Abstract
Directing cell movements within 3D channels is a key challenge in biomedical devices and tissue engineering. In two dimensions, closely spaced arrays of asymmetric teardrop islands can intermittently polarize cells and sustain their autonomous directional migration with no gradients. However, in 3D microchannels composed of linearly connected teardrop segments, negligibly low directional bias is observed. Rather than adopt teardrop shapes, cells evade morphological polarization by spreading across multiple teardrop segments, only partly filling each. We demonstrate here that cells can be forced to adopt the shape of individual segments by connecting the segments at an angle to minimize cell spreading across multiple segments. The resulting rhythmic polarization leads to significant directional bias for NIH3T3 fibroblasts, epithelial cells, and even cells whose intracellular signalling have been purposely altered to affect lamellipodia extension (Rac1) and cell polarity (Cdc42). This gradient-free approach to directing cell migration in 3D microchannels may find significant applications in tissue scaffolds and cell on a chip devices.
Introduction
Directing the movement of mammalian cells into porous three-dimensional (3D) scaffolds has important implications for cell biology and regenerative medicine. Cells move directionally in many physiological processes, such as tissue morphogenesis, embryonic development, angiogenesis, and wound healing.1–2 For example, cells migrate preferentially along fibrils during normal tissue development by contact guidance.3 The ability to control the directional cell movement in vitro would have important implications in the bottom up assembly of tissue patterns,4 wound healing, cells on chip devices,5 and motility based cell sorting.6 Cell migration is initiated when cells are polarized by gradients of extracellular cues, e.g., chemoattractants, resulting in the redistribution of cytoskeletal components and a polarized morphology characterized by a broad front and a narrow rear.7–10 Cytoskeletal components in the front lead to membrane protrusions called lamellipodia that attach to the substrate via focal adhesions and generate traction to pull the cell forward upon the release of focal contacts at the rear of the cell.1
Guiding cell movements in vitro solely with variations in substrate properties has the advantage of not requiring a steady gradient of diffusive chemokines or external shear. Experiments have shown that cells preferentially migrate toward stiffer regions of the substrate11–14 and toward higher surface concentrations of extracellular matrix (ECM).15–18 However, due to the finite range of gradients that cells respond to, gradient-based approaches are intrinsically limited in the range over which they can guide cell migration. To overcome this, we and others have designed gradient-free arrays of closely spaced cell-adhesive micropatterns to guide the path and direction of cell movement on flat surfaces.19–22 The asymmetric shape of the teardrop islands polarizes and promotes protrusion of lamellipodia from the broad end.7, 19–20 Continuous directional migration results when the gap between islands is set close enough to allow the extended protrusions to reach a neighbouring adhesive patch, but far enough to intermittently impose the teardrop morphology (Fig. 1A). However, the isolated teardrop patterns do not translate to continuous paths for cell migration in 3D microchannels.
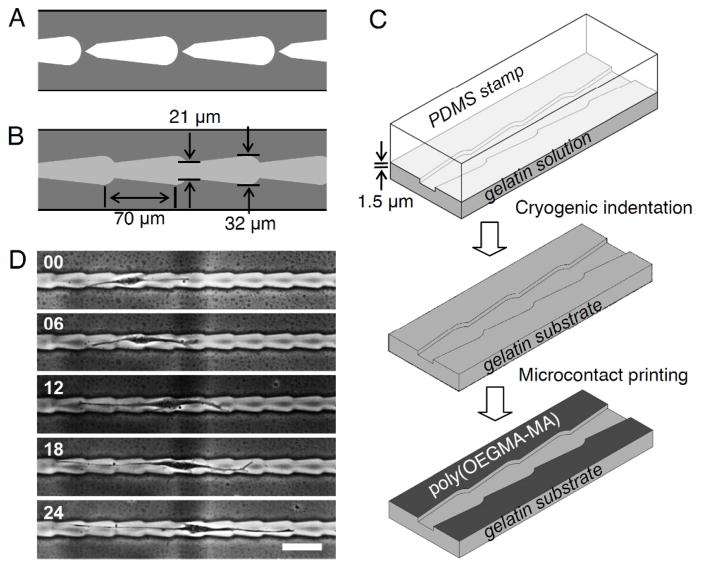
Fig. 1.
Schematic design of (A) 2D micropatterns consisting of closely spaced disjointed teardrop adhesive patterns and (B) 3D microchannel with continuous teardrop patterns. (C) Fabrication of gelatin substrates with continuous asymmetric microchannels. (D) Time-lapse images (in hours) show the random non-directional movement of NIH3T3 fibroblasts within linearly connected teardrop microchannels. Scale bars: 100 μm.
Based on insights gained from gradient-free directional cell movement on 2D micropatterned surfaces,19–22 we describe here an important advance in the design of continuous microchannel within biomaterials that can impose directional bias for a large number of individual cells with micrometer precision. The microchannels consist of repeated wider and narrower regions to initiate cell-symmetry breaking and trigger cell migration from the wider sections. We explored the feasibility of using connected teardrop microchannels (Fig. 1B and 1C) in guiding the movement of NIH3T3 fibroblasts (Fig. 1D) and observed that when the teardrops are connected, the directional bias, expressed as the ratio of right to left movements, was only 1.1 to 1. This negligibly low directionality arises from the reduced confinement in continuous microchannels wherein fibroblasts, for example, retain long tails spanning more than one segment and do not conform snugly to the shape of the individual teardrops. The key challenge is to intermittently constrain cells within individual teardrop segments within continuous microchannels. Here we report a new general design principle to overcome this challenge and enable sustained autonomous directional cell migration within 3D microchannels without chemical or mechanical gradients.
Experimental
Materials
Silicon wafers (n-type, Si:P, 76 mm) were used to prepare patterned silicon masters by standard photolithography process. Polydimethylsiloxane (PDMS, Dow Corning Corporation) was used to fabricate the replica stamp. Gelatin (G2500, porcine skin, Sigma Aldrich Inc.) was used as the culture substrate. Poly[oligo(ethyleneglycol methacrylate)-co-(methacrylic acid)] (poly(OEGMA-MA)) was synthesized by free radical polymerization (24 hours at 60°C) of methoxy polyethyleneglycol monomethacrylate (M-265, Scientific Polymer Products Inc.) and methacrylic acid (M-151, Scientific Polymer Products Inc.) and fluorescein-O-methacrylate (568864, Sigma Aldrich Inc.) with 0.1 wt% 2,2′-azobis (2-amidinopropane) dihydrochloride (992-11062, Wako Pure Chemical Industries, Ltd) as an initiator.23 Phalloidin (A122379, Alexa Fluor 488 phalloidin), DAPI (D1360, 4′,6-diamidino-2-phenylindole) and BODIPY TR ceramide (D7540, N-((4-(4,4-difluoro-5-(2-thienyl)-4-bora-3a, 4a-diaza-s-indacene-3-yl)phenoxy)acetyl)sphingosine) were obtained from Molecular Probes, Inc.
Fabrication of three-dimensional microchannels on gelatin substrates
The teardrop microchannels were designed in AutoCAD (Autodesk, Inc.) and printed on a chrome mask. The master patterns on the silicon wafer were generated by a standard photolithography process. The silicon master was coated with (tridecafluoro-1,1,2,2-tetrahydro-octyl)-1-trichlorosilane (T2492, United Chemical Technologies, Inc.) by vacuum evaporation. PDMS prepolymer (containing 10.0 wt% curing agent) was poured on the silicon master to mold the PDMS stamp as a replica and kept at 60°Cfor 2 hours. The cured PDMS stamp was detached from the silicon master and rinsed with 70% ethanol. Topographical patterns on the PDMS stamp were transferred to a gelatin by cryogenic indentation.24 Gelatin aqueous solution (15.0 w/w%) was spread on a glass slide and the PDMS stamp was applied to the gelatin. The PDMS stamp with gelatin construct was kept on a cold iron plate (−20°C) for 15 seconds before peeling the PDMS stamp. The moulded gelatin film was kept at room temperature for 20 minutes followed by crosslinking with 1.0 w/w% glutaraldehyde solution for 2 hours and blocking with 50 mM glycine solution overnight. The gelatin film was rinsed with purified water and dried at room temperature. The depth of grooves was measured using KLA-Tencor P15 Profilometer. The length and width of microchannel was obtained using a phase contrast microscope (TE2000-S, Nikon).
To suppress cell attachment outside the microchannels, we inked cell resistant poly(OEGMA-MA) using a flat PDMS stamp and brought it into contact with the plateau region of the gelatin film. Poly(OEGMA-MA) on the gelatin substrate was stabilized by aging at 60°C for a week prior to cell seeding. The printed regions remain cell resistant for more than a week in cell culture media (37°C).
Cell culture
NIH3T3 fibroblast cells and their mutants with constitutively activated Rac1, RhoA, and Cdc42 (CA-Rac1L61, CA-RhoAL63 and CA-Cdc42L61) were obtained from Yi Zheng (Cincinnati Children’s Hospital) and proliferated in our laboratory.25 Site-directed mutagenesis based on oligonucleotide mediated PCR was used to prepare the mutant cells. MCF10A human epithelial cells were purchased from American Type Culture Collection (ATCC). The fibroblasts were grown in a tissue culture flask (25 cm2) with Iscove’s Modified Dulbecco’s Medium (IMDM, GIBCO, Invitrogen Corporation) supplemented with 10.0 v/v% fetal bovine serum (FBS), 1.0 v/v% Penicillin-Streptomycin-Neomycin Antibiotic Mixture (PSN, GIBCO, Invitrogen Corporation). MCF10A cells were grown in IMDM with 5% horse serum and rhEGF (10 ng/ml). The cell culture was maintained in an incubator equilibrated with 5% CO2 at 37°C. Prior to seeding cells, gelatin films were cut into rectangular (25 × 45 mm) pieces and fixed on the tissue culture dish. The gelatin film was incubated with 1.0 v/v% PSN for 30 minutes, rinsed with phosphate buffered saline (PBS) solution, and the media. The cells were detached from the culture dish using 0.25 w/v% trypsin and 0.1 w/v% EDTA, and neutralized by IMDM solution containing 10 v/v% FBS. Cells were plated on the gelatin film at a density of 500 cells/cm2 with medium containing 10.0 v/v% FBS. The culture media was refreshed to mixture of Opti-MEM and IMDM (80:20 v/v%) with PDGF (5 ng/ml) for fibroblasts to promote cell migration 1 hour prior to tracking.26
Fluorescence microscopy
The cells were washed with PBS and fixed with 3.7% methanol-free formaldehyde (Thermo Fisher Scientific Inc.) for 10 minutes, and permeabilized with 0.2% Triton X-100 for 5 minutes. After rinsing with PBS the cells were incubated with phalloidin and DAPI to analyze F-actin filaments and nuclei. The Golgi apparatus was visualized with BODIPY TR ceramide. Photomicrographs of the stained cells were obtained using an inverted microscope (TE-2000s, Nikon) equipped with a digital CCD camera (SPOT, Diagnostic Instruments Inc.) and analyzed with Metamorph software (Universal Imaging).
Analysis of cell locomotion
Time-lapse images were captured using a Nikon TE-2000s inverted microscope with a SPOT digital CCD camera and SPOT software (Diagnostic Instruments, Inc). Cells were tracked 8 hours after seeding for 24 hours at 6 hours intervals. The directionality and speed of cell migration were analyzed from sequential time-lapse image of cells making no contacts with neighboring cells. Cells were considered to have “turned” when their nuclei and over 50% of their cell area have moved from one segment to the next segment. Net translocation is calculated by determining the net displacement toward the right as obtained by subtracting the total displacement toward the left from the total displacement toward the right for all the cells divided by the tracking time and the number of cells. More than 100 turns were analyzed for each pattern. For time-lapse video microscopy, living cells were visualized by Spot-Cam CCD camera on Nikon TE-2000 inverted microscope equipped with phase contrast optics. Temperature was controlled by an infrared heater equipped with temperature controller. Images were recorded at an interval of every minute to yield the associated locomotion.
Results and discussion
Our hypothesis is that when the teardrop segments are connected at an angle, cells occupying multiple teardrops would suffer acute bending and be coaxed to adopt the polarizing shape of individual teardrops. Just as the width of the teardrop segments, varying from 21 to 32 μm, must be adjusted according to the size of the cell nucleus (15 to 19 μm) of the NIH3T3 fibroblasts, the interconnection angle must be set judiciously. If the teardrops are too narrow at the constriction point, cells can become stuck. If the channels are too wide, cells will not adopt the shape of the microchannel resulting in no cytoskeletal alignment and morphological polarization. Likewise, if the teardrops are connected at too sharp an angle, cells may not traverse across, but if the angle is too gradual, directional bias would be lost.
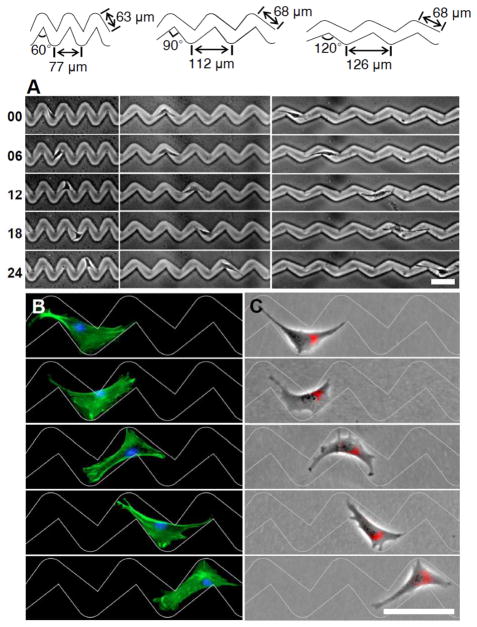
In redesigning the linear microchannels to adopt 60°, 90°, and 120° angles between teardrop segments as shown in Fig. 2A, our hypothesis was that these microchannels would prompt cells to avoid acute bending by adopting compact shapes that conform more closely to the individual teardrop segments thus promote morphological polarization and directional migration. The teardrop microchannels were fabricated in gelatin using soft lithography and cryogenic indentation techniques and the plateau regions outside the microchannels were coated with poly[oligo(ethyleneglycol methacrylate)-co-(methacrylic acid)] to limit attachment and spreading of cells exclusively within the microchannels.23–24, 27–28 Gelatin was chosen as a substrate here due to its wide application for tissue engineering and the microchannel designs presented here can be implemented on other biomaterials or cells on a chip devices.
Fig. 2.
Directional cell migration within teardrop microchannels. Time-lapse images (in hours) show the movement of (A) NIH3T3 fibroblasts. (B and C) Fluorescence images of NIH3T3 fibroblasts on the 2D teardrop micropatterns. The cells are stained to observe F-actin (green), nucleus (blue) and the Golgi apparatus (red) distribution during migration. Scale bars: 100 μm.
The cytoskeletal remodelling that occurs as fibroblasts traverse the 90° turns of connected teardrop microchannels is shown in Fig. 2B and Movie S1. Within linear channels, fibroblasts occupy multiple repeating units that reduce the efficacy of the asymmetric teardrop segments in setting the cell shape (Movie S2). In contrast, fibroblasts within the zigzag channels adapt to the shape of the teardrop segments, with their actin filaments aligned parallel to the confining boundaries and their broad ends directed towards the right. As the cells approach the 90° turns, the large lamellipodia protrusions spread to occupy the broad open area leading to a polarized triangular morphology with a wide leading edge and a narrow tail. Continued protrusion expands forwards at the junction as the cells traverse the turn. The repeating 90° turns reconfigure the cells to adopt polarized triangular morphologies that bias further translocation to the right. Lamellipodia extending in the backward direction are observed, but are consistently narrower than those at the front. Golgi apparatus typically localize toward the leading edge of the cell and in front of the nuclei of fibroblasts,29–30 although this has been shown to depend on cell type, environment, or confinement.31 Cell polarity as indicated by the relative position of the Golgi apparatus to the cell nucleus (Fig. 2C) show the consistent polarization induced by the teardrop segments on the fibroblasts as they migrate to the right with a significant right to left directional bias of 2:1 (Table 1). Measurements of cell spreading for more than 80 cells for each angle (Figure 3) show that most cells within the zigzag microchannels limit their spreading to one or two teardrop segment as cells transverse between the segments. However, within the straight 180° connected teardrop microchannels, cells span on average 3 segments, only partly filling each, and barely conforming to the polarizing shape of the individual teardrops.
Table 1.
Summary of the cell movement on microchannels with different angles
| Number of observed migration (turns) | Net translocation (μm/24 hours) | ||||
|---|---|---|---|---|---|
|
| |||||
| Cell types | Pattern | ⇨ | ⇦ | Directional ratio (right : left) | |
| NIH3T3 |
|
149 | 74 | 2.0 : 1 | 67 ± 3 |
|
|
99 | 51 | 1.9 : 1 | 100 ± 4 | |
|
|
200 | 105 | 1.9 : 1 | 139 ± 1 | |
|
|
111 | 100 | 1.1 : 1 | 39 ± 4 | |
|
| |||||
| NIH3T3-CA-RhoA |
|
98 | 59 | 1.7 : 1 | 57 ± 2 |
|
|
138 | 89 | 1.6 : 1 | 98 ± 8 | |
|
|
149 | 75 | 2.0 : 1 | 236 ± 14 | |
|
|
118 | 103 | 1.1 : 1 | 93 ± 4 | |
|
| |||||
| NIH3T3-CA-Rac1 |
|
60 | 34 | 1.8 : 1 | 48 ± 3 |
|
|
65 | 45 | 1.4 : 1 | 50 ± 2 | |
|
|
66 | 50 | 1.3 : 1 | 55 ± 9 | |
|
|
64 | 60 | 1.1 : 1 | 21 ± 5 | |
|
| |||||
| NIH3T3-CA-Cdc42 |
|
59 | 44 | 1.3 : 1 | 33 ± 3 |
|
|
90 | 58 | 1.6 : 1 | 70 ± 5 | |
|
|
104 | 58 | 1.8 : 1 | 132 ± 6 | |
|
|
71 | 67 | 1.1 : 1 | 18 ± 4 | |
|
| |||||
| MCF10A |
|
103 | 55 | 1.9 : 1 | 61 ± 1 |
|
|
95 | 73 | 1.3 : 1 | 49 ± 1 | |
|
|
91 | 60 | 1.5 : 1 | 89 ± 6 | |
|
|
74 | 67 | 1.1 : 1 | 42 ± 5 | |
Fig. 3.
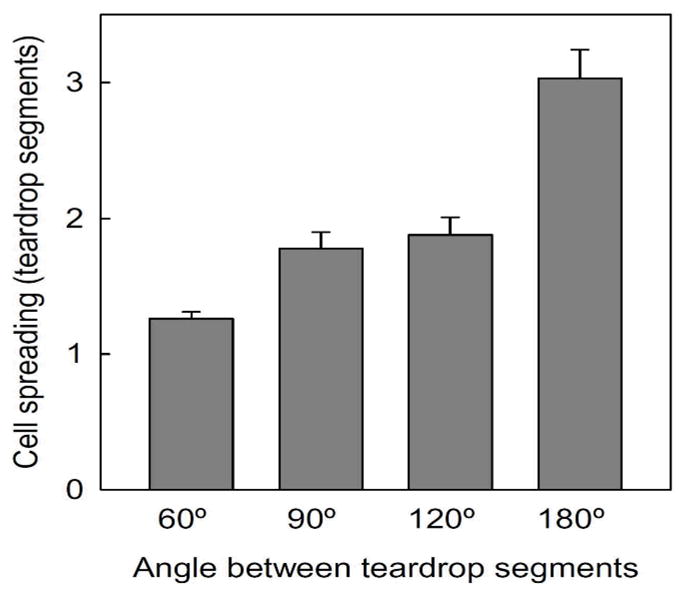
The angle between teardrop segments influences cell spreading within the microchannels. Data collected >80 cells. Error bars are mean ± S.E.
Effects of segment interconnection angle
In addition to providing directional guidance, the interconnection angle between teardrop segments can modulate the flux of the cellular traffic by the angle of their segment junctions. Varying the angle from 60 to 120° has minimal effect on the directional ratio (~2:1 right:left migration) of fibroblasts. However as described earlier, when the channels are completely straightened, i.e., 180°, the cells extend over multiple segments, are slow to release their rear, and the directional ratio drops precipitously to 1.1:1. For more gradual turns, each segment projects a longer displacement along the horizontal direction yielding greater net translocation. Although the 180° pattern has the longest displacement per segment the minimal directional bias yields the lowest net translocation. These results show that junction angles can modulate the speed of cell movement without significant altering directionality.
Effect of activated GTPases on cell locomotion
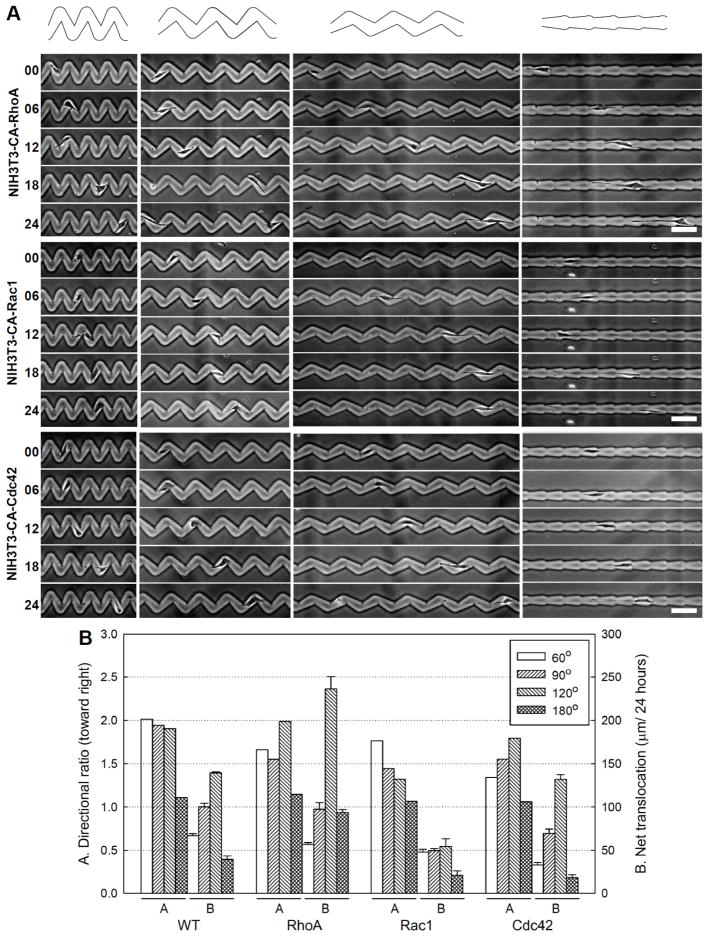
Wild type fibroblasts move along the straight 180° teardrop microchannels with long tails (Fig. 1D). The rear attachment of wild type fibroblasts do not release immediately as cells turn to the next segment. The observation of the slow retraction suggests that accelerating rear adhesion disassembly and retraction by alteration of intracellular signalling via RhoA can enhance the rate of cell movement.32–34 We examined this by investigating the migration of cells expressing constitutively activated RhoA. RhoA activation increased the net traversal rate in 120° and 180° microchannels (Fig. 4, Table 1). However, the net traversal rate is no faster on 60° and 90° microchannels suggesting that the sharp turns have caused the wild type fibroblasts to avoid acute bending of the cell body by retracting their rears as rapidly as cells expressing constitutively activated RhoA. The directional ratio for RhoA activated cells is similar or slightly reduced depending on the angle (Fig. 4B).
Fig. 4.
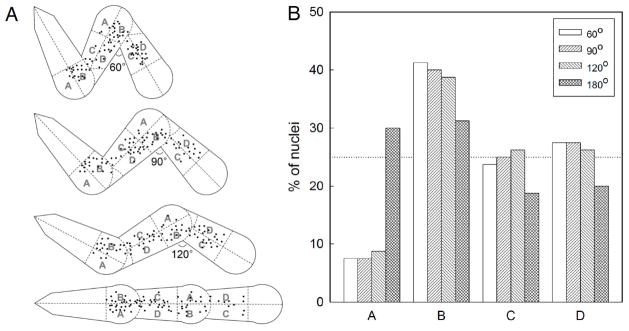
Cells move slower as their nucleus pass through the turns. (A) Centroid of cell nuclei measured at 6 hour intervals from 80 images for each pattern design. Each teardrop is divided into regions A, B, C, and D of equal area. (B) Percentage of nuclei in each region.
Global activation of intracellular signals that mediate lamellipodia extensions (Rac1) or regulate cell polarity (Cdc42) has been shown to reduce the directional persistence of migration and promote random migration.25, 33, 35 To examine whether the teardrop microchannels can direct movement of cells with intracellular signals mediating random migration, we investigated the movement of fibroblast cells expressing constitutively activated Rac1 or Cdc42 (Fig. 4A). Rac1 activation reduces the net traversal rate for all tested teardrop microchannels (Fig. 4B and Table 1). The decreased net traversal rate is likely induced by the formation of excessive peripheral lamellae and cytoskeletal reorganization in the cells expressing constitutively activated Rac1. The reduction in directional ratio demonstrates the important roles of the Rac1 and Cdc42 in directional movement. The observed preferential movement toward the right shows that these asymmetric microchannels can provide directional guidance even for cells with intracellular signalling that disrupt directional movement. These results imply that external physical confinement can overcome the loss of internally encoded persistence.
Traversal of nucleus through the microchannel
As pointed out earlier, the constriction at the turns of the microchannels must be sufficiently wide to allow the nucleus to traverse through. The turns are also the regions where the cell body experiences the highest bending stress. To examine the variation in speed as cells move along the teardrop microchannels, we plotted the centroids of the cell nuclei by spatially registering over 80 time-lapse images of cell movements for each pattern design (Fig. 5A). Each teardrop segment was divided into four equal area sections: A) outer blunt, B) inner blunt, C) inner tail, and D) outer tail. If the cells move with constant uniform speed along the microchannels, their nucleus would be equally likely to be located in each region, i.e., 25% in each region. A higher percentage indicates the cell nuclei resides in that region for a longer time thus implying a lower speed. In all microchannels, we observed a higher fraction of nuclei located in the inner blunt region (Fig. 5B) and only a small fraction in the outer blunt region. This is consistent with how the cells adapt their geometry to minimize bending while passing through the turns. On the linear (180°) channels, cell nuclei are preferentially found in the blunt sections (A+B). These results show that cells move slowest as they traverse or turn into the narrowest region of the teardrop segments.
Fig. 5.
Effects of Rho family GTPases on directional cell migration within teardrop gelatin microchannels. (A) Time-lapse images (in hours) show the movement of NIH3T3 fibroblast mutants with constitutively activated RhoA, Rac1, and Cdc42 in 60°, 90°, 120° and 180° microchannels. Scale bars: 100 μm. (B) Directional ratio and net translocation of cell migration. Graphs show the movement of wild type (WT) and NIH3T3 fibroblast mutants with constitutively activated RhoA, Rac1, and Cdc42. Data collected from >150 analyzed turns and >50 cells. Error bars are mean ± S.E.
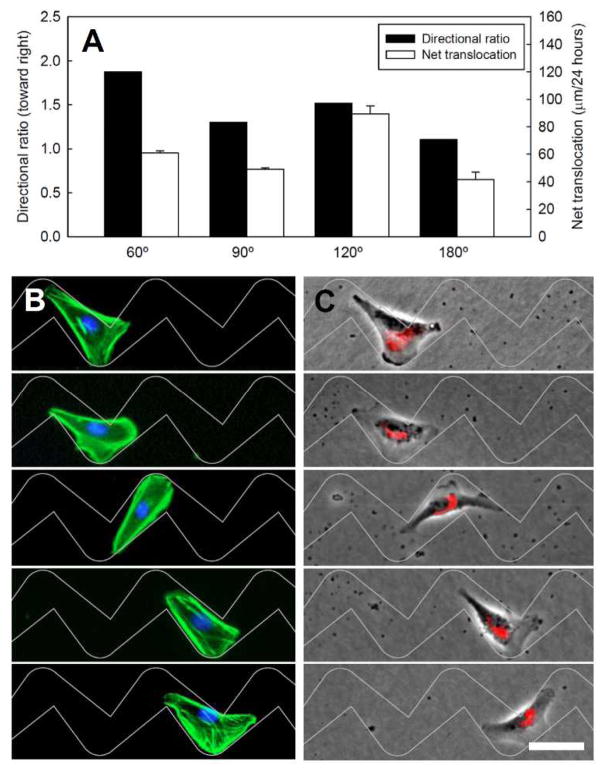
Mesenchymal and epithelial cell types have been shown to exhibit opposite directional bias on disjointed 2D micropatterns of teardrop islands.21–22 We observed that the MCF-10A epithelial cells migrate preferentially in the same direction as the fibroblasts within these microchannels (Fig. 6A, Movie S3). The actin filaments of MCF10A cells are more compact in shape and the protruding actin less pronounced compared to fibroblasts (Fig. 6B). The Golgi apparatus is consistently polarized toward the blunt end when the cells are confined in the individual teardrop segments. However, as MCF10A cells pass through the turns, the Golgi apparatus become oriented sideways unlike fibroblasts which remain polarized toward the right (Fig. 2C and 5C). This difference in polarization does not alter the directional preference in microchannels where cells are unable to move sideways. These observations highlight the difference in directional cell movement in disjointed 2D micropatterns where epithelial cells and fibroblasts exhibit opposite directional ratio.21–22 Within 3D microchannels as shown here, both cell types favour expanding channels and move toward the right from their blunt end.
Fig. 6.
(A) Directional ratio and net translocation of MCF10A cells for varying interconnections angles. (B and C) Fluorescence images of MCF10A epithelial cells on the 2D teardrop micropatterns that have 90° angle. The cells are stained to observe F-actin (green), nucleus (blue) and the Golgi apparatus (red) distribution during migration. Scale bar: 50 μm.
Uniform width linear microfluid-based chambers with or without chemoattractant gradient have been applied to mimic the 3D physical confinment experienced by dendritic cells or tumor cells in restrictive 3D space.36–37 These studies demonstrated that physical confinment can influence tumor cell migration and invasiveness. Persistent migration was observed for several hours within uniform width linear confinement only for cancer cells and dendritic cells.36–38 Cancer cell migration within uniform width linear confinement is independent of actin polymerization, myosin mediated contraction, and integrin based adhesion; while interfering with microtubule polymerization significantly reduces the directional persistance.36–38 The approach developed here for directing cell migration in 3D microchannels guide cells in a different mechanism and can be applied to different types of cells. Here, the intermittent morphological polarization achieved by limiting cell spreading within one or two asymmetric segments drives gradient-free net translocation persistently over durations limited only by cell viability, and along directions preset by the microchannel design.
Conclusions
The continuous microchannels described here provide a gradient-free approach to direct cell movement. Cell morphology within connected teardrop microchannels can be coaxed to the shape of individual teardrop segments by connecting the segments in a zigzag manner to minimize the likelyhood of cells extending across multiple segments. The intermittent polarization resulting from the teardrop cell morphology results in continuous directional cell migration without the need for chemical gradients. This approach is a significant advance from previous micropatterns that are limited to two-dimensional surface patches.21–22, 39–40
Cellular traffic within the zigzag microchannels can be tuned by changes in pattern design with minimal impact on directionality. The universality of this approach is demonstrated by the directional migration of NIH3T3 fibroblasts, epithelial cells, and even mutant fibroblasts whose intercellular signalling (Rac1 and Cdc42) have been purposely altered to promote random migration. This material based guidance is a powerful approach that may be incorporated in the design of porous scaffolds and biomedical devices using new advances in 3D prototyping technologies.41–42
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (R01EB010043) and the National Science Foundation (CBET0928219). We thank Ross Andrews for synthesizing the cell resistant poly(OEGMA-MA), Dr. Girish Kumar for helpful discussions, and Rebecca Moore for performing some preliminary studies.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Notes and references
- 1.Lauffenburger DA, Horwitz AF. Cells. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Nakatsuji N, Johnson KE. Nature. 1984;307:453–455. doi: 10.1038/307453a0. [DOI] [PubMed] [Google Scholar]
- 4.Mun K-S, Kumar G, Co CC, Ho C-C. Adv Healthcare Mater. 2012 doi: 10.1002/adhm.201200141. [DOI] [Google Scholar]
- 5.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 6.Co CC, Ho CC, Kumar G. Anal Chem. 2012;84:10160–10164. doi: 10.1021/ac302855m. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Dekker LV, Segal AW. Science. 2000;287:983. doi: 10.1126/science.287.5455.982. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. PNAS. 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin T, Zhang N, Long Y, Parent CA, Devreotes PN. Science. 2000;287:1034–1036. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- 10.Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey MT, Wang Y-l. Soft Matter. 2009;5:1918–1924. doi: 10.1039/b818104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni Y, Chiang MYM. Soft Matter. 2007;3:1285–1292. doi: 10.1039/b703376a. [DOI] [PubMed] [Google Scholar]
- 13.Saez A, Ghibaudo M, Buguin A, Silberzan P, Ladoux B. PNAS. 2007;104:8281–8286. doi: 10.1073/pnas.0702259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sochol RD, Higa AT, Janairo RRR, Li S, Lin L. Soft Matter. 2011;7:4606–4609. [Google Scholar]
- 15.Lin X, Helmke BP. Biophys J. 2008;95:3066–3078. doi: 10.1529/biophysj.107.127761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Ratner BD, Sage EH, Jiang S. Langmuir. 2007;23:11168–11173. doi: 10.1021/la701435x. [DOI] [PubMed] [Google Scholar]
- 17.Lo C-M, Wang H-B, Dembo M, Wang Y-l. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhoads DS, Guan JL. Exp Cell Res. 2007;313:3859–3867. doi: 10.1016/j.yexcr.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandere-Grzybowska K, Soh S, Mahmud G, Komarova Y, Pilans D, Grzybowski BA. Soft Matter. 2010;6:3257–3268. doi: 10.1039/B922647H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar G, Co CC, Ho CC. Langmuir. 2011;27:3803–3807. doi: 10.1021/la2000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar G, Ho CC, Co CC. Adv Mater. 2007;19:1084–1090. [Google Scholar]
- 22.Kushiro K, Chang S, Asthagiri AR. Adv Mater. 2010;22:4516–4519. doi: 10.1002/adma.201001619. [DOI] [PubMed] [Google Scholar]
- 23.Wang YC, Ho CC. FASEB J. 2004;18:525–527. doi: 10.1096/fj.03-0490fje. [DOI] [PubMed] [Google Scholar]
- 24.Ko YG, Co CC, Ho CC. Biomaterials. 2013;34:353–360. doi: 10.1016/j.biomaterials.2012.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R, Debreceni B, Jia B, Gao Y, Tigyi G, Zheng Y. J Biol Chem. 1999;274:29648–29654. doi: 10.1074/jbc.274.42.29648. [DOI] [PubMed] [Google Scholar]
- 26.De Donatis A, Comito G, Buricchi F, Vinci MC, Parenti A, Caselli A, Camici G, Manao G, Ramponi G, Cirri P. J Biol Chem. 2008;283:19948–19956. doi: 10.1074/jbc.M709428200. [DOI] [PubMed] [Google Scholar]
- 27.Kawai K, Suzuki S, Tabata Y, Ikada Y, Nishimura Y. Biomaterials. 2000;21:489–499. doi: 10.1016/s0142-9612(99)00207-0. [DOI] [PubMed] [Google Scholar]
- 28.Kumar G, Wang YC, Co C, Ho CC. Langmuir. 2003;19:10550–10556. [Google Scholar]
- 29.Magdalena J, Millard TH, Machesky LM. J Cell Sci. 2003;116:743–756. doi: 10.1242/jcs.00288. [DOI] [PubMed] [Google Scholar]
- 30.Nabi IR. J Cell Sci. 1999;112:1803–1811. doi: 10.1242/jcs.112.12.1803. [DOI] [PubMed] [Google Scholar]
- 31.Pouthas F, Girard P, Lecaudey V, Ly TBN, Gilmour D, Boulin C, Pepperkok R, Reynaud EG. J Cell Sci. 2008;121:2406–2414. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- 32.Cory GOC, Ridley AJ. Nature. 2002;418:732–733. doi: 10.1038/418732a. [DOI] [PubMed] [Google Scholar]
- 33.Etienne-Manneville S, Hall A. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 34.Nobes CD, Hall A. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irimia D, Toner M. Integr Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faure-Andre G, Vargas P, Yuseff MI, Heuze M, Diaz J, Lankar D, Steri V, Manry J, Hugues S, Vascotto F, Boulanger J, Raposo G, Bono MR, Rosemblatt M, Piel M, Lennon-Dumenil AM. Science. 2008;322:1705–1710. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- 39.Mahmud G, Campbell CJ, Bishop KJM, Komarova YA, Chaga O, Soh S, Huda S, Kandere-Grzybowska K, Grzybowski BA. Nat Phys. 2009;5:606–612. [Google Scholar]
- 40.Voldman J. Nat Phys. 2009;5:536–537. [Google Scholar]
- 41.Cicha K, Li Z, Stadlmann K, Ovsianikov A, Markut-Kohl R, Liska R, Stampfl J Appl Phys. 2011;110 064911-064911-604911-064915. [Google Scholar]
- 42.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.