Abstract
Introduction
H-RAS gene is a protooncogene encoding p21ras, a small protein with GTPase activity. This protein is a component of many signaling cascades, while mutations in H-RAS gene are often found in urinary bladder cancer and leads to continuous transmission of signals stimulating cancer cell growth and proliferation. The T81C polymorphism of H-RAS gene is a SNP, which, although does not seem to impair p21ras protein structure and function, may contribute to the development of bladder cancer.
Objectives
The aim of our study was to characterize the prevalence and clinical significance of T81C polymorphism in patients with diagnosed bladder cancer.
Materials and methods
132 patients with diagnosed urinary bladder cancer were included in this study. The control group consisted of 106 healthy individuals. The experimental material was DNA, isolated from tumor tissue and peripheral blood lymphocytes. T81C polymorphism was detected using the MSSCP method and DNA sequencing.
Results
In the examined DNA samples, frequent polymorphic variations were found in codon 27 of H-RAS gene. In order to assess the clinical relevance of the polymorphism, the results were compared with those for the control group. The homozygous CC variant occurred more frequently in bladder cancer patients than in healthy individuals.
Conclusions
DNA polymorphisms start to play an important role in evaluation of disease risk and progression. The occurrence of multiple variants of the same gene may contribute to differences in reactions to specific medications and sensitivity to carcinogens or DNA repair capacity. Our study demonstrated T81C polymorphism of H-RAS gene to have seemingly been associated with an increased risk of bladder cancer development.
Keywords: urinary bladder cancer, H-RAS gene, T81C polymorphism
INTRODUCTION
Mammalian cells contain three different protooncogenes of the RAS family: N-RAS (1p13.2), K-RAS (12p12.1), and H-RAS (11p15.5). They encode small proteins with GTPase activity, which participate in the transmission process of growth signals from cell membrane receptors to the cell nucleus [1, 2]. They bind with GTP during their active state and GDP during their inactive state. Mutated proteins of the RAS family are not suitable for hydrolysis with GTP, thus – regardless of extracellular signals – the growth and proliferation of cells is continuously stimulated.
The H-RAS (Harvey rat sarcoma viral oncogene homologue) protooncogene is located at the terminal part of the short arm of chromosome 11. It was discovered in 1980 as an oncogene of sarcoma viruses [3]. It consists of four encoding and one noncoding exon, the latter localized closer to the 5’ end (Fig. 1). A point mutation in one of the three hot spots (codons 12, 13, and 61) may result in continuous stimulation of proliferation and development of many types of cancer [4, 5].
Fig. 1.
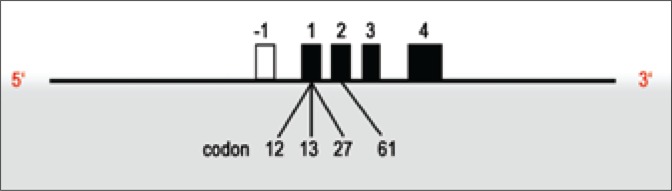
Structure of H-RAS gene. Black figures depict encoding exons.
In 1982, Reddy et al. [6] and, a year later, Capon et al. [7] confirmed the existence of the H-RAS gene mutations in a human bladder cancer cell line (T24). Further studies on urinary bladder cancer demonstrated that mutations in this gene were more frequent than mutations in the K-RAS or N-RAS genes [8, 9] and that these mutations were mainly responsible for the development of superficial tumors of low histopathological grade [10, 11 12]. However, the role of H-RAS gene mutations in the development of urinary bladder cancer still raises many controversies (mainly due to considerable discrepancies in the results of various research groups) regarding the incidence of identified point mutations in this gene (from 0 to 84%) [9, 13].
The H-RAS T81C polymorphism (rs12628) is a single nucleotide polymorphism (SNP) [14] localized in codon 27 (cDNA position: 81) and was described for the first time by Taparovsky et al. [15]. This polymorphism disturbs neither the structure nor function of the p21ras protein, since both CAT and CAC codons encode histidine (His27His); however, it demonstrates some relationship with increased susceptibility to the development of various types of cancer [14].
This study aimed to evaluate the incidence and clinical significance of the T81C polymorphism of H-RAS gene in a group of patients from Poland diagnosed with urinary bladder cancer.
MATERIAL AND METHODS
Study group
The study was performed in 132 patients from the Macroregion of Łódź, Poland with diagnosed urinary bladder cancer (22 females and 110 males; median age: 68 years; age range: 41-87 years). All cases were confirmed by histological evaluation. Transitional cell carcinoma (TCC) of the bladder was found in 121 samples, while histological evaluation confirmed dysplasia in three cases, and inflammation without evidence of tumor or abnormal epithelium in eight cases. However, in the latter 11 cases, TCC was found previously (patients with recurrent disease). The average observation period of the patients was 32 months. The characteristics of the study group are shown in Table 1.
Table 1.
Clinicopathological patient profiles
| No. of cases: N = 132 | |
|---|---|
| Gender: | |
| Male | 110 (83.3%) |
| Female | 22 (16.7%) |
| Smoking: | |
| Yes | 110 (83.3%) |
| No | 22 (16.7%) |
| Occupational exposure: | |
| Yes | 70 (53.0%) |
| No | 62 (47.0%) |
| Tumor Stage: | |
| Ta | 83 (62.9%) |
| T1 | 25 (18.9%) |
| T2 | 9 (6.8%) |
| T3 | 3 (2.3%) |
| T4 | 1 (0.8%) |
| Tumor Grade: | |
| LG | 81 (61.4%) |
| HG | 40 (30.3%) |
| Dysplasia LG* | 2 (1.5%) |
| Dysplasia HG* | 1 (0.8%) |
| Inflammation without evidence of tumor* | 4 (3.0%) |
| Normal epithelium* | 4 (3.0%) |
Patients with recurrent disease
Primary tumors were observed in 72 of the patients (54.5%), while the remaining patients (45.5%) demonstrated recurrent disease. Twenty-six patients (19.7%) were treated by chemotherapy. Recurrence of bladder cancer in our study group was noted in 29 cases (18 with primary tumor and 11 with recurrent disease). Death from bladder cancer was recorded in 14 cases (10.6%).
In order to assess the clinical significance of T81C polymorphism, the obtained results were compared with the results of the 106 healthy controls (19 women, 87 men; median age: 65; age range: 39-92 years) with the same ethnic background and from the same approximate age group. There were no significant differences in gender, age, smoking status and occupational exposure between the cancer-affected patients and controls.
Identification of genotypes
DNA isolated from tumor tissue (the study group) and peripheral blood lymphocytes (the study group and the control group) was used as the experimental material. Fragments of neoplastic tissue were obtained in the course of transurethral resection of bladder tumor (TURBT). They were frozen in liquid nitrogen and stored until DNA isolation in −70°C. DNA was isolated by means of a commercial kit for DNA isolation from biological remnants (Sherlock AX; A&A Biotechnology, Poland) and stored in temperature of 4-8°C. Blood samples were collected from healthy individuals into EDTA-K2 containing tubes and stored in refrigerator temperature until DNA isolation was performed using the MagNA Pure Compact device and MagNa Pure Compact Nucleic Acid Isolation Kit I (Roche, Germany).
Sequences of H-RAS (exon1) were amplified, using primers:
F: 5’AGGAGCGATGACGGAATATAAGC3’
R: 5’GGCTCACCTCTATAGTGGGGTCGTATT3’.
The obtained PCR reaction mixture (25 µl) contained 100 ng of genomic DNA, 2.5 µl of 10x concentrated reaction buffer (Novazym, Poland), 0.5 µl of dNTPs – 2.5 mM each (TaKaRa, Japan), 1 µl of each starter (5 pmol/µl), and 1.25U of Taq DNA polymerase (Novazym, Poland). All reactions contained a non-template control.
MSSCP (Multitemperature Single Strand Conformation Polymorphism) conditions
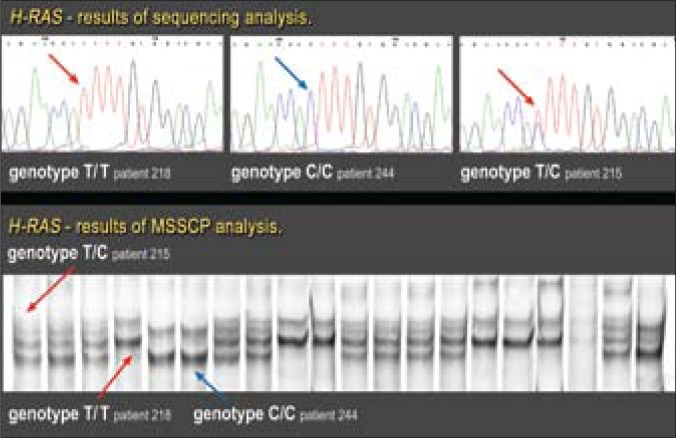
The T81C polymorphism in exon 1 of the H-RAS gene was detected using the MSSCP method and DNA sequencing (Fig. 2). The MSSCP technique was carried out in one of the following temperature profiles: 25° C for 30 minutes, 20°C for 20 minutes, or 15°C for 20 minutes. The details of MSSCP analysis were reported elsewhere [16]. PCR products were sequenced (Genomed sp. z o.o., Poland) to rule out or confirm the mutation or polymorphism identified during the MSSCP analysis.
Fig. 2.
Results of DNA sequencing and MSSCP analysis of exon 1 H-RAS gene (tumor tissue).
Statistical analysis
The study cases and controls were compared using Pearson's χ2 test. The first step in assessing the genotype results was to find out if deviations in the genotype frequencies were compliant with the Hardy-Weinberg equilibrium. The odds ratios (ORs) and their 95% confidence intervals (95% CIs) were calculated to estimate the association between genotypes and cancer occurrence. Variant TT of T81C polymorphism was a reference category in dichotomized variable analysis. A p-value <0.05 indicated statistical significance. Raw data of genotype frequencies (without adjustment) were used to estimate OR in meta-analysis. The extent of heterogeneity was estimated by the Cochran's χ2 test. The statistical analysis and meta-analysis were performed using STATISTICA 9.1 (StatSoft, Inc., Tulsa, OK, USA).
RESULTS
Exon 1 of the H-RAS gene was evaluated in each of the patients in search of polymorphism in codon 27. At the same time, no mutation was observed in codons 12 or 13 in any of the patients. In all studied cases, the results of T81C polymorphism in DNA from tumor tissue corresponded to the results of the T81C analysis in DNA obtained from the peripheral blood lymphocytes.
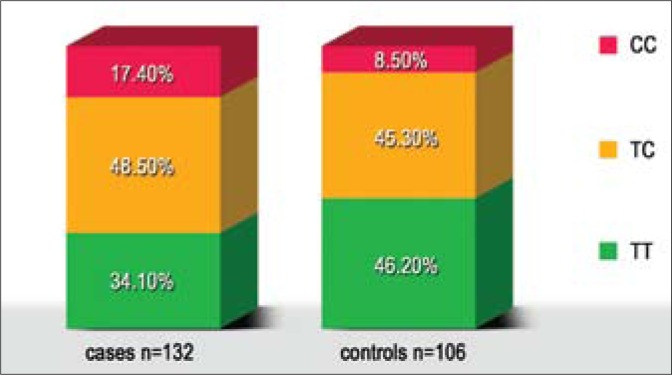
The incidence rates of the observed genotypes in both groups fulfilled the principles of Hardy-Weinberg Equilibrium (cases: p = 0.98; controls: p = 0.56). Thus, carrying out odds ratio (OR) calculation, it became possible to assess whether the occurrence of the polymorphic variants of H-RAS gene at codon 27 was in any way associated with the increased susceptibility to urinary bladder cancer formation. Detailed data of the incidence of particular genotypes and T81C polymorphism alleles in both studied groups are shown in the Table 2 and Figure 3.
Table 2.
The incidence H-RAS T81C polymorphism and allele distribution in the patient and control groups
| Genotype | Patients (n = 132) | Controls (n = 106) | OR (95% CI) | P – value |
|---|---|---|---|---|
| TT | 45 (34.1%) | 49 (46.2%) | 1.00 | – |
| TC | 64 (48.5%) | 48 (45.3%) | 1.45 (0.83–2.5) | 0.18 |
| CC | 23 (17.4%) | 9 (8.5%) | 2.78 (1.16–6.64) | 0.02 |
| CC against TC + TT | 109 (82.6%) | 97 (91.5%) | 2.27 (1.0–5.15) | 0.05 |
| TC + CC against TT | 87 (65.9%) | 57 (53.8%) | 1.66 (0.98–2.80) | 0.06 |
| Allele type | ||||
| T | 154 (58.3%) | 146 (68.9%) | 0.63 (0.43–0.92) | 0.02 |
| C | 110 (41.7%) | 66 (31.1%) | 1.58 (1.08–2.31) | 0.02 |
Fig. 3.
The incidence of particular variants of T81C polymorphism in bladder cancer cases and in healthy controls.
The C risk allele of H-RAS gene T81C polymorphism occurred more frequently in the studied group of patients (41.7%) than in the control group (31.1%) (OR = 1.58; 95% CI: 1.08 – 2.31; p = 0.02). When comparing particular polymorphism variants (CC variant with TC heterozygotes and TT wild variant), we found the CC risk genotype to be associated with a greater than double increase in the risk for urinary bladder cancer (OR = 2.27; 95% CI: 1.00 – 5.15; p = 0.05), a statistically significant difference was obtained.
A relationship between the occurrence of polymorphism variants and either tumor stage (p = 0.75) or grade (p = 0.61) was not demonstrated. Neither were any relationships between occupational exposure (p = 0.14), smoking status (p = 0.73), or disease recurrence (p = 0.24) with the incidence of polymorphic variants.
DISCUSSION
Environmental exposure (especially occupational) to chemical carcinogens and products of tobacco tar are regarded to be the main causes of urinary bladder cancer formation [10, 17, 18]. However, one should not forget about the genetic and epigenetic changes that follow the effects of exposure to the environmental factors and initiate the process of cancer development [11, 12]. The effects of these carcinogens may disturb a wide range of cellular processes (such as DNA repair, cellular cycle control, carcinogen metabolism, apoptosis, and signal transmission pathways) [19]. These disruptions lead to genomic defects and compromise cellular equilibrium, the consequence of which is the selection of clones of neoplastic cells. The existence of different variants of the same gene may trigger differences in reactions to certain drugs, in their sensitivity to carcinogenic activity and ability for DNA repair. In this way it leads to an increased susceptibility to cancer development, including urinary bladder neoplasms [19]. Therefore, DNA polymorphisms begin to play a more and more important role in procedures evaluating the risk of neoplastic disease, as well as those assessing cancer progression.
Studies of H-RAS gene exon 1, based on the MSSCP method, and DNA sequencing did not indicate mutations in codons 12 and 13 in our group of 132 patients. The available literature data demonstrate that H-RAS gene mutations may occur in urinary bladder neoplasms, but incidence rates vary. Our results are conformable with those of earlier studies, carried out by Karimianpour et al. [20] and by Johne et al. [14], which did not find protooncogene mutations in patients with urinary bladder cancer either. However, Przybojewska et al. [9] demonstrated the presence of such mutations in codon 12 in as many as 84% of examined patients. What is fairly surprising is the fact that the examined group of patients had also come from the Lodz Macroregion. The discrepancy between the presented results from our study and the results of the above-mentioned group of researchers may result from differences in employed research methods (the results were not confirmed by DNA sequencing).
Our studies have demonstrated that carriers of the CC homozygous variant of H-RAS T81C polymorphism, compared with TC heterozygotes and carriers of TT wild variants, are twice more at risk for urinary bladder cancer development (OR = 2.27; p = 0.02). So far only three research teams have published any data from studies of the polymorphism in case of urinary bladder cancer. Johne et al. [14] carried out studies on the German population, demonstrating the presence of the polymorphism in 13.5% of patients and in 7.1% of controls. Following conducted studies and analyses, they found out that the CC homozygous variant increased the risk for urinary bladder cancer development two-fold. The results, obtained by the group of researchers, are conformable with our results, unlike the data, published by Sanyal et al. [21] (studies on the Swedish population). They revealed that the prevalence of the CC risk allele was lower in the study group (1%) vs. the control group (5%). The analyses, carried out by that group of researchers, demonstrated that CC homozygotes were associated with a lower risk for urinary bladder cancer development (OR = 0.13). Pandith et al. [22] (Kashmir population) found that 5.7% of the studied cases were CC homozygotes and that variant did not occur in the control group. Therefore, they could not calculate OR for CC risk variant, when compared to either TC heterozygotes or TT wild variant carriers.
The contradictory character of reports on polymorphism in H-RAS gene codon 27 does not allow for any unequivocal determination of how big a relationship exists between the occurrence of the CC variant in environmentally/occupationally exposed subjects and the subsequent formation of urinary bladder cancer in these subjects. Therefore, further studies should be undertaken on much bigger groups of patients. It is also very clear that the ethnic background of the studied patient groups demonstrates pronounced effects on the incidence of particular variants of the T81C polymorphism.
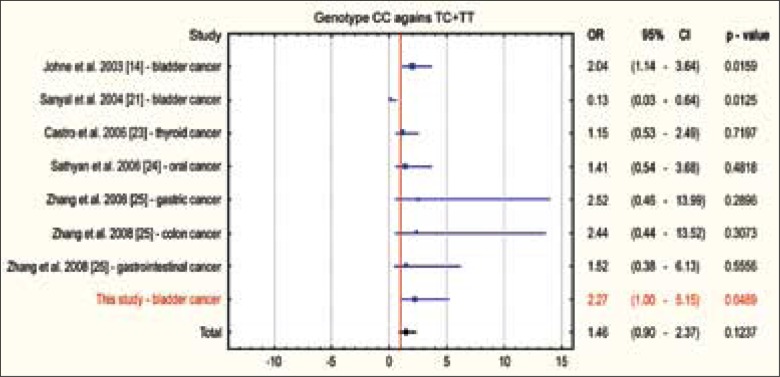
Meta-analysis is a research tool allowing the combination of results from various studies by the application of statistical methods. The meta-analysis presented in Fig. 4 was performed for earlier published studies on T81C polymorphism in cases of various neoplasms in man [14, 21, 23, 24, 25], including also the results of our study. The consolidated OR was 1.46 (95% CI: 0.90-2.37; p = 0.1237), showing that occurrence of the CC homozygous variant increases almost 1.5-times the risk for the development of various neoplasms in man.
Fig. 4.
Meta-analysis of T81C polymorphism in H-RAS gene for all cancers.
CONCLUSION
DNA polymorphisms start to play an important role in evaluation of disease risk and progression. The occurrence of multiple variants of the same gene may contribute to differences in reactions to specific medications and in their sensitivity to carcinogens or DNA repair capacity. Our study has shown that the T81C polymorphism, localized in the coding region of the H-RAS gene, may be a risk factor for bladder cancer development in the Polish population.
Acknowledgements
The reported study was carried out within Research Project No. 502-03/2-159-02/502-24-017 from the Medical University of Lodz. Our studies and applied procedures were approved by the Committee of Ethics at the Medical University of Łódź (Document No. RNN/215/10/KE).
REFERENCES
- 1.Takai Y, Sasaki T, Matozaki T. Small GTP- Binding Proteins. Physiol Rev. 2001;81(1):154–164. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- 3.Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim et Biophys Acta. 2007;1773(8):1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2(3):344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982;300:149–152. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- 7.Capon DJ, Chen EY, Levinson AD, et al. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983;302:33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- 8.Rabbani F, Cordon-Cardo C. Mutation of cell cycle regulators and their impact on superficial bladder cancer. Urol Clin North Am. 2000;27:83–102. doi: 10.1016/s0094-0143(05)70237-8. [DOI] [PubMed] [Google Scholar]
- 9.Przybojewska B, Jagiello A, Jalmuzna P. H-RAS, K-RAS, and N-RAS gene activation in human bladder cancers. Cancer Genet Cytogenet. 2000;121(1):73–77. doi: 10.1016/s0165-4608(00)00223-5. [DOI] [PubMed] [Google Scholar]
- 10.Pasin E, Josephson D, Mitra A, et al. Superficial bladder cancer: An update on etiology, molecular development, classification and natural history. Rev Urol. 2008;10(1):31–43. [PMC free article] [PubMed] [Google Scholar]
- 11.Goebell PJ, Knowles MA. Bladder cancer or bladder cancer? Genetically distinct malignant conditions of the urothelium. Urol Oncol. 2010;28(4):409–428. doi: 10.1016/j.urolonc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27(3):361–373. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 13.Boulalas I, Zaravinos A, Karyotis I, et al. Activation of RAS Family Genes in Urothelial Carcinoma. J Urol. 2009;181(5):2312–2319. doi: 10.1016/j.juro.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Johne A, Roots I, Brockmoller J. A single nucleotide polymorphism in the human HRAS proto-oncogene determines the risk of urinary bladder cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:68–70. [PubMed] [Google Scholar]
- 15.Taparowsky E, Suard Y, Fasano O, et al. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–765. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 16.Borkowska E, Traczyk M, Pietrusiński M, et al. The significance of P53 gene codons 72 and 213 polymorphisms in urinary bladder cancer in Central Poland. CEJUrol. 2010;63(1):9–13. [Google Scholar]
- 17.Colombel M, Soloway M, Akaza H, et al. Epidemiology, Staging, Grading and Risk Stratification of Bladder Cancer. Eur Urol Suppl. 2008;7:618–626. [Google Scholar]
- 18.Zlotta AR, Cohen SM, Dinney C, et al. BCAN Think Tank session 1: Overview of risks for and causes of bladder cancer. Urol Oncol. 2010;28(3):329–333. doi: 10.1016/j.urolonc.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Lin X, Dinney CP, et al. Genetic polymorphism in bladder cancer. Front Biosci. 2007;12:192–213. doi: 10.2741/2058. [DOI] [PubMed] [Google Scholar]
- 20.Karimianpour N, Mousavi-Shafaei P, Ziaee AA, et al. Mutations of RAS Gene Family in Specimens of Bladder Cancer. Urol J. 2008;5(4):237–242. [PubMed] [Google Scholar]
- 21.Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25(5):729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 22.Pandith AA, Shah ZA, Khan NP. HRAS T81C polymorphism modulates risk of urinary bladder cancer and predicts advanced tumors in ethnic Kashmiri population. Urol Oncol. 2011 doi: 10.1016/j.urolonc.2011.03.004. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Castro P, Soares P, Gusmao L, et al. HRAS 81 polymorphism is significantly associated with aneuploidy in follicular tumors of the thyroid. Oncogene. 2006;25:4620–4627. doi: 10.1038/sj.onc.1209491. [DOI] [PubMed] [Google Scholar]
- 24.Sathyan KM, Nalinakumari KR, Abraham T, Kannan S. Influence of single nucleotide polymorphisms in HRAS and cyclin D1 genes on oral cancer susceptibility. Oral Oncol. 2006;42:607–613. doi: 10.1016/j.oraloncology.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Jin M, Liu B, et al. Association between HRAS T81C genetic polymorphism and gastrointestinal cancer risk: A population- based case-control study in China. BMC Cancer. 2008;8:256–262. doi: 10.1186/1471-2407-8-256. [DOI] [PMC free article] [PubMed] [Google Scholar]