Abstract
We demonstrate the generation of systemically releasable anti-cancer drugs from multilayer nanofilms. Nanofilms designed to drug release profiles in programmable fashion are promising new and alternative way for drug delivery. For the nanofilm structure, we synthesized various unique 3-dimensional anti cancer drug incorporated DNA origami structures (hairpin, Y, and X shaped) and assembled with peptide via layer-by-layer (LbL) deposition method. The key to the successful application of these nanofilms requires a novel approach of the influence of DNA architecture for the drug release from functional nano-sized surface. Herein, we have taken first steps in building and controlling the drug incorporated DNA origami based multilayered nanostructure. Our finding highlights the novel and unique drug release character of LbL systems in serum condition taken full advantages of DNA origami structure. This multilayer thin film dramatically affects not only the release profiles but also the structure stability in protein rich serum condition.
Functional films containing DNA have received considerable attentions due to their numerous potential applications in sensing, diagnostics, and gene therapy1,2,3,4,5. From materials science and engineering point of view, DNA molecules have unique chemical and physical properties which are greatly useful for the design and construction of versatile nanomaterials with a variety of functions. Various DNA structures including 1-dimensional linear, 2-dimensional branch, and 3-dimensional dendritic DNA can be synthesized even though DNA in nature state only form linear or circular structures. Effective DNA based-systems are based on not only dealing with DNA but also containing additional therapeutics6,7,8,9,10. The primary objective of these systems is controlled delivery of DNA and tailor-incorporated drugs from thin films, hydrogels, scaffolds, and particles; however, the development of efficient and safe localized delivery systems remains a significant challenge for DNA based delivery systems. Recently, layer-by-layer (LbL) assembly deposition has emerged as a simple, versatile, and high-quality approach for the construction of functional thin films11,12,13,14,15,16. LbL multilayer films are assembled by alternate deposition of complementary forces such as positively and negatively charged objects. Various functional materials including proteins, graphenes, nanoparticles, and dendrimers can be easily incorporated within nanofilm structures with highly precise control. Here we describe the LbL assembly of multilayer films consisting of negatively charged branched DNA molecules with the positively charged natural peptide poly-L-Lysine (PLL), with distinct anti-cancer drug release profiles driven by changes in the three-dimensional DNA structures. Branched DNA molecules were designed and synthesized by efficient, ligase-mediated reactions in which each arm of the DNA molecule possessed a complementary sticky end with palindromic sequences8,17. The 3-dimensional DNA architectures that serve as building blocks for LbL assembly are hairpin-shaped (H-DNA), Y-shaped (Y-DNA), and X-shaped (X-DNA). Taking full advantage of the diverse stabilities of these DNA architectures, anti-cancer drug, Doxorubicin (Dox, a hydrophilic apoptosis-inducing anticancer drug) encapsulated in DNA molecules paired with PLL exhibited a distinctive Dox release profile under physiological conditions [fetal bovine serum (FBS), 37°C, 5% CO2].
Results
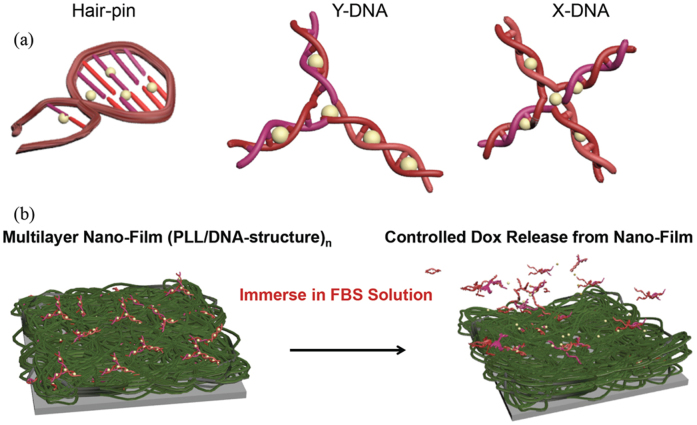
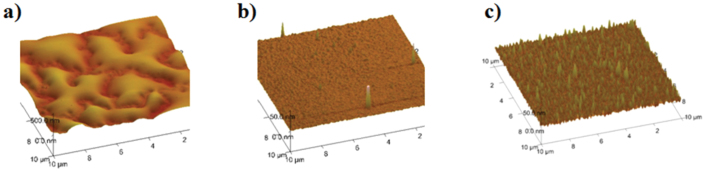
Fig. 1 shows schematics of various DNA structures of H-DNA, Y-DNA, and X-DNA and the preparation of multilayer films consisting of PLL and Dox-incorporated DNAs. DNA is highly biodegradable due to the presence of nucleases. The degraded products are nucleotides, naturally occurring metabolites in the body, making them non-toxic and biocompatible, ideal characteristics for a biomaterial. In addition, DNA can be designed and engineered in a highly controlled manner and high purity and reaction yields can be achieved with extremely precise control, allowing easy control of their physical and chemical properties, even though DNA in nature only have linear or circular forms2,18,19,20,21,22,23. DNA is thus a highly useful nanomaterial and building block for nano-assembly systems including multilayered structures. H-DNA, branched Y-DNA, and branched X-DNA were synthesized according to previously published methods24,25. The successful formation of Y-DNA and X-DNA was confirmed by gel electrophoresis (Fig. S1, supplementary information (SI)). Branched DNA can overcome the structural and topological limitations of conventional linear DNA. First, branched DNA can contain various functional moieties on each branch and are inherently multivalent. Furthermore, the binding affinities and stability of synthesized branched DNAs are controlled by their design17,24,25. Table 1 shows the DNA sequences of DNA used in the present study. Since Dox molecules are appropriate in size and chemical properties to fit between DNA base pairs, Dox can be incorporated into synthesized DNA structure by intercalation, which has been intensively studied before26,27. PLL, serving as counter building block with DNA, is natural homopolymer. It is a weak polyelectolyte with pKa value of around 9–1028,29,30, indicating electrostatic interaction between positively charged PLL and negatively charged DNA, which is main driving force for PLL and DNA to form nanofilm structure, can be modulated by external pH conditions after film construction. Assembly of thin films consisting of Dox-incorporated DNA and PLL was performed by the LbL deposition method. Table 2 shows the thickness of thin films composed of 20 bilayers as measured by a profilometer, indicating successful preparation of multilayered thin structures. It shows linear film growth with the number of bilayers and each (PLL/DNA) bilayer thickness is approximately 22, 26, and 27 nm for H-, Y-, and X-DNA, respectively. It is believed that the difference in film thickness of (PLL/H, Y, and X-DNA) arose from their different DNA dimension when considering synthesized DNAs have different length and the number of binding sites, yielding various DNA topologies. The length of one helical turn of double-stranded DNA (dsDNA) consists of ten base pairs is 3.4 nm and its width is 2 nm. Furthermore, the thin film surfaces were analyzed by atomic force microscopy (AFM) as shown in Fig. 2. Since synthesized DNA structures have different sizes and molecular properties, resulting in slightly different PLL adsorption behaviors, they also have different surface roughness and uniformity properties. AFM images, however, indicate the thin film structures were successfully and homogeneous created.
Figure 1. Schematic illustration of (a) hair-pin-, Y-, and X-shaped DNA, (b) multilayer assembly combination of Dox encapsulated DNA and PLL, and (c) experimental procedure.
Table 1. Oligonucleotide sequences of DNA building blocks.
| Building Block | Strand | Sequence |
|---|---|---|
| Y-DNA | Y01 | TGGATCCGCATGACATTCGCCGTAAG |
| Y02 | CTTACGGCGAATGACCGAATCAGCCT | |
| Y03 | AGGCTGATTCGGTTCATGCGGATCCA | |
| X-DNA | X01 | GCACCGATGAATAGCGGTCAGATCCGTACCTACTCG |
| X02 | CGAGTAGGTACGGATCTGCGTATTGCGAACGACTCG | |
| X03 | CGAGTCGTTCGCAATACGGCTGTACGTATGGTCTCG | |
| X04 | CGAGACCATACGTACAGCACCGCTATTCATCGGTGC | |
| Hairpin | Hairpin | TTTTTTCCTAGCGGTTGGTGTGGTTGGAGCTAGGTTTTTT |
| FTIC- ssDNA | Single | 5′-FITC-CTTACGGCGAATGACCGAATCAGCCT-3′ |
Table 2. Thickness of (PLL/DNA)n multilayer films measured by profilometer.
| (PLL/Hair-pin DNA)20 | (PLL/Y- DNA)20 | (PLL/X- DNA)20 | (PLL/FITC-ssDNA)20 | |
|---|---|---|---|---|
| Film Thickness | 445 ± 21 nm | 521 ± 49 nm | 534 ± 44 nm | 436 ± 20 nm |
Figure 2. Height mode AFM images of a) (PLL/hairpin)20, b) (PLL/Y-DNA)20, and c) (PLL/X-DNA)20.
Discussion
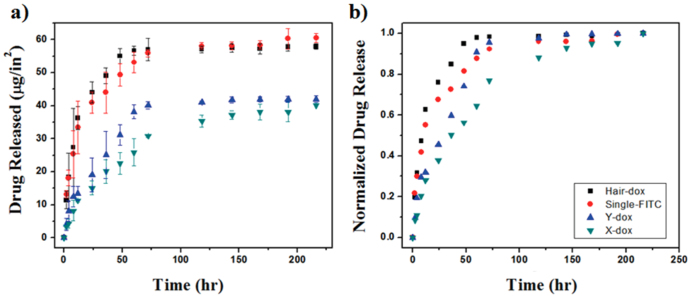
The release of Dox from (PLL/Dox-incorporated DNA)n multilayer films after incubation in FBS was systematically analyzed by photoluminescence (PL) spectroscopy as a function of incubation time after contact with FBS solution as shown in Fig. 3. Fluorescein isothiocyanate (FITC) labeled single-stranded DNA (ssDNA) was used for comparison with the synthesized DNAs. As described above, Since PLL is weak polyelectrolyte, when the nanofilm consist of PLL and DNA is immersed in FBS, variation in pH condition leads to change in electrostatic interaction between positively charged PLL and negatively charged DNA, which results in disassembly of nanofilm structure. Before measuring the PL intensity of released solutions, all DNAs (H-, Y-, and X-DNA) released from the multilayer thin film building block structures during incubation in FBS solution were clearly eliminated by column purification. Gel electrophoresis data shows the all DNAs were successfully separated from the FBS solution, as shown in SI Fig. S2. Therefore, the fluorescence intensity is derived from free Dox molecules separated from the DNAs rather than Dox-incorporated DNAs. Furthermore, it has been very well known that all the DNA structures without specific chemical treatments are collapsed in the presence of serum due to the nuclease degradation24,25,31. Therefore, Dox-incorporated DNA cannot be existed and affect to our PL data. The normalized cumulative release profiles show the effect of DNA shape on release profiles (Fig. 3b). X-DNA multilayer films show a slow release profile over a long duration with a low burst initial release in the early stage of incubation compare to other DNA types, resulting in sustained release of Dox molecules in a controlled manner over a broad range of time span. On the other hand, H-DNA multilayer film provided faster release and a larger initial burst even than multilayer films with ssDNA. We believe the difference in release kinetics is derived from different binding affinities between the DNA base pairs. We suggest 2 possible mechanisms for the release of Dox molecules from the (PLL/Dox-incorporated DNA)n multilayer films. First, degradation of multilayer films to the polymer chains (PLL) and Dox-loaded DNA due to the variation in pH condition may lead to the release of Dox-loaded DNA in the presence of FBS. In addition, the separation of Dox molecules from the DNA, originally combined through intercalation, can be another release mechanism; however, all the DNAs (H-, Y-, and X-DNA except FITC-labeled ssDNA) were clearly removed from the release solution by column purification before the photoluminescence intensity of released Dox was measured. Therefore, it was not possible for separated Dox-loaded DNA to influence the release profile data in Fig. 3. We believe the separation of Dox molecules from the DNA is the dominant reason for our detection of photoluminescence intensity even though both mechanisms may occur during incubation in serum. Furthermore, in the case of FITC labeled ssDNA, FITC cannot be freed from the DNA due to their strong covalent bonding between DNA and FITC molecules. If some Dox-DNAs still remain in the FBS even after purification of all Dox-DNAs, we believe that the free separation of Dox from the DNA determines the release kinetics because if the separated Dox-loaded DNA dominantly determine the release kinetics, ssDNA should show faster release profile than synthesized DNA structures due to its relatively simple geometrical shape compare with H-, Y-, and X-DNA resulting in relatively fast release from the thin film structure. H-, Y-, and X-DNA have a different number of binding sites (6, 13, and 18, respectively), resulting in different binding affinities. dsDNA structure is more loosely bound with fewer binding sites and more tightly bonded DNA structures limit the release of Dox molecules due to their geometrical restrictions and high stability. Therefore, Dox molecules can be easily freed from the inside of dsDNA structures as the number of binding sites decreases. The differing properties of synthesized DNAs including binding affinity produce drastic changes in the release profile of Dox from the PLL/Dox-loaded DNA multilayer structure during incubation in serum.
Figure 3.
a) The total amount and b) cumulative normalized release profile of Doxorubicin from DNA incorporated LbL films as a function of incubation time after rehydration in FBS (pH 7.4 at 37°C, 5% CO2).
More significantly, all the results presented so far were obtained in serum (FBS) medium. Even when drug release is achieved in simple model physiological conditions, such as phosphate buffered saline (PBS), this can be lost in relevant physiological conditions. Rather than the absolute drug release to condition, a most critical feature is the capacity of the nanofilm to avoid possible interactions in complex biological milieu in which they are applied. Although these studies focus on a release fashions from nano-structure, the mechanisms between building blocks and specific biological fluids should be understood in near future within in situ binding analysis.
In summary, we synthesized DNAs with various 3-dimensional shapes (H-, Y-, X-DNA) from ssDNA with unique sticky ends. An anticancer drug was encapsulated in these DNAs and assembled with biodegradable polymer with nanoscale control by LbL deposition method to produce a nanofilm for biodegradable drug delivery. The release profile of the model drug (Dox) from the nanofilm (PLL/Dox-incorporated DNA) under serum conditions was modulated by the unique shape of each 3-dimensional DNA structure. Our results here show that multilayer nanofilm released Dox by their unique DNA origami structure even in serum condition. Considering serum include lot's of proteins & chemicals, our approach provides significant advantages for controlling the release of biomolecules from nanosurfaces. We believe this approach can be extended to the development of bioactive materials loaded nanosurfaces with various functionalities, stabilities, and highly controllable release properties when considering great growth potential of both thin film fabrication for controlled drug release and DNA engineering.
Methods
Synthesis of Y-DNA and X-DNA
The Y-DNA was synthesized by mixing the equal molar amount of three corresponding oligonucleotide strands whose nomenclature is as follows: Y01, Y02 and Y03. Hybridization was performed by heating the mixture at 95°C for 2 min and then cooling it to desired temperature by the following protocol: 2 min at 65°C, 5 min 60°C, 30 sec of incubation every −1°C decrease from 60°C to 20°C. The resultant solution was then held at 4°C. Similarly, the four corresponding single oligonucleotides, X01, X02, X03 and X04, were mixed and then annealed by the above annealing protocol to form an X-DNA. All strands including modified strands were purchased from IDT (Integrated DNA Technologies, Coralville, Iowa).
Drug loading in Y-DNA and X-DNA
DNA building blocks solution (20μM) were mixed with Doxorubicin solution (final concentration 25μg/ml) and then incubated for overnight at 25°C. Free Doxorubicin was removed by desalting column (YM-3, Millipore).
Gel electrophoresis
Y-DNA, X-DNA and hairpin DNA were run in a 3% agarose gel at 90 V at 25°C in Tris-acetate-EDTA (TAE) buffer (40 nM Tris, 20 nM acetic acid and 1 mM EDTA, pH 8.0, Bio-Rad). The gel was then stained with 0.5 μg/ml of ethidium bromide in TAE buffer for 30 min.
Materials
PLL (Mw = 80 kDa) was purchased from Sigma (St. Louis, MO).
Preparation of Polyelectrolyte Solutions
PLL was dissolved in pH 6 in 100 mM sodium acetate buffer (NaOAc) at a concentration of 2.0 mg/mL. Solution pH values were adjusted to 6.0 with the addition of 0.10 M HCl or 0.10 M NaOH.
Film Construction and Charaterization
LbL multilayer films were assembled with a modified programmable home made slide stainer. Typically, films were constructed on a silicon wafer and glass slide with approximate size of 2 × 1.4 in2, which were cleaned initially with a Piranha solution (sulfuric acid/hydrogen peroxide 70/30 v/v%) and negatively charged by O2 plasma equipment (Femto Science). The substrate was then dipped in positively charged polyelectrolyte solutions (PLL solution in 100 mM NaOAc buffer, pH 6.0) for 10 min, followed by three sequential rinsing steps with pH-adjusted water (pH 6.0) for 1 min each. Then, the substrate is dipped in negatively charged DNA (0.05 mg/mL in 100 mM NaOAc buffer, pH 6.0) for 10 min and exposed to the same rinsing steps as described above to achieve the desired numbers of bilayer film. The prepared films were dried and stored in air. Film thickness was measured by profilometer (Dektak 150 surface profiler). The lab made 100,000–600,000 MW range column is used to eliminate all the remaining DNAs (H-, Y-, and X-DNA) released from the multilayer structures during incubation in FBS solution.
Release Experiment
In order to measure the release profiles of Dox from the multilayer films containing (Dox-DNAs/PLL)n, each sample containing multilayer film was immersed into 3 ml of fetal bovine serum (FBS, 5% CO2) solution at 37°C. After the controlled time, each sample was taken out and transferred to next FBS solutions. The concentration of released Dox buffer was measured by Photoluminescence (PL) Spectroscopy (plate reader (Tekan)) at room temperature. The normalization was performed by the total amount of released Dox during release experiment.
Author Contributions
J.H. designed research and J.H., Y.C. and J.B.L. performed research. J.H., Y.C. analyzed data. Y.C., J.H. wrote the main manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Info
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (No. 2013R1A1A1076126) for Jinkee Hong. And also supported by the Human Resources Development program (No. 20124010203260) of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) grant funded by the Korea government Ministry of Trade, Industry and Energy for Jong Bum Lee.
The authors declare no competing financial interests.
References
- Fu H. L., Cheng S. X., Zhang X. Z. & Zhuo R. X. Dendrimer/DNA complexes encapsulated functional biodegradable polymer for substrate-mediated gene delivery. J. Gene Med. 10, 1334–1342 (2008). [DOI] [PubMed] [Google Scholar]
- Eckardt L. H. et al. DNA nanotechnology: Chemical copying of connectivity. Nature 420, 286 (2002). [DOI] [PubMed] [Google Scholar]
- Sano T., Smith C. L. & Cantor C. R. Immuno-Pcr - Very Sensitive Antigen-Detection by Means of Specific Antibody-DNA Conjugates. Science 258, 120–122 (1992). [DOI] [PubMed] [Google Scholar]
- Zhang J. T., Chua L. S. & Lynn D. M. Multilayered thin films that sustain the release of functional DNA under physiological conditions. Langmuir 20, 8015–8021 (2004). [DOI] [PubMed] [Google Scholar]
- Elghanian R. et al. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277, 1078–1081 (1997). [DOI] [PubMed] [Google Scholar]
- Jiang Q. et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 134, 13396–13403 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties. Acs Nano 6, 8684–8691 (2012). [DOI] [PubMed] [Google Scholar]
- Roh Y. H. et al. DNAsomes: Multifunctional DNA-Based Nanocarriers. Small 7, 74–78 (2011). [DOI] [PubMed] [Google Scholar]
- Song L. et al. Efficient, pH-Triggered Drug Delivery Using a pH-Responsive DNA-Conjugated Gold Nanoparticle. Adv. Healthcare Mater. 2, 275–280 (2013). [DOI] [PubMed] [Google Scholar]
- Nishikawa M., Rattanakiat S. & Takakura Y. DNA-based nano-sized systems for pharmaceutical and biomedical applications. Adv. Drug Deliver. Rev. 62, 626–632 (2010). [DOI] [PubMed] [Google Scholar]
- Dierich A. et al. Bone formation mediated by synergy-acting growth factors embedded in a polyelectrolyte multilayer film. Adv. Mater. 19, 693–697 (2007). [Google Scholar]
- Decher G. Fuzzy nanoassemblies: Toward layered polymeric multicomposites. Science 277, 1232–1237 (1997). [Google Scholar]
- Blacklock J. et al. Gene delivery in vitro and in vivo from bioreducible multilayered polyelectrolyte films of plasmid DNA. Biomaterials 30, 939–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. et al. Graphene Multilayers as Gates for Multi-Week Sequential Release of Proteins from Surfaces. Acs Nano 6, 81–88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori S. S. & Rubner M. F. pH-dependent thickness behavior of sequentially adsorbed layers of weak polyelectrolytes. Macromolecules 33, 4213–4219 (2000). [Google Scholar]
- Cho Y., Lee J. B. & Hong J. Tunable growth factor release from nano-assembled starch multilayers. Chem. Eng. J. 221, 32–36 (2013). [Google Scholar]
- Roh Y. H. et al. Engineering DNA-based functional materials. Chem. Soc. Rev. 40, 5730–5744 (2011). [DOI] [PubMed] [Google Scholar]
- LaBean T. H. et al. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 122, 1848–1860 (2000). [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- Ding B. Q. et al. Gold Nanoparticle Self-Similar Chain Structure Organized by DNA Origami. J. Am. Chem. Soc. 132, 3248–3249 (2010). [DOI] [PubMed] [Google Scholar]
- Sakamoto K. et al. Molecular computation by DNA hairpin formation. Science 288, 1223–1226 (2000). [DOI] [PubMed] [Google Scholar]
- Tinland B., Pluen A., Sturm J. & Weill G. Persistence length of single-stranded DNA. Macromolecules 30, 5763–5765 (1997). [Google Scholar]
- Andersen E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–75 (2009). [DOI] [PubMed] [Google Scholar]
- Li Y. G. et al. Controlled assembly of dendrimer-like DNA. Nature Mater. 3, 38–42 (2004). [DOI] [PubMed] [Google Scholar]
- Lee J. B. et al. Multifunctional nanoarchitectures from DNA-based ABC monomers. Nature Nanotech. 4, 430–436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S. Citation Classic - Structural Considerations in the Interaction of DNA and Acridines. CC/Life Sci. 19–19 (1984). [DOI] [PubMed] [Google Scholar]
- Richards A. D. & Rodger A. Synthetic metallomolecules as agents for the control of DNA structure. Chem. Soc. Rev. 36, 471–483 (2007). [DOI] [PubMed] [Google Scholar]
- Westwood M., Roberts D. & Parker R. Enzymatic degradation of poly-L-lysine-polygalacturonic acid multilayers. Carbohyd. Polym. 84, 960–969 (2011). [Google Scholar]
- Gergely C. et al. Human serum albumin self-assembly on weak polyelectrolyte multilayer films structurally modified by pH changes. Langmuir 20, 5575–5582 (2004). [DOI] [PubMed] [Google Scholar]
- Hong J., Kim B. S., Char K. & Hammond P. T. Inherent Charge-Shifting Polyelectrolyte Multilayer Blends: A Facile Route for Tunable Protein Release from Surfaces. Biomacromolecules 12, 2975–2981 (2011). [DOI] [PubMed] [Google Scholar]
- Conway J. W., McLaughlin C. K., Castor K. J. & Sleiman H. DNA nanostructure serum stability: greater than the sum of its parts. Chem. Commun. 49, 1172–1174 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Info