis. 2005;35:153–157. doi: 10.1016/j.bcmd.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci. 2009;66:2913–2932. doi: 10.1007/s00018-009-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z, Huang X. Rab32 is important for autophagy and lipid storage in Drosophila. PLoS One. 2012;7:e32086. doi: 10.1371/journal.pone.0032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Yan H, Li WL, Xu JJ, Zhu SQ, Long X, Che JP. D2 dopamine receptor antagonist raclopride induces non-canonical autophagy in cardiac myocytes. J Cell Biochem. 2013;114:103–110. doi: 10.1002/jcb.24306. [DOI] [PubMed] [Google Scholar]
- Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta. 2013;1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zou W, Yang Y, Chai Y, Chen B, Cheng S, et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol. 2012;197:27–35. doi: 10.1083/jcb.201111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains M, Florez-McClure ML, Heidenreich KA. Insulin-like growth factor-I prevents the accumulation of autophagic vesicles and cell death in Purkinje neurons by increasing the rate of autophagosome-to-lysosome fusion and degradation. J Biol Chem. 2009;284:20398–20407. doi: 10.1074/jbc.M109.011791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains M, Zaegel V, Mize-Berge J, Heidenreich KA. IGF-I stimulates Rab7-RILP interaction during neuronal autophagy. Neurosci Lett. 2011;488:112–117. doi: 10.1016/j.neulet.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol. 2003;23:6494–6506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Li F, Ranek MJ, Wei N, Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41:458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- Egami Y, Kiryu-Seo S, Yoshimori T, Kiyama H. Induced expressions of Rab24 GTPase and LC3 in nerve-injured motor neurons. Biochem Biophys Res Commun. 2005;337:1206–1213. doi: 10.1016/j.bbrc.2005.09.171. [DOI] [PubMed] [Google Scholar]
- Kim CJ, Shimakage M, Kushima R, Mukaisho K, Shinka T, Okada Y, et al. Down-regulation of drs mRNA in human prostate carcinomas. Hum Pathol. 2003;34:654–657. doi: 10.1016/s0046-8177(03)00240-5. [DOI] [PubMed] [Google Scholar]
- Tambe Y, Yamamoto A, Isono T, Chano T, Fukuda M, Inoue H. The drs tumor suppressor is involved in the maturation process of autophagy induced by low serum. Cancer Lett. 2009;283:74–83. doi: 10.1016/j.canlet.2009.03.028. [DOI] [PubMed] [Google Scholar]
- Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as "Curecumin": from kitchen to clinic. Biochem Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Han J, Pan XY, Xu Y, Xiao Y, An Y, Tie L, et al. Curcumin induces autophagy to protect vascular endothelial cell survival from oxidative stress damage. Autophagy. 2012;8:812–825. doi: 10.4161/auto.19471. [DOI] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- He H, Dai F, Yu L, She X, Zhao Y, Jiang J, et al. Identification and characterization of nine novel human small GTPases showing variable expressions in liver cancer tissues. Gene Expr. 2002;10:231–242. doi: 10.3727/000000002783992406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor O, Nativ O, Stein A, Novak L, Lehavi D, Shiboleth Y, et al. Molecular analysis of transitional cell carcinoma using cDNA microarray. Oncogene. 2003;22:7702–7710. doi: 10.1038/sj.onc.1207039. [DOI] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell. 2007;13:496–510. doi: 10.1016/j.devcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tao X, Jia L, Cheng KW, Lu Y, Yu Y, et al. Knockdown of RAB25 promotes autophagy and inhibits cell growth in ovarian cancer cells. Mol Med Rep. 2012;6:1006–1012. doi: 10.3892/mmr.2012.1052. [DOI] [PubMed] [Google Scholar]
- Gupta SC, Kismali G, Aggarwal BB. Curcumin, a component of turmeric: from farm to pharmacy. BioFactors. 2013;39:2–13. doi: 10.1002/biof.1079. [DOI] [PubMed] [Google Scholar]
- Vara D, Salazar M, Olea-Herrero N, Guzman M, Velasco G, Diaz-Laviada I. Anti-tumoral action of cannabinoids on hepatocellular carcinoma: role of AMPK-dependent activation of autophagy. Cell Death Differ. 2011;18:1099–1111. doi: 10.1038/cdd.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chiu J, Zhang H, Qi T, Tang Q, Ma K, et al. Autophagic cell death induced by resveratrol depends on the Ca(2+)/AMPK/mTOR pathway in A549 cells. Biochem Pharmacol. 2013;86:317–328. doi: 10.1016/j.bcp.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Bernard A, Popelka H, Klionsky DJ. A unique hairpin-type tail-anchored SNARE starts to solve a long-time puzzle. Autophagy. 2013;9:813–814. doi: 10.4161/auto.24359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifki OF, Bodemann BO, Battiprolu PK, White MA, Hill JA. RalGDS-dependent cardiomyocyte autophagy is required for load-induced ventricular hypertrophy. J Mol Cell Cardiol. 2013;59:128–138. doi: 10.1016/j.yjmcc.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]