Abstract
While it is known that rare copy-number variants (CNVs) contribute to risk for some neuropsychiatric disorders, the role of CNVs in bipolar disorder is unclear. Here, we reasoned that a contribution of CNVs to mood disorders might be most evident for de novo mutations. We performed a genome-wide analysis of de novo CNVs in a cohort of 788 trios. Diagnoses of offspring included bipolar disorder (n = 185), schizophrenia (n= 177), and healthy controls (n= 426). Frequencies of de novo CNVs were significantly higher in bipolar disorder as compared with controls (OR= 4.8 [1.4,16.0], p= 0.009). De novo CNVs were particularly enriched among cases with an age at onset younger than 18 (OR= 6.3 [1.7,22.6], p= 0.006). We also confirmed a significant enrichment of de novo CNVs in schizophrenia (OR= 5.0 [1.5,16.8], p= 0.007). Our results suggest that rare spontaneous mutations are an important contributor to risk for bipolar disorder and other major neuropsychiatric diseases.
Introduction
Bipolar disorder (BD, also known as manic-depressive illness) is a severe mood disorder consisting of episodes of mania and depression. The lifetime prevalence of bipolar disorder in the general population is ~1% and the illness is associated with considerable morbidity and a high lifetime risk of suicide (Merikangas et al., 2011).
Genes play an important role in risk for BD. The rate of concordance for monozygotic twins is 40%, compared with a 5% rate in dizygotic twins (Kendler et al., 1995, Kieseppä et al., 2004 and McGuffin et al., 2003), and risk among the first-degree relatives of individuals with BD is ten-fold greater than risk among the general population (Barnett and Smoller, 2009). However, as with other psychiatric disorders, the genetics of BD is complex, probably due to a high degree of genetic heterogeneity and considerable phenotypic heterogeneity of clinical populations (Potash et al., 2007).
Genetic risk factors with individually large effects are likely to be rare. Association-based methods to identify common genetic risk alleles in BD have met with limited success. Early studies implicated a few common variants with modest effects (Baum et al., 2008 and Ferreira et al., 2008). Robust support for one of these loci, the L-type calcium channel CACN1AC, has been obtained in a recent meta-analysis of 11,974 patients and 51,792 controls, along with new evidence for a second locus, ODZ4 (Sklar et al., 2011). However, the paucity of significant findings in very large samples of cases and controls suggests that the contribution of common genetic variants to heritability of BD is limited.
Alternative approaches that focus on rare genetic variants are needed. One genetic approach that has been used effectively to overcome some of the problems of heterogeneity is the genome-wide analysis of rare copy-number variants (CNVs). Studies from our group (McCarthy et al., 2009, Sebat et al., 2007, Vacic et al., 2011 and Walsh et al., 2008) and from multiple independent groups (International Schizophrenia Consortium, 2008, Pinto et al., 2010, Stefansson et al., 2008 and Xu et al., 2008) have now firmly established that rare CNVs contribute to genetic risk for schizophrenia (SCZ) and autism spectrum disorder (ASD) and, in particular, that spontaneous (de novo) CNVs are important risk factors in the sporadic form of these disorders (Levy et al., 2011, Marshall et al., 2008, Sanders et al., 2011, Sebat et al., 2007 and Xu et al., 2008).
Observations of a similar nature have been made in studies of BD. CNV loci at 16p11.2 (McCarthy et al., 2009) and 3q29 (Clayton-Smith et al., 2010, Mulle et al., 2010 and Quintero-Rivera et al., 2010) confer risk for multiple psychiatric disorders, and two studies have found preliminary evidence implicating both in BD (McCarthy et al., 2009 and Quintero-Rivera et al., 2010). Two studies have demonstrated an enrichment of rare CNVs in patients with bipolar disorder (Priebe et al., 2011 and Zhang et al., 2009) as compared with healthy controls. In both studies, the greatest enrichment was observed in subjects with an earlier disease onset, defined as an age at onset (AAO) < 18 and < 21 in Zhang et al. (2009) and Priebe et al. (2011), respectively. However, two subsequent studies did not support these findings (Grozeva et al., 2010 and McQuillin et al., 2011). Thus, the role of copy-number variation in conferring risk for bipolar disorder remains in question (Grozeva et al., 2010 and Zhang et al., 2009).
Some of the earliest conclusive evidence for the role of rare CNVs in psychiatric disorders has come from family-based studies that examined the genomic burden of spontaneously occurring (de novo) CNVs (Marshall et al., 2008, Sebat et al., 2007 and Xu et al., 2008). De novo CNVs have consistently shown the strongest association with risk for autism (Itsara et al., 2010, Levy et al., 2011, Pinto et al., 2010 and Sanders et al., 2011) or schizophrenia (Xu et al., 2008), with a 5- to 10-fold enrichment in patients as compared with controls. We reasoned that if rare highly penetrant CNVs contribute to risk for bipolar disorder, the genetic effect would be most evident for de novo mutations. We further reasoned that the contribution of de novo CNVs to risk of bipolar disorder would be greatest in patients with an earlier disease onset (AAO ≤ 18). We screened for de novo CNVs ≥ 10 kb in size in blood-derived DNAs from 788 subject-mother-father trios including subjects with diagnoses of bipolar disorder and schizophrenia, and normal healthy controls. Here, we show that rare de novo copy-number mutations are significantly enriched in bipolar disorder and in schizophrenia.
Results
Our study sample included 788 subject-mother-father trios with confirmed parentage. DNA from all subjects was derived from whole blood. Details of the subjects are described in the Supplemental Experimental Procedures (available online). Diagnoses of subjects included bipolar disorder (n = 185, including 107 with an age at onset ≤ 18), schizophrenia (n = 177), and healthy controls (n = 426). While the primary disease focus of this study was BD, the inclusion of an additional schizophrenia cohort served first to replicate the one previous study of de novo CNVs in SCZ (Xu et al., 2008) and second to enable a valid comparison of patterns of de novo CNVs in BD with another disorder. In addition, a small set of autism spectrum disorder (ASD) trios (n = 45), all of which had been included in a previous study (Sebat et al., 2007) and three of which carried known de novo CNVs, were included as a “positive control” to confirm the sensitivity of our methods for detecting de novo events.
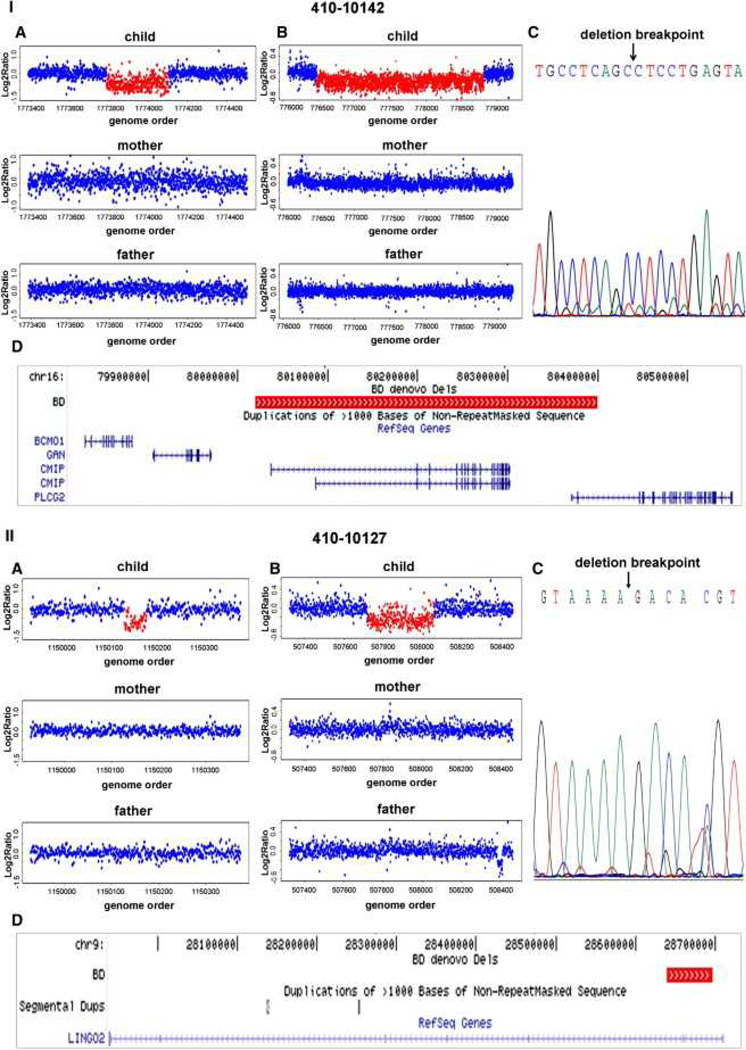
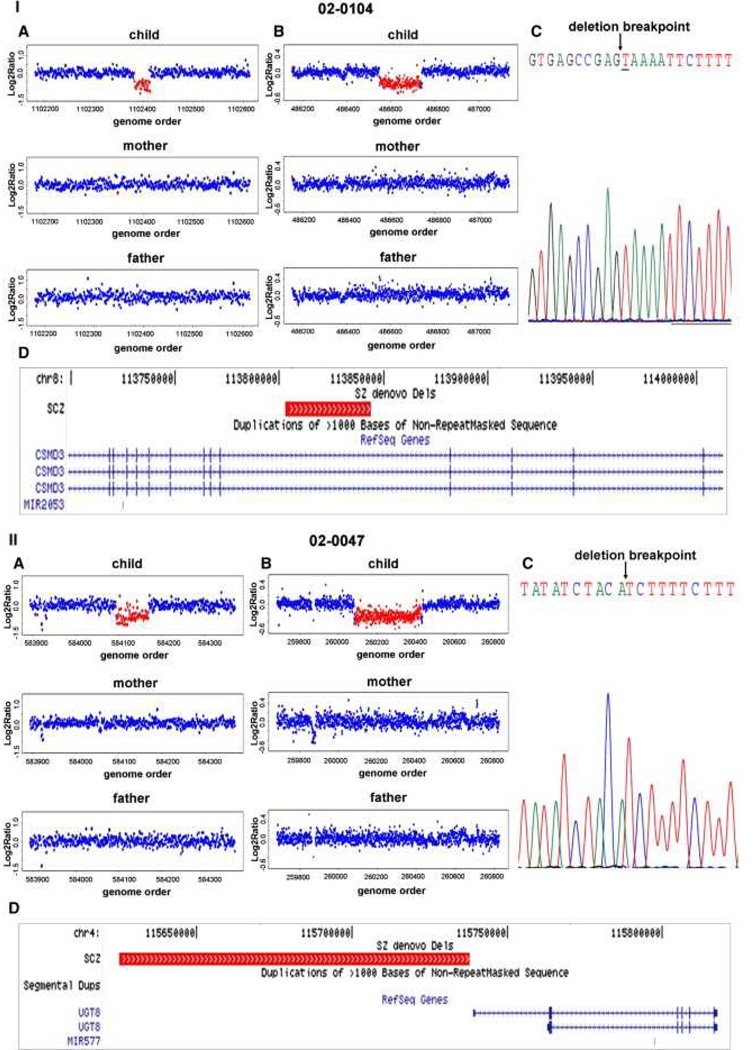
We performed high-resolution genome-wide copy-number scans, using the NimbleGen HD2 array comparative genomic hybridization (CGH) platform, on all subjects and their biological parents. Data processing and CNV detection were performed as described in Experimental Procedures. CNV call sets were filtered based on probe ratio (≤0.8 and ≥ 1.2), number of probes (≥10), frequency (<1%), and confidence score (Supplemental Experimental Procedures, Tables S1 and S2, and Figures S1 and S2). Rare CNVs that were present in subjects and not in their parents were subsequently validated and fine mapped using a custom tiling-resolution CGH array (Oxford Gene Technology) (Table S3. Custom Tiling array CGH Validation of Putative De Novo CNVs and Document S1. Figures S1–S3; Tables S1, S2, S4–S8, and S10; and Supplemental Experimental Procedures). Results for the genome-wide scans, tiling array validations, and breakpoint sequencing are illustrated by four examples: a deletion involving CMIP and PLCG2 genes (Figure 1I) and an exonic deletion of LINGO2 gene (Figure 1II) detected in subjects with a diagnosis of BD, and an intronic deletion of CSMD3 gene (Figure 2I) and a deletion adjacent to UGT8 gene (Figure 2II) detected in subjects with a diagnosis of SCZ.
Figure 1. Detection, Validation, and Breakpoint Sequencing of De Novo CNVs in BD.
Representative examples of microarray data and sequencing results are provided for deletions detected in two subjects with diagnoses of BD, 410-10142 (panel I) and 410-10127 (panel II).
(A) De novo CNVs were identified from whole-genome scans of the child, mother, and father, using the NimbleGen HD2 platform.
(B) CNV validation and breakpoint refinement was performed by analysis of the trio using a custom Agilent microarray with dense probe coverage of the target region (~200 bp spacing).
(C) Deletion breakpoints were determined by PCR and Sanger sequencing.
(D) UCSC genome browser tracks of known genes are shown to scale with a track for de novo deletions displayed in red.
Figure 2. Detection, Validation, and Breakpoint Sequencing of De Novo CNVs in SCZ.
Representative examples of microarray data and sequencing results are provided for deletions detected in two subjects with diagnoses of SCZ, 02-0104 (panel I) and 02-0047 (panel II).
(A) De novo CNVs were identified from whole-genome scans of the child, mother, and father, using the NimbleGen HD2 platform.
(B) CNV validation and breakpoint refinement was performed by analysis of the trio using a custom Agilent microarray with dense probe coverage of the target region (~200 bp spacing).
(C) Deletion breakpoints were determined by PCR and Sanger sequencing.
(D) UCSC genome browser tracks of known genes are shown to scale with a track for de novo deletions displayed in red.
A total of 23 de novo CNVs were detected and validated in our study sample, including fourteen deletions and nine duplications (Table 1). De novo CNVs ranged in size from 15.1 to 7,178 kb, with a median size of 112 kb, and contained a median of two genes.
Table 1.
De Novo CNVs Detected in This Study
| Diagnosis | Sample ID | Age at Onset (yrs) |
Sex | Cytoband | Start | End | CNV Size (bp) |
CNV Type | Genes | Breakpoint Sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| Bipolar disorder | 420-10015 | 15 | F | 3q212 | 126663504 | 126720022 | 56519 | deletion | SNX4 | |
| Bipolar disorder | 400-10117 | 21 | F | 3q29 | 197417247 | 198249463 | 832217 | deletion | 25 genes | SegDup |
| Bipolar disorder | 410-10124 | 11 | M | 5q15.1 | 17508759 | 17561308 | 52550 | deletion | SegDup | |
| Bipolar disorder | 410-10145 | 11 | F | 5q31.2 | 138051454 | 138254688 | 203235 | duplication | CTNNA1; LRRTM2 | |
| Bipolar disorder | 5459 | 17 | F | 9p24.1 | 5264155 | 7119082 | 1854928 | duplication | 15 genes | SegDup |
| Bipolar disorder | 5459 | 17 | F | 9p23 | 9855970 | 9927360 | 71391 | duplication | PTPRD | |
| Bipolar disorder | 410-10127 | 8 | M | 9p21.1 | 28639667 | 28696714 | 57048 | deletion | LINGO2 | Unique |
| Bipolar disorder | bbn-106-01 | 23 | M | 16p11.2 | 29512728 | 30124017 | 611290 | duplication | 38 genes | SegDup |
| Bipolar disorder | 410-10145 | 11 | F | 16q22.3 | 73021657 | 73057216 | 35560 | duplication | AL833498;GLG1 | SegDup |
| Bipolar disorder | 410-10142 | 7 | F | 16q23.2 | 80019981 | 80399720 | 379740 | deletion | CMIP;BC040927; PLCG2 | SINE |
| Control | SSC02546 | F | 13q33.1 | 101035498 | 101078525 | 43028 | deletion | ITGBL1 | ||
| Control | SSC00206 | M | 18p11.31-p11.23 | 6479110 | 7904147 | 1425038 | duplication | 6 genes | ||
| Control | SSC02714 | F | 19q13.12 | 42608251 | 42647120 | 38870 | deletion | ZNF569 | SINE | |
| Control | SSC00695 | M | 19q13.33 | 54915631 | 54930771 | 15141 | deletion | |||
| Schizophrenia | 4822 | 21 | M | 1p36.33 | 755082 | 1266721 | 511640 | deletion | 31 genes | SegDup |
| Schizophrenia | 02-0047 | 20 | F | 4q26 | 115625575 | 115737638 | 112064 | deletion | Unique | |
| Schizophrenia | 02-0135 | 22 | F | 4q35.2 | 187892312 | 191152733 | 3260422 | deletion | 7 genes | SegDup |
| Schizophrenia | 3236 | 17 | M | 6q22.3 | 17639894 | 17988098 | 348205 | duplication | CAP2;FAM8A1; NUP153;KIF13A | |
| Schizophrenia | 02-0135 | 22 | F | 7q36.1-q36.3 | 151641965 | 158820241 | 7178277 | duplication | 41 genes | |
| Schizophrenia | 02-0022 | 12 | M | 8p21.2 | 25028902 | 25046800 | 17899 | deletion | ||
| Schizophrenia | 02-0104 | 22 | F | 8q23.3 | 113802932 | 113843685 | 40754 | deletion | CSMD3 | SINE |
| Schizophrenia | 6574 | 26 | M | 12q24.23 | 117065514 | 117132798 | 67285 | deletion | PEBP1;TAOK3 | |
| Schizophrenia | 4295 | 24 | M | 22q11.21 | 18869860 | 20006782 | 1136923 | duplication | 32 genes | SegDup |
All 23 de novo CNVs discovered and validated in 788 trios are sorted by diagnosis and genomic location. Diagnosis denotes affected or unaffected disease status. "SampleID" is a unique identifier for each subject. Age at onset and sex of subjects are listed. Cytological band and start and end coordinates are listed for hg18 build. CNV length ("CNV Size") in base pairs for deletions and duplications ("CNV Type") is displayed. For each de novo CNV, all overlapping annotated UCSC Known Genes are listed, and the presence of flanking segmental duplications within 15 kb of a CNV boundary is indicated (SegDup). Breakpoint sequences were determined for an additional five deletions, and breakpoints consisted of either unique sequence or short interspersed nuclear element (SINE) repeats.
About one-third (8/23) of de novo CNVs in our study were flanked by segmental duplications (SDs) at one (6/23) or at both boundaries (2/23). This class of CNV most likely occurred by nonallelic homologous recombination (NAHR) (Lupski, 1998). By contrast, the majority of de novo CNVs were not flanked by SDs. Breakpoint sequences were obtained for five deletions (Table 1). Junction sequences of three out of five deletions were in short interspersed nuclear element (SINE) repetitive elements and two deletions had unique sequences at their breakpoints. A 1 bp insertion occurred at one of the breakpoints (see underlined base in Figure 2I). Notably, the median size of SD and non-SD-mediated de novo CNVs was 722 kb and 67 kb respectively, consistent with previous studies that have found differences in CNV size related to the underlying mutational mechanism (Itsara et al., 2010 and Stefansson et al., 2008).
De novo CNVs were significantly associated with BD and SCZ (Table 2). The rate of de novo mutation in controls was 0.9% (4/426). This rate is consistent with estimates from previous studies ranging from 0.5% to 3% (Conrad et al., 2010, Itsara et al., 2010, Levy et al., 2011, Sebat et al., 2007 and Xu et al., 2008). The observed rate of de novo CNVs in bipolar disorder subjects was 4.3% (8/185), a significant enrichment compared with controls (p = 0.009, OR = 4.8 [1.4,16.0]). De novo CNVs were also detected at a significantly higher rate (8/177, 4.5%) in schizophrenia subjects than in controls (p = 0.007, OR = 5.0 [1.5,16.8]). These results provide significant evidence for an association of de novo mutation with bipolar disorder and confirm earlier reports of a high rate of de novo copy-number mutation in schizophrenia (Xu et al., 2008).
Table 2.
De Novo CNVs Are Significantly Associated with Bipolar Disorder and Schizophrenia
| Sample | # Trios | # De Novo CNVs |
# Subjects with De Novo CNVs |
Rate of Subjects with De Novo CNVs |
Odds Ratio [95% CI] |
p Value | Dels:Dups (Ratio) |
|---|---|---|---|---|---|---|---|
| Bipolar | 185 | 10 | 8 | 0.043 | 4.8[1.4,16.0] | 0.009 | 5:5 (1) |
| AAO ≤ 18 | 107 | 8 | 6 | 0.056 | 6.3[1.7,22.6] | 0.006 | |
| AAO > 18 | 76 | 2 | 2 | 0.026 | 2.9[0.5,15.8] | 0.22 | |
| Schizophrenia | 177 | 9 | 8 | 0.045 | 5.0[1.5,16.8] | 0.007 | 6:3 (2.0) |
| AAO ≤ 18 | 59 | 2 | 2 | 0.034 | 3.7[0.7,20.7] | 0.158 | |
| AAO > 18 | 112 | 7 | 6 | 0.054 | 6.0[1.7,21.5] | 0.007 | |
| Controls | 426 | 4 | 4 | 0.009 | 3:1 (3) |
The total number of de novo CNVs detected and the number subjects carrying a de novo event are listed. Cases are further stratified based on age at onset (AAO) of BD and SCZ. p values were computed using a Fisher's exact test of the number of subjects with and without de novo CNVs in cases and controls.
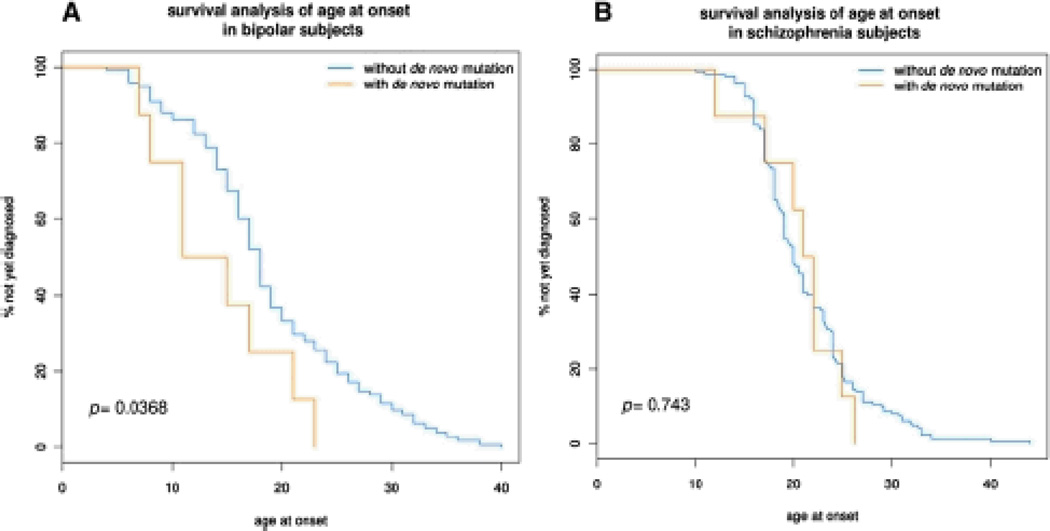
We investigated the influence of age at onset on the frequency of de novo mutations. After stratifying patients by age at onset ≤ 18, we observed a significantly higher rate of de novo CNVs in early-onset BD (p = 0.006, OR = 6.3 [1.7,22.6], Table 2). This difference was also nominally significant (p = 0.03) based on a survival analysis comparing AAO in subjects with or without a de novo CNV (Figure 3A). By contrast, we did not observe an effect of AAO on the frequency of de novo mutations in schizophrenia (Table 2, Figure 3B).
Figure 3. Survival Analysis of De Novo CNVs and Age at Onset in BD and SCZ.
We performed a survival analysis/Kaplan-Meier test to analyze the effect of de novo mutations on age at onset in BD(n = 185, panel A) and SCZ (n = 177, panel B). This test determines whether “time to diagnosis” differs systematically between patients who have de novo mutations and those who do not. The test is formalized by performing the Mantel-Haenszel test on the survival curves and reporting the resulting p value.
We further reasoned that frequencies of de novo CNVs might be influenced by the presence or lack of a family history of mental illness, a hypothesis based on earlier findings by our group and others that de novo CNVs occur more frequently in sporadic cases of ASD (Marshall et al., 2008 and Sebat et al., 2007) and schizophrenia (Xu et al., 2008). We stratified subjects based on evidence of positive family history, defined as having a first-degree relative with a diagnosis of bipolar I, bipolar II, major depression, schizophrenia, schizoaffective disorder, autism, or intellectual disability. In BD and SCZ cohorts, rates of de novo mutation were not higher in sporadic cases as compared with subjects with a positive family history (Table S5).
While de novo CNVs might have a stronger effect size, it is quite plausible that inherited CNVs could also contribute to risk for BD. Notably, inherited CNVs detected in this study included variants at loci that have been previously linked to schizophrenia (International Schizophrenia Consortium, 2008 and Stefansson et al., 2008), including a duplication at 1q21.1 in a subject with bipolar disorder and a duplication and a deletion at 15q13.3 detected in subjects with bipolar disorder and schizophrenia, respectively (Document S2, bed file). Therefore, we examined the burden of rare inherited CNVs overlapping with genes in BD, SCZ, and controls, and subjects were stratified based on family history. We observed a trend of enrichment for large (≥500 kb) inherited duplications in familial cases of bipolar disorder (OR = 1.77, p = 0.03, Table 3). We did not observe an enrichment of deletions in familial bipolar disorder. Likewise, we did not observe a significant enrichment of deletions or duplications in sporadic bipolar disorder or in schizophrenia (Table 3). These results are consistent with a role for inherited CNVs in familial BD, particularly for large duplications; however, data from a much larger sample are needed to draw firm conclusions.
Table 3.
Mutational Burden Analysis of Genic Rare Inherited CNVs In BD, SCZ, and Controls
| CNV Size | CNV Type | Sample Group | # Trios | # CNVs | # CNVs per Genome |
Case/ Control Ratio |
Wilcoxon p Value |
|---|---|---|---|---|---|---|---|
| ≥ 100 kb | Deletions & Duplications | Controls | 426 | 237 | 0.56 | ||
| Familial BD | 107 | 71 | 0.66 | 1.19 | 0.65 | ||
| Sporadic BD | 78 | 28 | 0.36 | 0.65 | 0.36 | ||
| Familial SCZ | 44 | 23 | 0.52 | 0.94 | 0.92 | ||
| Sporadic SCZ | 97 | 46 | 0.47 | 0.85 | 0.43 | ||
| ≥ 100 kb | Deletions | Controls | 426 | 75 | 0.18 | ||
| Familial BD | 107 | 18 | 0.17 | 0.96 | 0.90 | ||
| Sporadic BD | 78 | 10 | 0.13 | 0.73 | 0.50 | ||
| Familial SCZ | 44 | 7 | 0.16 | 0.90 | 0.97 | ||
| Sporadic SCZ | 97 | 18 | 0.19 | 1.05 | 0.55 | ||
| ≥ 100 kb | Duplications | Controls | 426 | 162 | 0.38 | ||
| Familial BD | 107 | 53 | 0.50 | 1.30 | 0.42 | ||
| Sporadic BD | 78 | 18 | 0.23 | 0.61 | 0.62 | ||
| Familial SCZ | 44 | 16 | 0.36 | 0.96 | 0.91 | ||
| Sporadic SCZ | 97 | 28 | 0.29 | 0.76 | 0.22 | ||
| ≥ 500 kb | Deletions & Duplications | Controls | 426 | 40 | 0.09 | ||
| Familial BD | 107 | 14 | 0.13 | 1.39 | 0.06 | ||
| Sporadic BD | 78 | 5 | 0.06 | 0.68 | 0.82 | ||
| Familial SCZ | 44 | 8 | 0.18 | 1.94 | 0.53 | ||
| Sporadic SCZ | 97 | 9 | 0.09 | 0.99 | 0.33 | ||
| ≥ 500 kb | Deletions | Controls | 426 | 13 | 0.03 | ||
| Familial BD | 107 | 2 | 0.02 | 0.61 | 0.95 | ||
| Sporadic BD | 78 | 2 | 0.03 | 0.84 | 0.73 | ||
| Familial SCZ | 44 | 1 | 0.02 | 0.74 | 0.42 | ||
| Sporadic SCZ | 97 | 3 | 0.03 | 1.01 | 0.62 | ||
| ≥ 500 kb | Duplications | Controls | 426 | 27 | 0.06 | ||
| Familial BD | 107 | 12 | 0.11 | 1.77 | 0.03 | ||
| Sporadic BD | 78 | 3 | 0.04 | 0.61 | 0.88 | ||
| Familial SCZ | 44 | 7 | 0.16 | 2.51 | 0.22 | ||
| Sporadic BCZ | 97 | 6 | 0.06 | 0.98 | 0.30 |
We Tested for differences in global CNV burden between cases (BD or SCZ) and controls by comparing CNV rate, i.e., average number of CNVs per genome. CNV burden was tested only for large (≥ 100 kb) rare inherited CNVs that impacted one or more annotated UCSC known genes and significance was estimated using Wilcoxon test. Uncorrected p values are listed for each test. Analyses were further stratified according to CNV type (deletions and duplications separately), CNV size (≥ 100 kb and ≥ 500 kb), and family history of BD and SCZ cases.
We sought additional genetic evidence for the loci at which we found de novo CNVs by performing follow-up analyses of the 23 de novo CNV regions in additional cohorts and families. We performed an analysis of CNVs in SNP genotyping data from multiple case-control studies, including the Bipolar Genome Study (BiGS) and Molecular Genetics of Schizophrenia (MGS) study (see Supplemental Experimental Procedures). De novo CNV regions were tested for association with BD and SCZ using a permutation-based method described previously (Vacic et al., 2011) (see Supplemental Experimental Procedures). No significant associations were detected in bipolar disorder (Table S6A). In schizophrenia, three genomic regions were significant (Table S6B), all corresponding to CNVs that have been previously implicated in schizophrenia at 3q29 (Mulle et al., 2010), 7q36.3 (Vacic et al., 2011), and 16p11.2 (McCarthy et al., 2009).
Previous studies have reported that rare CNVs associated with neuropsychiatric disorders are enriched for genes involved in neurodevelopment (Walsh et al., 2008 and Zhang et al., 2009). Here we examined whether genes impacted by de novo CNVs in SCZ and BD are enriched in specific functional categories. Pathway enrichment analysis was performed on the sets of genes overlapping with de novo CNVs in SCZ, BD, and controls (see Experimental Procedures). Enrichment of functional classes of genes was tested using the DAVID software (http://david.abcc.ncifcrf.gov/), followed by two additional permutation-based tests to correct for the known bias of CNVs toward large genes (Raychaudhuri et al., 2010), one implemented as a case-only analysis and a second implemented as a case-control analysis in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/cnv.shtml#burden2).
Based on the primary analysis using DAVID, eight functional categories were enriched among de novo CNVs in SCZ (p < 0.05). Three were also found to be enriched based on permutation tests of de novo CNVs in cases and controls, including “neural tube development,” “hindbrain development,” and “response to ethanol” (Table 4). Genes involved in neurodevelopment were not enriched among de novo CNVs in BD (Table 5) or in controls (Table S7). We then extended our analysis of schizophrenia to a large independent data set of rare (>100 kb) CNVs from 8,290 cases and 7,431 controls. Eight categories were tested in the case-control sample using the PLINK-CNV parametric test, and two categories, “synapse” and “Kelch-type beta propeller,” were significantly enriched among rare variants in cases (Table 4). The case-control data set did not provide significant support for an enrichment of genes related to neural tube development or hindbrain development. While the strength of evidence supporting specific pathways differs between data sets, we do find evidence consistent with our earlier observations that there is an enrichment of “neurodevelopmental” and “synaptic” genes among rare CNVs in SCZ (Walsh et al., 2008).
Table 4.
Gene Set Enrichment Analysis of De Novo CNVs in Schizophrenia
| Category | Database ID | Term | CNV Genes | De Novo CNVs (This Study) | Rare CNVs (from Vacic et al., 2011) |
||
|---|---|---|---|---|---|---|---|
| DAVID | Permutation | PLINK-CNVa | PLINK-CNV | ||||
| GOTERM_BP_FAT | GO:0021915 | neural tube development | MNX1, SHH, DVL1 | 0.029 | 0.003 | 0.045 | 0.612 |
| GOTERM_BP_FAT | GO:0030902 | hindbrain development | EN2, SDF4, SHH | 0.023 | 0.006 | 0.045 | 0.831 |
| GOTERM_BP_FAT | GO:0045471 | response to ethanol | PEBP1, SDF4, SHH | 0.026 | 0.006 | 0.016 | 0.207 |
| GOTERM_BP_FAT | GO:0051146 | striated muscle cell differentiation | AGRN, SHH, DVL1 | 0.047 | 0.025 | 0.153 | 0.040 |
| INTERPRO | IPR015915 | Keich-type beta propeller | KLHL17, LZTR1, KLHL22 | 0.031 | 0.027 | 0.164 | 0.002 |
| GOTERM_CC_FAT | GO:0019717 | Synaptosome | SNAP29, PEBP1, DVL1 | 0.037 | 0.032 | 0.122 | 0.140 |
| GOTERM_CC_FAT | GO:0045202 | Synapse | SNAP29, P2RX6, KLHL17, PEBP1, AGRN, DVL1 | 0.009 | 0.043 | 0.145 | 0.001 |
| GOTERM_BP_FAT | GO:0007267 | cell-cell signaling | ISG15, P14KA, PEBP1, AGRN, VIPR2, SHH, DVL1 | 0.029 | 0.110 | 0.222 | 0.040 |
For genes within de novo CNVs, eight functional categories were identified as enriched (p value < 0.05) by primary analysis using DAVID. We further tested enrichment of these eight categories in de novo CNVs using a permutation-based test for enrichment (case only) and using the pathway enrichment test implemented in PLINK-CNV (case-control). Three out of eight categories were found to be enriched among de novo CNVs in SCZ by all three tests (p < 0.05, shown in bold), including "neural tube development," "hindbrain development," and "response to ethanol." We also tested association of the eight categories in an independent case-control data set consisting of rare CNVs (> 100 kb in size) from 8,290 SCZ cases and 7,431 controls using PLINK-CNV enrichment test. Two categories, "synapse" and "kelch-type beta propeller," were significantly enriched in cases relative to controls (after correction for eight tests; p values shown in bold). p values from PLINK-CNV were derived from a two-sided parametric test.
For analysis of de novo CNVs using PLINK-CNV, a one-sided empirical p value was derived based on permutation.
Table 5.
Gene Set Enrichment Analysis of De Novo CNVs in Bipolar Disorder
| Category | Database ID | Term | CNV Genes | DAVID | De Novo CNVs (This Study) |
Rare Cnvs (from BiGS) |
|
|---|---|---|---|---|---|---|---|
| Permutation | PLINK-CNVa | PLINK-CNV | |||||
| GOTERM_BP_FAT | GO:0042129 | regulation of T cell proliferation | CORO1A, CD274, PDCD1LG2, SPN | 0.001 | 0.002 | 0.039 | 0.743 |
| GOTERM_BP_FAT | GO:0006644 | phospholipid metabolic process | CDIPT, PIGZ, PIGX, PLCG2, PCYT1A | 0.002 | 0.004 | 0.024 | 0.304 |
| GOTERM_BP_FAT | GO:0008360 | regulation of cell shape | ALDOA, CORO1A, TAOK2 | 0.010 | 0.009 | 0.195 | 0.004 |
| GOTERM_CC_FAT | GO:0005783 | endoplasmic reticulum | SEZ6L2, ERMP1, CDIPT, PIGZ, PIGX, ASPHD1, PCYT1A, MLANA, FAM57B | 0.028 | 0.039 | 0.058 | 0.400 |
| PANTHER_PATHWAY | P00005 | Angiogenesis | PAK2, MAPK3, PLCG2 | 0.032 | 0.046 | 0.014 | 0.565 |
| GOTERM_CC_FAT | GO:0031224 | intrinsic to membrane | GLG1, ERMP1, CDIPT, TM4SF19, LRRC33, TMEM219, ZDHHC19, FAM57B, LINGO2, SEZ6L2, KIAA1432, PAK2, PRRT2, LRRTM2, C3ORF43, C9ORF46, OSTALPHA, MLANA, SPN, PTPRD, PIGZ, TAOK2, PIGX, MFI2, GDPD3, PDCD1LG2, CD274, ASPHD1, C16ORF54, KCTD13, MVP | 0.007 | 0.052 | 0.061 | 0.627 |
| GOTERM_BP_FAT | GO:0007010 | cytoskeleton organization | ALDOA, CORO1A, PAK2, TAOK2, PPP4C | 0.033 | 0.121 | 0.309 | 0.014 |
For genes within de novo CNVs, seven functional categories were identified as enriched (p value < 0.05) by primary analysis using DAVID. We further tested enrichment of these eight categories in de novo CNVs using a permutation-based test for enrichment (case only) and using the pathway enrichment test implemented in PLINK-CNV (case-control). Three categories were found to be enriched among de novo CNVs in BD by all three tests (p < 0.05, shown in bold), including "regulation of T-cell proliferation," "phospholipid metabolic process," and "angiogenesis." We also tested association of the seven categories in an independent case control data set consisting of rare CNVs from 2,777 BD cases and 3,508 controls using PLINK-CNV enrichment test. One category, "regulation of cell shape," was significantly enriched in cases relative to controls (after correction for seven tests; p values shown in bold). p values from PLINK-CNV were derived from a two-sided parametric test.
For analysis of de novo CNVs using PLINK-CNV a one-sided empirical p value was derived based on permutation.
Discussion
Here we find strong evidence implicating rare de novo CNVs in genetic risk for bipolar disorder. We also confirm previous findings that de novo CNVs occur at a significantly increased rate in individuals with schizophrenia (Xu et al., 2008). Based on our study, a contemporaneous study (Kirov et al., 2011), and an earlier study by our coauthor (Xu et al., 2008), we estimate that the overall frequency of de novo CNVs > 10 kb is approximately 4% in BD and 5%–10% in SCZ.
Two previous case-control studies have observed an enrichment of rare CNVs in bipolar disorder and in subjects with an early age at onset. However, the observed effects were small (OR ~ 1.3) and results from two other studies (Grozeva et al., 2010 and McQuillin et al., 2011) did not support these findings. In our present study, which focused on the detection of de novo CNVs using a family-based design, we observe a large effect (OR > 4). This is consistent with other family-based studies of autism (Levy et al., 2011, Marshall et al., 2008, Sanders et al., 2011 and Sebat et al., 2007) and schizophrenia (Xu et al., 2008 and Xu et al., 2009) that have found a strong and robust genetic effect for de novo mutations and a weaker genetic effect for inherited variants. The much greater effect size for de novo CNVs as compared with inherited variants is consistent with de novo mutations having a much higher proportion of risk alleles relative to neutral alleles.
The high-density microarray platform used in this study (2.1 million probes) provides good ascertainment of CNVs > 10 kb, a substantial improvement in sensitivity over earlier studies of schizophrenia and autism. De novo CNVs of intermediate size (10–100 kb) were detected a rate of 3/426 (0.7%) in controls and at a rate of 3/177 (1.7%) in schizophrenia and 5/185 (2.7%) in bipolar disorder. Based on these observations, intermediate-size CNVs may contribute to risk for these disorders; however, they occur in a small fraction (2%–3%) of cases, and the relative effect size is smaller than for large de novo CNVs. We conclude that a small fraction of the heritability in schizophrenia can be found among intermediate sized de novo structural variants.
The effect of de novo CNV on age at onset in bipolar disorder was nominally significant. These preliminary findings and similar results from previous studies (Priebe et al., 2011 and Zhang et al., 2009) suggest that individuals with an early onset of mania might constitute a subclass of bipolar disorder in which there is a greater contribution from rare alleles of large effect. Also consistent with this notion is a previous study (Grigoroiu-Serbanescu et al., 2001), which found that segregation of early-onset BD in families was consistent with major gene effects, while familial segregation of late-onset BD was consistent with a multifactorial etiology.
The observed rate of de novo CNVs in cases of bipolar disorder or schizophrenia with a positive family history of mental illness was similar to the rate in sporadic cases. These results contrast with earlier findings by our group and others in autism (Marshall et al., 2008 and Sebat et al., 2007) and schizophrenia (Xu et al., 2008) documenting a higher rate of CNVs in sporadic cases as compared with subjects who have a positive family history. These early observations have not been universally replicated (Kirov et al., 2011 and Pinto et al., 2010). The reason for the inconsistency is not clear. Possibly, the initial findings were incorrect. Alternatively, variation in the observed effect could occur by chance or it could potentially be explained by methodological differences between cohorts in how mental illness in first-degree relatives is ascertained.
Pathway analysis of genes impacted by de novo CNVs in SCZ lends support to independent findings from our group (Walsh et al., 2008) and others (Kirov et al., 2011) that rare CNVs in SCZ are enriched for genes that are related to synaptic function and other genes involved in neurodevelopment. By contrast, categories that were found to be enriched among de novo CNVs in BD were related to cell proliferation and shape and phospholipid metabolism (Table 5), the biological relevance of which is far from obvious. Greater knowledge of the specific genes involved in these disorders is needed to determine how these pathways might relate to the pathophysiology of disease.
Our findings establish a contribution of rare CNVs and spontaneous mutation to risk for bipolar disorder. This can only be regarded as a starting point for studies of rare alleles in BD and SCZ. A larger fraction of the heritability must lie among different classes of alleles and will probably include rare and de novo point mutations and small insertions or deletions (indels). Indeed, preliminary studies of a small number of families have found evidence that the exomic burden of de novo point mutations is increased in schizophrenia (Girard et al., 2011 and Xu et al., 2011). While the degree of enrichment of exonic point mutations in schizophrenia (0.73/exome in cases as compared with 0.32/exome in controls in the combined sample) is modest compared to the effect size for de novo CNVs, these results are nevertheless intriguing. It is conceivable that the overall contribution of de novo CNVs and point mutations to disease risk could be substantial. High-throughput sequencing in a much larger number of trios will be needed to determine the total contribution of de novo mutation to risk for BD and SCZ in the population.
Procedures
Experimental Procedures
The institutional review board of all participating institutions approved this study and written informed consent from all subjects was obtained.
Microarray Data Processing and Segmentation
We performed high-resolution genome-wide copy-number scans, using the Nimblegen HD2 2.1 M array CGH platform, on all subjects and their biological parents. Complete details for microarray intensity data processing, CNV discovery, and quality control (QC) measures for sample hybridizations are provided in Supplemental Experimental Procedures. In brief, dual-color microarray hybridizations were performed at the service laboratory of Roche NimbleGen according to the manufacturer's specifications. Raw intensity data were normalized in a two step process, first involving “spatial” normalization which is an adjustment for regional variation in probe intensities across the surface of the array, and second involving “invariant set normalization,” which normalizes the distribution of intensities for test and reference samples. CNV detection from the Log2 probe ratios was performed using two segmentation algorithms, HMM Seg and Genome Alteration Detection Analysis (GADA).
In addition, probe ratio data was used to identify and genotype common copy-number polymorphisms (CNPs) using automated correlation- and clustering-based methods (see Supplemental Experimental Procedures). Stringent QC filters were applied to arrays and CNV calls to ensure that the ascertainment of CNVs was consistent between subjects and their parents (see Supplemental Experimental Procedures and Table S1).
Rare CNV Determination
We determined the population frequency of CNVs detected in our study sample by comparison with CNV calls (based on ≥ 50% reciprocal overlap of its CNV length) from a larger reference population of 4,081 unrelated subjects analyzed in our laboratory using the same array platform. Unrelated subjects consisted of 3,309 population controls, 604 subjects with diagnosis of schizophrenia, 154 subjects with mood disorders, and 14 subjects with a diagnosis of ASD (Table S2). CNVs that were detected in > 1% of the reference population were excluded.
Rare CNVs were further filtered by three metrics: (1) Confidence score (CS), (2) segmental duplication (SD) content, and (3) overlap with validated common copy-number loci. CS was calculated using MeZOD, an outlier-detection-based method published previously (McCarthy et al., 2009), where the rare CNV call is assigned a p value based on the distribution of probe ratios across the reference population. Thresholds for CS were then adjusted within each size class of CNV to achieve a 5% rate of mendelian inconsistency across all size classes (Figure S1). Second, we removed rare CNVs which had > 70% overlap with known SDs from the UCSC hg18 Human Genome browser annotations. A SD filter is helpful because it eliminates regions where the exact location, boundaries, and patterns of inheritance of the CNV calls are often too difficult to determine from array CGH data due to the complexity of the local genomic architecture. This final rare CNV call sets consisted of 3,856 CNVs in 788 offspring including BD, SCZ, and controls and in 45 ASD subjects (Table S1).
Parentage Testing
Before examining the parent-child transmission of CNVs in trios, we first confirmed parentage of all trios included in CNV analysis. We used genotypes from 486 CNPs (see Supplemental Experimental Procedures) to test relatedness. For each pair within a trio (i.e., mother-child, father-child, and mother-father), genetic relatedness was tested by the Glaubitz Relationship Score (GRS) (Glaubitz et al., 2003). Based on this test, first-degree relative pairs (i.e., mother-child and father-child) were clearly distinguishable from the distribution of GRS scores for unrelated individuals (i.e., mother-father pairs) as shown in Figure S2. We applied a threshold of > 0.37 to define relatedness. Thirty-two families failed parentage testing, and the remaining 788 trios were included in our analysis. Pairwise relationship of all subjects in 788 trios was confirmed using a second relatedness testing method, Graphical Representation of Relationships (GRR, http://www.sph.umich.edu/csg/abecasis/GRR/).
Identification of De Novo CNVs
The identification of rare de novo mutations from CNV data on families is nontrivial. While CNV calls that show mendelian patterns of inheritance (which is the overwhelming majority of CNVs in the genome) are quite reliable, the fraction of CNV calls that are present in offspring and not in parents are enriched for technical errors, in particular false-positive calls (in the offspring) and false-negative calls (in parents). In addition, the enrichment of such errors is greater for smaller CNVs. To address these sources of error, we designed a set of algorithms for de novo CNV identification in families. The false-positive CNV call rate was controlled (maintained at 5%) for a wide range of CNV sizes by adaptive filtering of confidence scores (CSs), as described above. In order to minimize the number of false-negative calls in parents, the CNV region was directly genotyped using the MeZOD, and a confidence score was used to assign genotypes to the parents. Rare CNVs in children were called inherited if the CS was ≤ 0.04 in either of the two biological parents and de novo if CS was > 0.04 in both biological parents. We identified 145 putative de novo CNVs in 788 subjects including BD, SCZ, and controls.
Custom Tiling Array Validation of Putative De Novo CNVs
All putative de novo CNVs detected in our whole-genome scans were independently validated on second custom tiling array platform.
Array CGH
A custom Agilent 1M array was designed with dense coverage (average probe spacing of 200 bp) of all putative de novo CNV regions. Samples were coded and hybridizations were done in random order to avoid any plate effects. Two-color hybridizations were performed with two micrograms of sample and reference DNA (CHP-SKN-1) and hybridized to the array at the Oxford Gene Technology service laboratory (Cambridgeshire, UK).
Array Data Processing
Raw intensity data were normalized by Oxford Gene Technology service lab using Agilent's recommended normalization method. Experiments with poor derivative log2 ratio spread (DLRS > 0.2) were repeated. We received normalized intensity data on all samples from Oxford Gene technology in one batch. Probe Log2 Ratios were then standardized within each array.
CNV Detection
Detection of rare CNVs was performed using MeZOD as follows. For each CNV region that was defined in our whole-genome scans, we computed the median Z score of tiling array probes in each individual. The median of a region was then standardized vertically across all individuals. We then assign deletion genotypes using a Z score threshold of ≤ −2 and duplication genotypes using a Z score threshold of ≥ +2. Positive CNV calls were further verified by manual inspection of log2 ratios in the subject, mother, and father. Representative examples of validated de novo deletions are shown in Figure 1 and Figure 2. The details of the number of putative de novo CNVs identified in BD, SCZ, and controls and their validation by tiling array CGH are described in Table S3. The rates of validations are presented in Table S4. The overall validation rate of putative de novo CNVs was 16% (23/145). As expected, the validation rate was highest for CNVs > 100 kb in size and lowest (3%) for CNVs that were < 20 kb in size.
We evaluated the performance of our de novo CNV calling method by: i) analyzing a small set of 45 ASD trios included in our previous CNV study (Sebat et al., 2007) and, ii) by comparing results on validated control de novo CNVs identified by our group with results from a recent study(Levy et al., 2011) by Mike Wigler's group. In 45 ASD trios we detected and validated all 3 de novo CNVs that were identified in our previous study and in addition, we identified one novel de novo CNV 38 kb in size (Table S8). We compared our list of validated control de novo CNVs with de novo CNVs reported by (Levy et al., 2011) in the same 426 control trios using an entirely different informatics approach to identify de novo CNVs. Both groups identified four validated de novo CNVs in controls and therefore observed an identical rate (0.9%) of de novo CNVs in 426 controls. Three out of four de novo events overlapped between two groups. One de novo event that was unique to each group was < 20 kb in size. These comparisons showed that our method had high sensitivity in identifying de novo CNVs in different size classes.
Breakpoint Sequencing
CNV boundaries were estimated based on the probe (log2) ratio information from tilling array CGH. PCR primers were then designed to amplify the breakpoints of five de novo deletions. PCR was performed on genomic DNA from all members of the trio. PCR products were sequenced by the Sanger method using both forward and reverse primers specific for each de novo deletion.
Gene Set Enrichment Analysis of De Novo CNVs
We examined whether genes impacted by de novo CNVs in SCZ, BD, and controls were enriched for specific functional categories. In addition, functional categories found to be enriched within each diagnostic group were interrogated in rare CNVs from large independent cases control data sets including 8,290 SCZ, 2,777 BD, and 7,431 controls.
For gene set enrichment analysis, we used 39 de novo CNVs including nine in SCZ, ten in BD, four in our controls, and an additional 16 CNVs detected in a previous study by Levy et al. (2011) in an independent set of control subjects using the same array platform. We prefer to use only de novo CNVs as a control set. Naturally occurring variants in the population do not make the ideal control set for this analysis because the gene content of these CNVs is shaped by natural selection and is not likely to be representative of random mutation.
Gene set enrichment analyses was performed on the sets of genes impacted by de novo CNVs in SCZ, BD, and controls. The primary step was performed using “DAVID Bioinformatics Resources 6.7” website (http://david.abcc.ncifcrf.gov/) using Gene Ontology terms—biological processes (GO_BP), cellular components (GO_CC), and molecular functions (GO_MF)—including KEGG, BioCarta, BBID, and Panther pathway databases and by excluding pathway results containing < 3 CNV genes. We selected the nonredundant pathways from DAVID with p value < 0.05 for further analysis by permutation-based test.
Based on analysis using DAVID, eight categories were enriched among de novo CNVs in SCZ (Table 4), seven categories were enriched among de novo CNVs in BD (Table 5), and nine categories were enriched among de novo CNVs in controls (Table S7).
The enrichment test performed within the DAVID software does not correct for certain biases of CNVs toward certain functional classes of genes and large genes in particular. In order to correct for these biases we applied two permutation-based tests to the pathways found to be enriched by DAVID. First, we performed a case-only permutation-based test by constructing empirical null distributions that took the CNV size distribution and gene number into account. We randomly placed 10,000 sets of CNVs (same number of events, size distribution) throughout the genome. Placement on any autosome was allowed, but we sampled such that placement on chromosomes was weighted in proportion to the total number of de novo CNVs observed on the respective chromosome. We controlled for the number of genes impacted by CNVs by discarding individual permutations that intersected with more or less than the number of genes impacted in the observed data (± 10 genes). This procedure led to 1,000–2,000 permutations of null hypothesis CNV sets each for the bipolar, schizophrenia, and control de novo CNV sets. Significance for each of the query pathways was assessed by counting the number of pathway genes impacted by each null hypothesis CNV set, thus leading to a null distribution against which we could compare the number of observed hits and calculate enrichment p values.
Second we applied a case-control CNV enrichment test implemented in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) to the eight gene sets (pathways) associated with SCZ de novo CNVs and seven gene sets associated with BD de novo CNVs. For analysis of de novo CNVs using PLINK we report the one-sided empirical p value based on 10,000 permutations. A category was defined as “enriched” if nominally significant (p < 0.05) by all case-only and case-control permutation tests. Pathway enrichment analysis was also applied to de novo CNVs in control, and significance was based on a single (control only) permutation test.
We extended our analysis of pathways identified in this study (Table 4 and Table 5, and S7) to rare CNVs from large case-controls studies of SCZ and BD. These included rare CNVs from 8,290 SCZ cases and 7,431 controls from Vacic et al. (2011), a combined sample of three studies from our group, MGS (Levinson et al., 2011), and the International Schizophrenia Consortium (International Schizophrenia Consortium, 2008), and a BD case-control data set consisting of 2,777 cases and 3,508 controls from BiGS study (Smith et al., 2009). Pathway enrichment was assessed using the case-control CNV enrichment test implemented in PLINK.
Supplementary Material
Acknowledgments
We thank all patients and their families for their participation in this genetic study. Special thanks to James Watson for helpful discussions and support. This study was supported by a gift from Ted and Vada Stanley to the Cold Spring Harbor Laboratory, a gift to J.S. from the Beyster family foundation, NIH grants to J.S. (MH076431, HG04222), D.L.L. (MH071523), and M.K. (MH061399), grants to J.S. and D.L.L. from NARSAD, grants to A.C. and M.G. from the Wellcome Trust (072894/Z/03/Z) and Science Foundation Ireland (08INIB1916), and grants to D.L.L. from the Sidney R. Baer, Jr. Foundation and Essel Foundation. We thank the Genetic Association Information Network (GAIN), Molecular Genetics of Schizophrenia (MGS), and the Bipolar Genome Study (BiGS) for providing data for this study. We thank Roche NimbleGen and Oxford Gene Technology for their expert technical assistance. We thank Lilia M. Iakoucheva and Roser Corominas for helpful discussions.
Contributor Information
Dheeraj Malhotra, Beyster Center for Genomics of Psychiatric Diseases, University of California, San Diego, La Jolla, CA 92093, USA, Department of Psychiatry, University of California, San Diego, La Jolla, CA 92093, USA,, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Shane McCarthy, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Jacob J. Michaelson, Beyster Center for Genomics of Psychiatric Diseases, University of California, San Diego, La Jolla, CA 92093, USA, Department of Psychiatry, University of California, San Diego, La Jolla, CA 92093, USA
Vladimir Vacic, Department of Computer Science, Columbia University, New York, NY 10027, USA, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Katherine E. Burdick, Mount Sinai School of Medicine, New York, NY 10029, USA
Seungtai Yoon, Seaver Autism Center, Mount Sinai School of Medicine, New York, NY 10029, USA, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Sven Cichon, Department of Genomics, Life and Brain Center, University of Bonn, D-53127 Bonn, Germany, Institute of Human Genetics, University of Bonn, D-53127 Bonn, Germany, Institute of Neuroscience and Medicine (INM-1), Research Center Julich, D-52425 Julich, Germany.
Aiden Corvin, Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine and Department of Psychiatry, Trinity College Dublin, Dublin 2, Ireland.
Sydney Gary, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Elliot S. Gershon, Department of Psychiatry and Behavioral Neuroscience, University of Chicago, Chicago, IL 60637, USA
Michael Gill, Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine and Department of Psychiatry, Trinity College Dublin, Dublin 2, Ireland.
Maria Karayiorgou, Department of Psychiatry, Columbia University, New York, NY 10032, USA.
John R. Kelsoe, Department of Psychiatry, University of California, San Diego, La Jolla, CA 92093, USA, Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA 92093, USA, Veterans Affairs San Diego Healthcare System, San Diego, CA 92161, USA
Olga Krastoshevsky, McLean Hospital, Belmont, MA 02478, USA.
Verena Krause, McLean Hospital, Belmont, MA 02478, USA.
Ellen Leibenluft, Section on Bipolar Spectrum Disorders, Emotion and Development Branch, NIMH Building 15K - MSC 2670, Bethesda, MD 20892, USA.
Deborah L. Levy, McLean Hospital, Belmont, MA 02478, USA
Vladimir Makarov, Seaver Autism Center, Mount Sinai School of Medicine, New York, NY 10029, USA, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Abhishek Bhandari, Beyster Center for Genomics of Psychiatric Diseases, University of California, San Diego, La Jolla, CA 92093, USA, Department of Psychiatry, University of California, San Diego, La Jolla, CA 92093, USA, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
Anil K. Malhotra, Zucker Hillside hospital, North Shore Long Island Jewish health system, Glen Oaks, NY 11004, USA
Francis J. McMahon, Genetic Basis of Mood and Anxiety Disorders, National Institute of Mental Health, NIH, Convent Drive MSC 3719, Bethesda, MD 20892, USA
Markus M. Nöthen, Department of Genomics, Life and Brain Center, University of Bonn, D-53127 Bonn, Germany, Institute of Human Genetics, University of Bonn, D-53127 Bonn, Germany, German Center for Neurodegenerative Diseases (DZNE), D-53175 Bonn, Germany
James B. Potash, Department of Psychiatry, University of Iowa, Carver College of Medicine, Iowa City, IA 52242, USA
Marcella Rietschel, Central Institute of Mental Health, University of Heidelberg J5, 68159 Mannheim, Germany.
Thomas G. Schulze, Department of Psychiatry and Psychotherapy, George-August-University Gottingen, von-Siebold-Str.5 370075 Gottingen, Germany
Jonathan Sebat, Beyster Center for Genomics of Psychiatric Diseases, University of California, San Diego, La Jolla, CA 92093, USA, Department of Psychiatry, University of California, San Diego, La Jolla, CA 92093, USA, Department of Cellular Molecular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093, USA, Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA 92093, USA, Stanley Institute for Cognitive Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 12824, USA.
References
- Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum AE, Hamshere M, Green E, Cichon S, Rietschel M, Noethen MM, Craddock N, McMahon FJ. Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Mol. Psychiatry. 2008;13:466–467. doi: 10.1038/mp.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, Giblin C, Smith RA, Dunn C, Willatt L. Familial 3q29 microdeletion syndrome providing further evidence of involvement of the 3q29 region in bipolar disorder. Clin. Dysmorphol. 2010;19:128–132. doi: 10.1097/MCD.0b013e32833a1e3c. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Wellcome Trust Case Control Consortium. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, et al. Welcome Trust Case Control Consortium. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Glaubitz JC, Rhodes OE, Dewoody JA. Prospects for inferring pairwise relationships with single nucleotide polymorphisms. Mol. Ecol. 2003;12:1039–1047. doi: 10.1046/j.1365-294x.2003.01790.x. [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Martinez M, Nöthen MM, Grinberg M, Sima D, Propping P, Marinescu E, Hrestic M. Different familial transmission patterns in bipolar I disorder with onset before and after age 25. Am. J. Med. Genet. 2001;105:765–773. doi: 10.1002/ajmg.10047. [DOI] [PubMed] [Google Scholar]
- Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK, St Clair DM, Young AH, Ferrier N, Farmer AE, et al. Wellcome Trust Case Control Consortium. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch. Gen. Psychiatry. 2010;67:318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Pedersen NL, Neale MC, Mathé AA. A pilot Swedish twin study of affective illness including hospital- and population-ascertained subsamples: results of model fitting. Behav. Genet. 1995;25:217–232. doi: 10.1007/BF02197180. [DOI] [PubMed] [Google Scholar]
- Kieseppä T, Partonen T, Haukka J, Kaprio J, Lönnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am. J. Psychiatry. 2004;161:1814–1821. doi: 10.1176/ajp.161.10.1814. [DOI] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signaling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry. 2011 doi: 10.1038/mp.2011.154. in press. Published online November 15, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Zhang N, Mowry BJ, Olincy A, Amin F, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am. J. Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, et al. Wellcome Trust Case Control Consortium. Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- McQuillin A, Bass N, Anjorin A, Lawrence J, Kandaswamy R, Lydall G, Moran J, Sklar P, Purcell S, Gurling H. Analysis of genetic deletions and duplications in the University College London bipolar disorder case control sample. Eur. J. Hum. Genet. 2011;19:588–592. doi: 10.1038/ejhg.2010.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Jin R, He JP, Kessler RC, Lee S, Sampson NA, Viana MC, Andrade LH, Hu C, Karam EG, et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch. Gen. Psychiatry. 2011;68:241–251. doi: 10.1001/archgenpsychiatry.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, Sobreira NL, Valle D, Rudd MK, Satten G, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am. J. Hum. Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potash JB, Toolan J, Steele J, Miller EB, Pearl J, Zandi PP, Schulze TG, Kassem L, Simpson SG, Lopez V, et al. NIMH Genetics Initiative Bipolar Disorder Consortium. The bipolar disorder phenome database: a resource for genetic studies. Am. J. Psychiatry. 2007;164:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- Priebe L, Degenhardt FA, Herms S, Haenisch B, Mattheisen M, Nieratschker V, Weingarten M, Witt S, Breuer R, Paul T, et al. Genome-wide survey implicates the influence of copy number variants (CNVs) in the development of early-onset bipolar disorder. Mol. Psychiatry. 2011 doi: 10.1038/mp.2011.8. in press. Published online March 1, 2011. [DOI] [PubMed] [Google Scholar]
- Quintero-Rivera F, Sharifi-Hannauer P, Martinez-Agosto JA. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: case report and review. Am. J. Med. Genet. A. 2010;152A:2459–2467. doi: 10.1002/ajmg.a.33573. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Korn JM, McCarroll SA, Altshuler D, Sklar P, Purcell S, Daly MJ International Schizophrenia Consortium. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno- De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Jr, Rietschel M, Blackwood D, et al. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg HJ, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol. Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. GROUP. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, Gogos JA, Karayiorgou M. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc. Natl. Acad. Sci. USA. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Qian Y, Alliey-Rodriguez N, Kelsoe JR, Greenwood T, Nievergelt C, Barrett TB, McKinney R, Schork N, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol. Psychiatry. 2009;14:376–380. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.