Abstract
Objectives
The A148T polymorphism of CDKN2A gene is observed in various neoplasms with the incidence rate of 3-35%, however, rather little is known either about the frequency of its occurrence or of its significance in urinary bladder carcinoma.
Materials and methods
DNA was isolated from blood of 156 patients with urinary bladder carcinoma (130 men). In histopathology, 84 cases were classified as G1, 42 as G2, and 30 as G3. The clinical stage was in 81 cases estimated at Ta and in 75 cases at T1-T4. A148T polymorphism was detected by the MSSCP technique and by sequencing.
Results
A148T polymorphism was identified in 9/156 urinary bladder carcinoma cases (only in men). The obtained results were compared with the polymorphism incidence for the Polish population, estimated by Debniak et al. The occurrence in the group of the bladder cancer patients turned out higher (5.77%) from that in the control group (2.89%) (G test, table 2×2: NBLADDER CANCER = 156, NCONTROL = 1210, G = 4.298, p <0.05).
Conclusion
Summing up and taking into account the analysis of clinical parameters and the age of the disease occurrence, the A148T polymorphism of CDKN2A gene was identified in the study group only in men, in whom the disease was diagnosed above the age of 60, while the diagnosed neoplasms were in the majority of cases characterized by higher clinical stages and higher grades of malignancy. This has been the first study that attempted to show a potential association between A148T alterations and an increased risk for bladder cancer development.
Keywords: bladder cancer; CDKN2A, A148T polymorphism
INTRODUCTION
All genetic variations originate from mutations, which are defined as changes in DNA sequence. Mutations can affect either germline cells (the cells which produce gametes) or somatic cells (all the cells other than germline cells). In human genetics, the term “mutation” has often been reserved for DNA sequence changes that cause genetic diseases and are consequently relatively rare. DNA sequence variants, which are more common in populations (i.e., such sequence variants in which two or more alleles at locus demonstrate frequencies >1%), are said to be polymorphic (“having many forms”) [1]. Such loci (plural of locus) are termed polymorphisms, although nowadays, alleles with frequency <1% are often called polymorphisms as well.
Urinary bladder cancer is a common malignant disease in Poland [2]. Many genetic and epigenetic alterations are supposed to contribute directly or indirectly to bladder tumor induction [3, 4]. Attempts are undertaken to classify tumors with regards to the molecular characteristics of changes in neoplastic cells in order to be able to identify the relationship between the type of these changes and clinical characteristics. Every type of cancer has biological subsets that differ in clinical behavior and response to treatment. Personalized cancer medicine is defined as treatment based on the molecular characteristics of a tumor from individual patient. Nowadays this branch of medicine has great potential in the therapy of many types of cancer. Somatic mutations of P53, Rb, FGFR3, CDKN2A, and Ras genes belong to the group of mutations that most frequently accompany the occurrence of urinary bladder carcinoma [3, 4, 5]. Some of the constitutive changes of the genes (germline), e.g., A72P polymorphism of P53 gene, were also analyzed with regards to the higher risk of neoplasm development [6]. There are also reports, indicating correlations between constitutive mutations of CDKN2A gene and breast cancer, melanoma cancer, nervous system tumours and squamous cell carcinoma of the head/neck region [7, 8, 9], but rather little is known either about the frequency of its occurrence or of its significance in urinary bladder carcinoma. Most frequently described polymorphisms of CDKN2A gene are the substitution of alanine to threonine in the second exon (A148T- rs 3731249) and the two, which occur in the 3′ untranslated region of CDKN2A: NT 500c >g and Nt540c >t [7].
The CDKN2A gene (OMIM 600160), localized at chromosome 9p21 encodes p16INK4a cyclin-dependent kinase inhibitor and p14AR activator of p53 [3]. Both gene products have an independent first exon (1-alpha exon and 1-beta exon, respectively), but share exons 2 and 3 and are translated in different reading frames. The genes are involved in the negative control of cell proliferation; p16INK4a produces a G1 cell-cycle arrest by inhibiting the phosphorylation process of retinoblastoma protein (RB), and p14ARF acts both at G1/S phases in a p53-dependent manner via binding and inhibiting MDM2 protein. There are three major mechanisms at the base of genetic inactivation of CDKN2A gene, including deletions of both alleles, deletion of one allele and mutation of the other allele or deletion or mutation of the first allele and hypermethylation of the second allele. Unlike other suppressor genes, which most often undergo inactivation in result of point mutations, deletions are the most frequent mechanisms by which CDKN2A gene is inactivated. Regarding urinary bladder carcinoma, homozygotic deletions at 9p21 locus (CDKN2A) are found in approximately 20-30% of cases, while the loss of heterozygosity may be observed even in 60% of cases [3, 8].
The aim of our study was to characterize A148T polymorphism of CDKN2A gene in bladder cancer patients.
MATERIAL AND METHODS
DNA from 84 subjects was analyzed, all of them patients of the Department of Urology, The J.P. II Regional Hospital in Bełchatów, and from 72 patients of the “Urologists M. Rozniecki and Partners” Department of Urology in Łask with diagnosed tumor of the urinary bladder (130 men and 26 women, 32-86 year old, the mean age: 62.3 years).
The patients were informed about the study goal and provided written consents to participate. A detailed medical history was obtained from all the enrolled participants, taking into account the effect of risk factors on neoplastic disease progression. All the identified neoplasms were assessed by two histopathologists for staging and grading according to WHO Histopathological Classification [10]. The mean observation period per individual patient was 32 months (16-59). The number of recurrence cases amounted to 59/156 (38%).
See Tables 1 and 2 for data of the analyzed material.
Table 1.
Clinical data of the bladder cancer group (the dark blue color designates the cases in which A148T polymorphism of CDKN2A gene was found)
| No. | DNA | Gender/Age | Stage/Grade | Recurrence (time after diagnosis in months) | No. | DNA | Gender/ Age | Stage/ Grade | Recurrence |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 151 | M/44 | Ta/G1 | Yes/18 | 81 | 141 | M/56 | T1/G2 | No |
| 2 | 166 | M/66 | Ta/G1 | Yes/5,12 | 82 | 153 | M/43 | /T1G2 | No |
| 3 | 168 | M/51 | Ta/G1 | No | 83 | 154 | M/81 | T1/G2 | Yes |
| 4 | 169 | M/47 | Ta/G1 | Yes/8,18 | 84 | 162 | M/73 | T1/G2 | Yes |
| 5 | 171 | M/65 | Ta/G1 | No | 85 | L1 | M/62 | T1/G3 | No |
| 6 | 175 | M/32 | Ta/G1 | No | 86 | L2 | M/82 | Ta/G2 | No |
| 7 | 178 | M/66 | Ta/G1 | No | 87 | L3 | M/57 | T1/G2 | Yes/1 |
| 8 | 182 | M/84 | Ta/G1 | No | 88 | L4 | M/53 | Ta/G1 | Yes/1 |
| 9 | 187 | M/82 | Ta/G1 | No | 89 | L5 | M/77 | T1/G2 | Yes/3 |
| 10 | 188 | M/54 | Ta/G1 | Yes/16 | 90 | L6 | M/62 | Ta/G2 | Yes/6 |
| 11 | 189 | M/63 | Ta/G1 | Yes/17,19 | 91 | L7 | F/59 | Ta/G1 | No |
| 12 | 190 | M/70 | Ta/G1 | Yes/11 | 92 | L8 | M/73 | T3/G2 | No |
| 13 | 192 | M/67 | Ta/G1 | No | 93 | L9 | M/78 | Ta/G1 | No |
| 14 | 193 | M/83 | Ta/G1 | No/D | 94 | L10 | F/68 | T1/G2 | Yes/1 |
| 15 | 195 | M/60 | Ta/G1 | Yes/9,17 | 95 | L11 | M/61 | T1/G2 | No |
| 16 | 196 | M/77 | Ta/G1 | No | 96 | L12 | M/60 | Ta/G1 | No |
| 17 | 197 | M/76 | Ta/G1 | No | 97 | L13 | M/41 | Ta/G1 | Yes/4 |
| 18 | 198 | M/53 | Ta/G1 | Yes/3 | 98 | L14 | M/59 | Ta/G1 | No |
| 19 | 203 | M/61 | Ta/G1 | No | 99 | L15 | M/77 | Ta/G2 | No |
| 20 | 204 | M/75 | Ta/G1 | Yes/15 | 100 | L16 | F/82 | Ta/G1 | No |
| 21 | 206 | M/71 | Ta/G1 | No | 101 | L17 | M/85 | Ta/G3 | No |
| 22 | 209 | M/58 | Ta/G1 | No | 102 | L18 | M/68 | T1/G3 | Yes/3 |
| 23 | 210 | M/58 | Ta/G1 | Yes/4 | 103 | L19 | M/57 | Ta/G1 | Yes/7 |
| 24 | 211 | M/54 | Ta/G1 | No | 104 | L20 | F/61 | T1/G1 | No |
| 25 | 219 | M/55 | Ta/G1 | No | 105 | L21 | F/83 | Ta/G1 | No |
| 26 | 220 | M/67 | Ta/G1 | No | 106 | L22 | M/62 | T2/G2 | Yes/3 |
| 27 | 221 | F/53 | Ta/G1 | No | 107 | L23 | M/41 | Ta/G1 | Yes |
| 28 | 226 | M/52 | Ta/G1 | No | 108 | L24 | M/63 | Ta/G2 | Yes/3 |
| 29 | 228 | M/67 | Ta/G1 | No | 109 | L25 | M/67 | Ta/G3 | Yes |
| 30 | 234 | M/67 | Ta/G1 | No | 110 | L26 | F/57 | Ta/G1 | No |
| 31 | 238 | M/73 | Ta/G1 | No | 111 | L27 | M/53 | T1/G2 | No |
| 32 | 235 | M/52 | Ta/G1 | No | 112 | L28 | M/73 | T1/G3 | Yes |
| 33 | 240 | M/83 | Ta/G1 | No | 113 | L29 | F/54 | T1/G2 | No |
| 34 | 241 | M/78 | Ta/G1 | Yes/12 | 114 | L30 | F/50 | Ta/G1 | No |
| 35 | 242 | M/61 | Ta/G1 | No | 115 | L31 | M/61 | Ta/G1 | Yes/2 |
| 36 | 247 | M/59 | Ta/G1 | Yes/6 | 116 | L32 | F/44 | Ta/G1 | No |
| 37 | 249 | M/61 | Ta/G1 | No | 117 | L33 | M/81 | Ta/G1 | No |
| 38 | 251 | F/73 | Ta/G1 | Yes/3,9 | 118 | L34 | M/66 | Ta/G2 | Yes/6 |
| 39 | 255 | M/56 | Ta/G1 | No | 119 | L35 | M/67 | Ta/G1 | Yes/24 |
| 40 | 165 | M/55 | Ta/G2 | Yes/5,9,14/D | 120 | L36 | M/52 | T1/G2 | Yes/9 |
| 41 | 170 | M/74 | Ta/G2 | No | 121 | L37 | M/79 | Ta/G1 | No |
| 42 | 184 | M/60 | Ta/G2 | Yes/1 | 122 | L38 | M/62 | Ta/G1 | Yes/3 |
| 43 | 232 | M/73 | Ta/G2 | No | 123 | L39 | M/67 | T1/G3 | Yes/12 |
| 44 | 245 | M/73 | Ta/G2 | Yes/5 | 124 | L40 | M/70 | Ta/G1 | Yes/3 |
| 45 | 212 | M/80 | T1/G1 | No | 125 | L41 | M/74 | Ta/G1 | Yes/1 |
| 46 | 214 | F/45 | T1/G1 | No | 126 | L42 | M/58 | Ta/G1 | Yes/3 |
| 47 | 222 | M/71 | T1/G1 | No/Dacc | 127 | L43 | F/69 | Ta/G1 | Yes/2 |
| 48 | 223 | M/70 | T1/G1 | No/Dint | 128 | L44 | M/68 | Ta/G1 | Yes/2 |
| 49 | 244 | M/77 | T1/G1 | No | 129 | L45 | M/62 | Ta/G1 | Yes/3 |
| 50 | 248 | M/62 | T1/G1 | No/Dint | 130 | L46 | M/72 | Ta/G1 | Yes/3 |
| 51 | 173 | M/86 | T1/G2 | No | 131 | L47 | M/54 | Ta/G2 | Yes/3 |
| 52 | 179 | M/84 | T1/G2 | Yes/20 | 132 | L48 | F/62 | T1/G1 | No |
| 53 | 202 | M/71 | T1/G2 | Yes | 133 | L49 | F/60 | Ta/G1 | Yes/3 |
| 54 | 218 | M/72 | T1/G2 | Yes/4/D | 134 | L51 | F/58 | Ta/G1 | No |
| 55 | 227 | M/80 | T1/G2 | No/D | 135 | L52 | F/78 | Ta/G1 | Yes/1 |
| 56 | 231 | M/57 | T1/G2 | No | 136 | L53 | F/77 | T1/G3 | No |
| 62 | 237 | M/60 | T1/G2 | Yes/3/D | 137 | L54 | M/67 | T1/G3 | Yes/3 |
| 57 | 252 | M/83 | T1/G2 | No | 138 | L56 | F/75 | T1/G1 | No |
| 58 | 205 | M/53 | T1/G3 | No | 139 | L57 | M/58 | T1/G1 | No |
| 59 | 215 | M/84 | T1/G3 | Yes/6/D | 140 | L58 | M/64 | T1/G1 | No |
| 60 | 243 | M/72 | T1/G3 | No/D | 141 | P2 | M/66 | Ta/G2 | No |
| 61 | 256 | M/68 | T1/G3 | No | 142 | P3 | F/76 | Tis/G3 | No |
| 62 | 217 | M/83 | T2/G1 | Yes/1/D | 143 | P4 | M/75 | T1/G1 | Yes |
| 63 | 185 | K/76 | T2/G2 | No/D | 144 | P5 | F/57 | T1/G2 | Yes |
| 64 | 191 | M/82 | T2/G2 | No/D | 145 | P6 | M/72 | T2/G2 | No |
| 65 | 199 | M/82 | T2/G2 | No/D | 146 | P7 | M/64 | T2/G3 | No |
| 66 | 200 | M/55 | T2/G2 | No/D | 147 | P8 | M/72 | T2/G3 | No |
| 67 | 208a | F/78 | T2/G2 | No/D | 148 | P9 | M/76 | T1/G1 | No |
| 68 | 177 | M/65 | T2/G3 | No/D | 149 | P11 | M/49 | T2/G3 | No |
| 69 | 180 | M/76 | T2/G3 | No/D | 150 | P13 | F/45 | T1/G1 | No |
| 70 | 183 | F/61 | T2/G3 | Yes/12 | 151 | P20 | M/54 | T1/G1 | Yes |
| 71 | 201 | M/74 | T2/G3 | No/D | 152 | P23 | M/66 | T2/G3 | Yes |
| 72 | 186 | M/67 | T2/G3 | No | 153 | P24 | M/76 | T1/G1 | Yes |
| 73 | 213 | M/77 | T2/G3 | No | 154 | P25 | M/72 | T2/G2 | Yes |
| 74 | 230 | F/88 | T2/G3 | No/D | 155 | P26 | M/65 | T2/G3 | Yes |
| 75 | 239 | F/62 | T2/G3 | No | 156 | P28 | F/61 | T2/G2 | Yes |
| 76 | 253 | M/73 | T2/G3 | No | |||||
| 77 | 254 | M/72 | T2/G3 | No | |||||
| 78 | 208 | F/75 | T3/G3 | No/D | |||||
| 80 | 207 | M/81 | T4/G3 | No |
D - death, Dacc - death in accident, Dint - death of intrinsic cause
Table 2.
Division of studied material with regards to tumor staging and grading
| Tumor stage | Tumor grade | ||||
|---|---|---|---|---|---|
| Primary tumors | Recurrent tumors | Primary tumors | Recurrent tumors | ||
| Tis | 1 | ||||
| Ta | 53 | 28 | G1 | 61 | 23 |
| T1 | 33 | 9 | G2 | 28 | 14 |
| T2 | 18 | 7 | G3 | 23 | 7 |
| T3-T4 | 7 | ||||
| Total | 112 | 44 | 112 | 44 | |
DNA isolation
Blood samples (5 ml), collected onto EDTA3K (potassium versenate), were used as study material. A Roche MagNA Pure Compact automatic workstation was used to isolate DNA from 1 ml of anticoagulated blood (out of all samples) by means of a Nucleic Acid Isolation Kit I-Large Volume (cat. No. 03 730 972 001, Roche Diagnostic GmbH). The volume and purity of obtained DNA were determined by absorbance (A) measurements directly in aqueous solutions. The measurements were performed by an ND-1000 spectrophotometer in UV light (measurement of UV adsorbed by DNA bases). Nucleic acid concentration was determined by comparing measurement results for blind trial (distilled water) and absorbance levels of studied sample at wavelength of 260 nm (this wave length corresponds to the maximal absorption for nucleic acids). Purity was determined by calculation of A280/A260 coefficient. Absorbance at λ = 280 nm is characteristic for proteins and reflects protein- derived contaminations. The coefficient values above 1.7 were regarded satisfactory.
PCR conditions
PCR amplifications were performed as follows: 1 μl of template DNA (approx. 100 ng), 2.5 μl of 10 × reaction buffer (700 mM Tris-HCl, pH 8.3, 166 mM (NH4)2SO4, 25 mM MgCl2), 0.5 μl of 2,5 mM dNTPs (TaKaRa), 1 μl of each primer pair (5 pmol/ μl), 1.25 U of Taq DNA polymerase (Novazym), and 18.75 μl of distilled water in a total volume of 25 μl were used. The used primers included: 5′CCCAAGCTTGCATGGAGCCGGCGGCG3′ and 5′CGGGATCCCTTTCAATCGGGGATGT3′. The applied PCR procedure: preliminary denaturation for 10 minutes at 95°C, followed by 40 cycles of 30-second denaturation in 95°C, 30-second annealing in 58°C, and 30-second elongation in 72°C. The final elongation lasted 5 minutes at 72°C. All the reactions were referred to non-template controls. PCR reactions were carried out on BioRad iQ5 and LabCycler SensoQuest thermocyclers. PCR products of leukocytes (10 μl) were electrophoresed on 2.0% agarose gel and stained with ethidium bromide. The gels were evaluated by visual inspection in UV light.
MSSCP (Multitemperature Single Strand Conformation Polymorphism) conditions
The search for A148T polymorphism in exon 2 of CDKN2A gene was carried out by the MSSCP technique in two different temperature profiles (the first one at 23°C, for 120 minutes and the second one at 37°C for 40 minutes, 23°C for 25 minutes, and at 15°C for 25 minutes) and by the sequencing technique. Details of the MSSCP method were described in earlier reports [11]. PCR products were sequenced to rule out / confirm mutation or polymorphism, identified during electrophoresis (Genomed sp. z o.o.).
Statistics
The G test, based on the likelihood ratio, was applied. Like the chi-square (χ2) test, G test is used to assess the significance of differences in contingency tables. Both tests lead to the same conclusions when each cell of the contingency table has a sufficiently high number (O >10) of observations. However, while the chi-square test cannot be used for lower numbers in any cell, no such restrictions occur for G test. A null hypothesis of no effect cannot be rejected when G statistics is higher than the critical value G0,05 = 3.841 for 1 degree of freedom (table 2×2) [12].
RESULTS
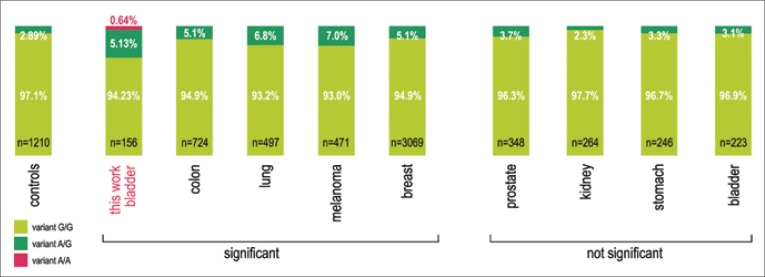
Common polymorphic variants of CDKN2A gene were found in our study at codon 148 in exon 2 (Ala148Thr) in 9 cases (Tab. 3, Fig. 1). All the confirmed cases were first identified by the MSSCP technique and then verified by sequencing. A full conformability of results was attained in both techniques. The obtained frequencies were compared with the control group from the Polish population, examined by Debniak and colleagues, and a significant difference was found in the prevalence of A148T polymorphism in the bladder cancer group (G test, Table 2 ×2: NBLADDER CANCER = 156, NCONTROL = 1210, G = 4.298, p <0.05) [13]. No differences were observed in the frequencies of Nt500c >g alleles or in the frequencies of Nt540c >t alleles, either in the bladder cancer group or in the reference control group. We used the data for the control group, published by other authors, because they were obtained in studies on a large number of subjects (i.e., 1210) for the Polish population, and by researchers, who are unquestionable authorities in the studies on hereditary predispositions to neoplasm development, including the A148T variant of CDKN2A gene.
Table 3.
Frequencies of CDKN2A variants in controls and in the bladder cancer group
| CDKN2A | Debniak et.al (n=1210) controls | This study (n=156) | G test (table 2×2) |
|---|---|---|---|
| A148T | A/A 0 (0%) A/G 35 (2.89%) G/G 1175 (97.1%) Allele A frequency 1.5% | A/A 1 (0.64%) A/G 8 (5.13%) G/G 147 (94.23%) Allele A frequency 3.2% | G= 4.298 p<0.05 |
Fig. 1.
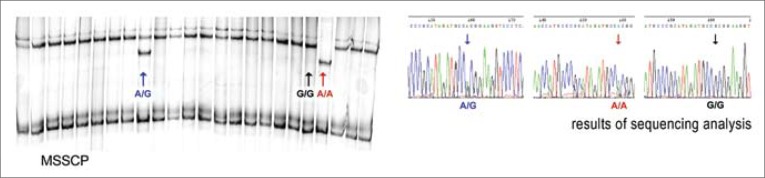
Results of electrophoresis (MSSCP) of CDKN2A gene exon 2 and of sequentioning of particular codon 148 variants. The black arrow designates the G/G (Ala/Ala) homozygotic system, the blue arrow – the heterozygotic A/G (Thr/Ala) system and the red arrow – the homozygotic A/A (Thr/Thr) system.
Fig. 2.
The incidence rates of particular variants in codon 148 of CDKN2A gene in various neoplasms, found in the Polish population. The occurrence of adenine instead of guanine in codon 148 is associated with the change of the coded amino acid - alanine (CGG) - to threonine (CGA).
n- the number of studied subjects controls - a control group for the Polish population, consisting of 500 newborns and 710 adult; „significant” - designates a group of neoplasms, in which the percent of the threonine-encoding variant was statistically significantly higher than in the control population; „not significant” - designates a group of neoplasms in which the distribution of incidence of the threonine-encoding variant is not different from that in the general population.
Based on the study by Dębniak et al. 2006 [31].
DISCUSSION
CDKN2A gene inactivation is one of the genetic events, leading to urinary bladder cancer development [3]. According to published reports, 14-39% of homozygotic deletions are found in urinary bladder cancer, as well as approximately 12% of hemizygotic deletions of the gene and about 3% of mutations [5, 14]. For comparison, in renal cancer homozygotic deletions are observed even in 56%, while hypermethylation in 23% of the diagnosed cases [15]. However, because CDKN2A gene inactivation plays the key role in the molecular mechanism of urinary bladder cancer development, as designed by Goebell et al. in 2010, our interest focused on the constitutional mutations of that gene.
The A148T variant is broadly cited as a common polymorphism, following functional studies, which indicate that these amino acid substitutions do not affect the ability of P16 to precipitate with CDK4/ CDK6 proteins [13]. Variable frequencies of CDKN2A germline muta tions and polymorphism have been reported in different collections of melanoma families in Sweden, Poland, Great Britain, and Brazil [16–20]. The incidence rates of these changes are also higher in cases of liver carcinoma, nervous system tumors, and pancreatic or breast cancers. See Table 5 for exemplary data.
Table 5.
Genotypes and allele frequencies of the A148T allele in different populations
| Geographic origin | Study population | Genotype | Frequency of A148T gene CDKN2A polymorphism | Reference | |
|---|---|---|---|---|---|
| 1. | North-Western Italy | Melanoma families n = 14 | – | 5/14 (35.7%) | Chiorzo et al. (1999) [22] |
| 2. | French and Italian population | Italian melanoma patients n = 119 Italian controls n = 121 French melanoma patients n = 500 French controls n = 143 |
A/A G/A G/G A/A G/A G/G A/A G/A G/G A/A G/A G/G |
6.72% 1 (0.84%) 14 (11.76%) 104 (87.40%) 6.2% 1 (0.83%) 13(10.74%) 107 (88.43%) 3.4% 1 (0.2%) 32 (6.4%) 467 (93.4%) 4.2% 0 (0.0%) 12 (8.4%) 131 (91.6%) |
Spica et al. (2006) [24] |
| 3. | UK | Atypical nevus/Familial melanoma n = 488 Population-based sample n = 599 |
A/A G/A G/G A/A G/A G/G |
0/488 (0.0%) 24/488 (4.9%) 464/488 (93.0%) 0/599 (0.0%) 31/599 (5.2%) 568/599 (94.8%) |
Bertram et al. (2002) [23] |
| 4. | Sweden | Hereditary melanoma n = 108 | – | 9.3% in patients sample | Erlandson et al. (2007) [25] |
| 5. | Spain | Patients with more than one primary cutaneous melanoma n = 104 Spanish controls n = 110 |
– | 13.40% 5.45% |
Puig et al. (2005) [26] |
| 6. | Spain | Breast cancer and cutaneous melanoma n = 31 | – | 2/31 (6.4%) | Nagore et al. (2009) [27] |
| 7. | Poland | Larynx cancer n = 390 | – | 17/390 (4.4%) | Kiwerska et al. (2007) [28] |
| 8. | Sweden (Stockholm County) | Bladder cancer n = 167 | – | 10/167 (6%) | Sakano et al. (2003) [29] |
In Poland, the incidence of CDKN2A variant - an alanine to threonine substitution at codon 148 (A148T) - has been estimated for approximately 3 to 3.5% of the population [13, 21]. Recent studies have shown that A148T polymorphism is in linkage disequilibrium with a second alteration in the CDKN2A promoter (P-493 variant), which appears to affect gene expression [13]. In the Polish control population, the frequency of A148T changes in 1,210 control samples (500 newborns and 710 adults) was assessed by Debniak et al. at 2.89%. The same research team studied the incidence of A148T variant of CDKN2A gene (see their report of 2007) in the Polish population of patients with breast cancer (young women), colon, lung, melanoma, prostate, bladder, kidney, stomach, pancreas and thyroid cancers (see Figure 1 for a partial presentation of results) [13, 30, 31]. Significant differences were found for the first four neoplasms in the above list, indicating that the carrier state of the variant may be associated with an increased risk of tumor development. The results, obtained for a group of 223 urinary bladder carcinoma cases (3.1%) did not differ from those for the control group [13]. It should be emphasized that those patients came from the Pomeranian Region while in our study patients from the Lodz Voivodeship (central Poland) prevailed (131/156). The results of our study (5.77%) indicate that, regarding the Polish population (Łódź Macroregion), A148T variant seems to be associated with an increased bladder cancer risk. However, it should eventually be justified to refer to the comparison of results in the study group with those in the control group, representative for the studied region. The frequency of variant A148T found by us in patients with bladder cancer is similar to the results published in the work of Sakano et al. (6%) for patients with bladder cancer in the population of Stockholm county in Sweden [29]. However, the result cannot be compared with a general population as the authors do not specify what is the frequency of this variant in the population of Swedes.
The obtained results should by all means be confirmed in a study on a larger population, representing the Łódź Macroregion, especially that the results of studies on other neoplasms differ among populations or even within the same population, when differentiated for example by age (the groups of patients below and above the age of 50 years). Breast cancer can be an example, the results for which were different from those in the control group in the group of women below 50, while no differences in the incidence rates were observed between the control group and the group of women above 50 [31]. In case of the studied group, 8 out of 9 A148T polymorphism cases of CDKN2A gene were identified in men at the age above 60 years at diagnosis (data specified in Table 4). No recurrence was observed in 7 out of 9 diagnosed cases. In 6/9 cases, there were neoplasms in high clinical stage; while in 5/9 cases there were tumors with high malignancy grade (G2-G3). Summing up and taking into account the analysis of clinical parameters and the age of disease occurrence, A148T polymorphism of CDKN2A gene was found in the studied group only in men, in whom the disease was diagnosed at the age above 60 years, while the diagnosed neoplasms were in the majority of cases characterized by higher clinical stage and higher grade of malignancy, as well as by a low percentage of recurrence. Nevertheless, this has been the first study that attempted to show a potential association between A148T alterations and an increased risk for bladder cancer development.
Table 4.
Prevalence of A148T variant of CDKN2a gene vs. clinical data
| N (%) | A148T (+) N (%) | |
|---|---|---|
| Sex: | ||
| Male | 130 (83.3) | 9 (6.9) |
| Female | 26 (16.7) | 0 (0.0) |
| Age: | ||
| ≤60 | 45 (28.8) | 1 (2.2) |
| ≥61 | 111 (71.2) | 8 (8.1) |
| Grade: | ||
| I | 84 (53.8) | 4 (4.8) |
| ≥II | 72 (46.2) | 5 (6.9) |
| Stage: | ||
| Ta | 81 (51.9) | 3 (3.7) |
| ≤T1 | 75 (48.1) | 6 (8.0) |
| Local recurrence: | ||
| Positive | 59 (37.8) | 2 (3.3) |
| Negative | 97 (62.2) | 7 (7.2) |
CONCLUSION
Studies on genetic predispositions to neoplasm development allow to distinguish the group of subjects with an increased risk for neoplastic changes vs. the general population. The more we learn about human genetics and human variations, the more apparent it becomes that our individual make-up has a noticeable impact on the risk of cancer development and finally on the effectiveness of therapeutic medications. A148T variant of CDKN2A gene seems to be associated with an increased risk for developing bladder cancer. Nevertheless, according to Debniak et al. (2005), it does not imply that A148T polymorphism is associated with an increased risk of the diseases alone but it rather suggests the existence of some interactive modifiers that, when grouped together, may trigger the malignancy process.
The study was approved by the Ethics Committee of the Medical University of Łódź (Permission no RNN/66/02/KE). This work was supported by the State Committee for Scientific Research, Poland (KBN grant No 2P05C 076 30).
REFERENCES
- 1.Jorde LB, Carey JC, Bamshad MJ. Medical Genetics. 4th Edition. Mosby Inc, Elsevier Inc; 2010. Genetic variations: its origin and detection; pp. 26–27. Chapt. 3. [Google Scholar]
- 2.Wojciechowska U, Didkowska J, Zatoński W. The National Register of Neoplasms, The M. Skłodowska -Curie of Oncology. Warsaw: 2006. Nowotwory złośliwe w Polsce w 2006 roku. [Google Scholar]
- 3.Goebell PJ, Knowles MA. Bladder cancer or bladder cancers? Genetically distinct malignant conditions of the urothelium. Urol Oncol. 2010;28(4):409–428. doi: 10.1016/j.urolonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Bryan RT, Hussain SA, James ND, et al. Molecular pathways in bladder cancer: Part 1. BJU International. 2005;95:485–490. doi: 10.1111/j.1464-410X.2005.05325.x. [DOI] [PubMed] [Google Scholar]
- 5.Berggren P, Kumar R, Sakano S, et al. Detecting homozygous deletions in the CDKN2A(p16INK4a)/ARF(p14ARF) gene in urinary bladder cancer using real-time quantitative PCR. Clin Cancer Res. 2003;9(1):235–242. [PubMed] [Google Scholar]
- 6.Borkowska E, Traczyk M, Pietrusiński M, et al. The significance of P53 gene codons 72 and 213 polymorphisms in urinary bladder cancer in Central Poland. CEJUrol. 2010;63(1):9–13. [Google Scholar]
- 7.MacKie RM, Andrew N, Lanyon WG, Connor JM. CDKN2A germline mutations in U.K. patients with familial melanoma and multiple primary melanomas. J Invest Dermatol. 1998;111:269–272. doi: 10.1046/j.1523-1747.1998.00267.x. [DOI] [PubMed] [Google Scholar]
- 8.Smeds J, Berggren P, Ma X, et al. Genetic status of cell cycle regulators in squamous cell carcinoma of the oesophagus: the CDKN2A (p16INK4a and p14ARF) and p53 genes are major targets for inactivation. Carcinogenesis. 2002;23(4):645–655. doi: 10.1093/carcin/23.4.645. [DOI] [PubMed] [Google Scholar]
- 9.Cobbers JMJL, Wolter M, Reifenberger J, et al. Frequent inactivation of CDKN2A and rare mutation of TP53 in PCNLS. Brain Pathol. 1998;8:263–276. doi: 10.1111/j.1750-3639.1998.tb00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann T, Knuchel-Clarke R, Hartmann A, et al. EAU-EBU Update Series 4. 2006. Clinical implications of the 2004 WHO Histological Classification on non-invasive tumours of the urinary bladder; pp. 83–95. [Google Scholar]
- 11.Borkowska E, Binka-Kowalska A, Constantinou M, et al. P53 mutations in urinary bladder cancer patients from Central Poland. J Appl Genet. 2007;48(2):177–183. doi: 10.1007/BF03194676. [DOI] [PubMed] [Google Scholar]
- 12.Zar JH. Biostatistical analysis. Englewood Cliffs, New Jersey: Prentice-Hall, Inc; 1984. [Google Scholar]
- 13.Debniak T, Scott RJ, Huzarski T, et al. CDKN2A common variants and their association with melanoma risk: a population-based study. Cancer Res. 2005;65(3):835–839. [PubMed] [Google Scholar]
- 14.Aveyard JS, Knowles MA. Measurement of relative copy number of CDKN2A/ARF and CDKN2B in bladder cancer by real-time quantitative PCR and multiplex ligation-dependent 6probe amplification. JMD. 2004;6(4):356–365. doi: 10.1016/S1525-1578(10)60532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scraml P, Struckmann K, Bednar R, et al. CDKN2A mutation analysis, protein expression, and deletion mapping of chromosome 9p in conventional clearcell renal carcinoma. Am J Pathol. 2001;158(2):593–600. doi: 10.1016/s0002-9440(10)64001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monzon J, Liu L, Brill H, et al. CDKN2A mutations in multiple primary melanomas. N Engl J Med. 1998;338(13):879–887. doi: 10.1056/NEJM199803263381305. [DOI] [PubMed] [Google Scholar]
- 17.Castellano M, Pollock PM, Walters MK, et al. CDKN2A/p16 is inactivated in most melanoma cell lines. Cancer Res. 1997;57(21):4868–4875. [PubMed] [Google Scholar]
- 18.Levanat S, Situm M, Crnić I, et al. Alterations in CDKN2A locus as potential indicator of melanoma predisposition in relatives of non-familial melanoma cases. Croat Med J. 2003;44(4):418–424. [PubMed] [Google Scholar]
- 19.Wang H, Presland RB, Piepkorn M. A search for CDKN2A/p16INK4a mutations in melanocytic nevi from patients with melanoma and spouse controls by use of laser-captured microdissection. Arch Dermatol. 2005;141(2):177–180. doi: 10.1001/archderm.141.2.177. [DOI] [PubMed] [Google Scholar]
- 20.Berwick M, Orlow I, Hummer AJ, et al. The prevalence of CDKN2A germline mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev. 2006;15(8):1520–1525. doi: 10.1158/1055-9965.EPI-06-0270. [DOI] [PubMed] [Google Scholar]
- 21.Lamperska KM, Przybyła A, Kycler W, Mackiewicz A. The CDKN2A common variants: 148 Ala/Th rand 500 C/G In 3′UTR, and their association with clinical course of melanoma. Acta Bioch Polonica. 2007;54(1):119–124. [PubMed] [Google Scholar]
- 22.Chiorzo P, Ciotti P, Mantelli M, et al. Characterization of ligurian melanoma families and risk of occurrence of other neoplasia. Int J Cancer. 1999;83:441–448. doi: 10.1002/(sici)1097-0215(19991112)83:4<441::aid-ijc2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 23.Bertram CG, Gaut RM, Barrett JH, et al. An assessment of the CDKN2A variant Ala148Thr as nevus/melanoma susceptibility allele. J Invest Dermatol. 2002;119:961–965. doi: 10.1046/j.1523-1747.2002.01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Spica T, Portela M, Gerard B, et al. The A148T variant of the CDKN2A gene is not associated with melanoma risk in the French and Italian populations. J Invest Dermatol. 2006;126:1658–1660. doi: 10.1038/sj.jid.5700293. [DOI] [PubMed] [Google Scholar]
- 25.Erlandson A, Appelqvist F, Wennberg AM, et al. Novel CDKN2A mutations detected in Western Swedish Families with hereditary malignant melanoma. J Invest Dermatol. 2007;127:1465–1467. doi: 10.1038/sj.jid.5700718. [DOI] [PubMed] [Google Scholar]
- 26.Puig S, Malvehy J, Badenas C, et al. Role of the CDKN2A locus in patients with multiple primary melanomas. J Clin Oncol. 2005;23(13):3043–3051. doi: 10.1200/JCO.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 27.Nagore E, Montoro A, Garcia-Casado Z, et al. Germline mutations in CDKN2A are infrequent in female patients with melanoma and breast cancer. Melanoma Res. 2009;19(4):211–214. doi: 10.1097/CMR.0b013e3283281057. [DOI] [PubMed] [Google Scholar]
- 28.Kiwerska K, Rydzanicz M, Kram A, et al. Mutational analysis of CDKN2A gene in a group of 390 larynx cancer patients. Mol Biol Rep. 2010;37:325–332. doi: 10.1007/s11033-009-9731-z. [DOI] [PubMed] [Google Scholar]
- 29.Sakano S, Berggren P, Kumar R, et al. Clinical course of bladder neoplasms and single nucleotide polymorphism in the CDKN2A gene. Int J Cancer. 2003;104:98–103. doi: 10.1002/ijc.10919. [DOI] [PubMed] [Google Scholar]
- 30.Dębniak T, Scott RJ, Huzarski T, et al. CDKN2A common variant and multiorgan cancer risk - a population based study. Int J Cancer. 2006;118:3180–3182. doi: 10.1002/ijc.21760. [DOI] [PubMed] [Google Scholar]
- 31.Dębniak T. Some molecular and clinical aspects of genetic predisposition to malignant melanoma and tumours of various site origin. Hered Cancer Clin Pract. 2007;5(2):97–116. doi: 10.1186/1897-4287-5-2-97. [DOI] [PMC free article] [PubMed] [Google Scholar]