Abstract
The aim of the study is to explore the role of miR-194 in mediating the effect of high-K (HK) intake on ROMK channel. Northern blot analysis showed that miR-194 was expressed in kidney and that HK intake increased while low-K intake decreased the expression of miR-194. Real-time PCR analysis further demonstrated that HK intake increased the miR-194 expression in the cortical collecting duct. HK intake decreased the expression of intersectin 1 (ITSN1) which enhanced With-No-Lysine Kinase (WNK)-induced endocytosis of ROMK. Expression of miR-194 mimic decreased luciferase reporter gene activity in HEK293 T cells transfected with ITSN-1–3′UTR containing the complementary seed sequence for miR-194. In contrast, transfection of miR-194 inhibitor increased the luciferase activity. This effect was absent in the cells transfected with mutated 3′UTR of ITSN1 in which the complimentary seed sequence was deleted. Moreover, the inhibition of miR-194 expression increased the protein level of endogenous ITSN1 in HEK293T cells. Expression of miR-194 mimic also decreased the translation of exogenous ITSN1 in the cells transfected with the ITSN1 containing 3′UTR but not with 3′UTR-free ITSN1. Expression of pre-miR-194 increased K currents and ROMK expression in the plasma membrane in ROMK-transfected cells. Coexpression of ITSN1 reversed the stimulatory effect of miR-194 on ROMK channels. This effect was reversed by coexpression of ITSN1. We conclude that miR-194 regulates ROMK channel activity by modulating ITSN1 expression thereby enhancing ITSN1/WNK-dependent endocytosis. It is possible that miR-194 is involved in mediating the effect of a HK intake on ROMK channel activity.
Keywords: K secretion, collecting duct, WNK, Kir1.1, endocytosis
micrornas are highly conserved endogenous small RNA molecules and they play a role in regulating the expression of the target protein (5). For the generation of microRNA, pri-microRNA is trimmed by RNase III Drosha into pre-microRNA which is exported into the cytoplasma and further processed by dicer into a mature microRNA. One strand of the mature microRNA interacts with 3′UTR of the target mRNA through imperfect base pairing in the miRISC (microRNA-induced silencing complex) thereby inhibiting the translation of the target gene. A large body of evidence has demonstrated that microRNAs are highly expressed in the kidney and play important roles in the regulation of renal function under physiological conditions and in causing pathological development (4, 7, 24, 25, 33). Several microRNAs have been demonstrated to play a role in regulation of salt-sensitive hypertension (22, 30). Our previous study also suggested that microRNAs played a role in the regulation of K secretion.
We showed that the expression of miR-802 was enhanced by high dietary K intake and that an increase in miR-802 expression stimulated ROMK channels by downregulation of calveolin-1 (20). To our interest, an increase in dietary K intake enhances miR-194 transcription in the kidney. Since 3′UTR of intersectin 1 (ITSN1) contains the target site of miR-194, it is possible that miR-194 may regulate the expression of ITSN1. Moreover, it has been demonstrated that ITSN1 is required for mediating With-No-Lysine Kinase (WNK)-induced endocytosis of ROMK (8). Therefore, it is conceivable that a decrease in the expression of ITSN1 may be also involved in mediating the stimulatory effect of high-K (HK) intake on ROMK channel activity because decreasing ITSN1 is expected to inhibit the endocytosis of ROMK channels. Thus, the aim of the present study is to explore whether miR-194 regulates ROMK channel activity by targeting ITSN1.
METHODS
Animal preparation.
Male C57BL/6 mice (4–5 wk old) were purchased from Jackson Laboratory. Animals were fed with a normal-K (NK) diet (1% K), a low-K diet (<0.001%; LK), or a HK diet (5%; Harlan Laboratory, Madison, WI) for 7 days before use. Mice were killed by cervical dislocation and two kidneys were removed immediately after death. The protocol was approved by an independent animal user committee of New York Medical College.
Northern blot.
The total RNA and microRNA from the kidney were harvested with miRNeasy mini kit (Qiagen) for the preparation of Northern blot. Briefly, tissue (50 mg) of the mouse cortex and outer medulla (OM) was lysed with 700 μl QIAzol lysis reagent and was homogenized with QIAshredder (Qiagen). The tissue lysate was mixed with 140 μl chloroform and centrifuged for 15 min at 12,000 rpm. The sample was diluted by adding 100% ethanol (150% of the lysate volume), vortexed, and loaded into the mini-spin column. After centrifugation, the column was washed and the total RNA sample was eluted in 30 μl RNase-free water. The quality of the RNA was determined by measuring the ratio of A260/A280. After confirming the quality of the RNA, we carried out electrophoresis. For the pre-run, the denatured 15% PAGE gel was conducted at 400 V (40 mA) for 1 h in 0.5× Tris, Borate, EDTA (TBE) buffer in Protean III tank (Bio-Rad). The RNA sample was denatured by adding the equal volume of the buffer containing 10 mM EDTA (pH 8.0), 0.05% xylene cyanol and bromophenol blue in deionized formamide, and incubated for 20 min at 65°C. The denatured RNA sample was subjected to electrophoresis at 200 V in 1× TBE buffer until bromophenol blue reached the edge. After being rinsed, the RNA was wet-transferred into positive-charged nylon membrane and crosslinked with UV Stratalinker 1800 (Stratagene). The mmu-miR-194 probe was prepared by labeling 5′ 32P to DNA oligo, TCCACATGGAGTTGCTGTTACA (bold font, the seed sequence). Briefly, the membrane was prehybridized at 35°C for 30 min in 10 ml hybrid buffer (50% deinonized formamide, 5× Denhardt's solution, 0.5% SDS, 2 mg/100 ml boiled salmon sperm DNA, 250 mM NaCl, 50 mM Na-phosphate, 10 mM EDTA, pH 7.4). The membrane was then incubated with 10 pmol 32P-labeled oligo probe overnight at 37°C and was followed by the membrane being washed with 2× SSC containing 0.1% SDS three times. The miR-194 band was detected by placing the membrane on an X-ray film which is in a cassette with the intensify screen.
Western blot.
Renal cortex and OM were dissected, the tissue was homogenized, and the proteins were resolved by electrophoresis on 8% SDS-polyacrylamide gels. After proteins were transferred to nitrocellulose membranes, the membrane was blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) and incubated overnight with the primary antibody at 4°C. The membrane was then rinsed with 0.05% Tween-20-TBS three times followed by incubation for 30 min with the respective second antibody. After three washes, the membranes were scanned by Odyssey infrared imaging system (LI-COR) at a wavelength of 680 or 800 nm.
Real-time PCR to detect miR-194 expression in the cortical collecting duct.
The RNA of the dissected cortical collecting duct (CCD) was isolated with RNAqueous-Micro kit (Ambion). The cDNA was generated with Affinity Script RT enzyme from Stratagene (La Jolla, CA). Briefly, 1 μl random primer and 100 ng RNA or single tubule lysate were annealed at 95°C by adding 1 μl DTT, dNTP, and enzyme. The mixture was incubated for 1 h at 65°C. The primer used for detecting mouse pre-miR-194 in the connecting tubule (CNT)/CCD was ordered from Qiagen (miScript primer assay for SBYR Green method) and it targeted mouse mature miR-194 (5′-UGUAACAGCAACUCCAUGUGGA).
Before conducting the single tubule PCR, we validated the RT-PCR reaction with the mRNA purified from the kidney as temple. The miR-194 primers (2.5 μl, 12.5 nM) were mixed with 2 μl cDNA (200 ng) and 12.5 μl 2× SYBR Green master. MxPro3000 (Stratagene) was used to carry out the experiments and we used 2−ΔΔCT to analyze the comparative expression level of miR-194. U6 was used as a control gene and forward and reversal sequence were “CTCGCTTCGGCAGCACATA” and “ATATGGAACGCTTCACGAATT,” respectively.
Cell culture and transfection.
HEK293T cells were used in the present study and culture methods were described previously (1, 19). For each transfection experiment, 2 μg of plasmid DNA per 35-mm dish were mixed with 200 μl Opti-MEM culture medium (GIBCO) containing 6 μl of TransIT-293 (Mirus, Madison, WI). We used flag-tagged ITSN1-3′UTR, UTR-free flag-tagged ITSN1 cloned into pTracerCMV vector which contains green fluorescent protein (GFP) driven by a different promoter, and GFP-tagged ROMK cloned into pcDNA3.1. To overexpression of miR-194, we also inserted pre-miR-194 into BamHI and HindIII of vector pSilenceCMV to generate the miR-194 expressing vector-pSilenceCMV-194. The primers used to amplify pre-miR-194 from human gDNA were synthesized from IDT (Coralville, IA) and their sequences were ACTTGGATCCCTTATTTAGAACATGAA (sense) and GGCCGAAGCTTATACCTTTATATTGTAG (antisense).
For biochemical experiments, cells were harvested 24 or 48 h after transfection. The methods for cell lysis and sample preparation have been previously described (19).
Luciferase assay.
To obtain the full length of human ITSN1 3′UTR, the sequences used as forward and reversed primers are CCGCTCGAGATCATATGTTGTCCATCCC and ATTTGCGGCCGCCACGGAACTAATTCAAAAGG, respectively. To generate mutant ITSN1 3′UTR, in which miR-194 seed sequence was deleted, we used the primer (reversed) whose sequence was ATTTGCGGCCGCCAACAAGGTACAGCTGATA. The full length of ITSN1-3′UTR (1,478 bp) was inserted into XbaI site of pGL3 vector (Promega; downstream of a constitutively active luciferase cassette) to form ITSN1-3′UTR construct. HEK cells were transfected with the ITSN1-3′UTR construct or a mutant of ITSN1-3′UTR and with commercially available miR-194 mimic, inhibitor, or scrambled oligonucleotide (Qiagen). The luciferase activity was measured 24 h after transfection by Dual-Glo luciferase assay system with Glomax 20/20 luminometer (Promega).
Biotinylation.
The HEK293T cells were transfected with GFP-ROMK1+negative control nucleotides (nt), GFP-ROMK1+ miR-194 mimic, GFP-ROMK1+miR-194 +ITSN1 for 24 h. The cells were gently washed and incubated with 2 mg/ml EZ-Link Sulfo-NHS-SS-Biotin in 1× PBS for 1 h at 4°C. The biotin labeling was ended with 100 mM glycine in 1× PBS to absorb the unbound biotin. The cells were washed twice with 1× TBS and lysed with RIPA buffer (100 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate) in the presence of a protease inhibitor cocktail and phosphatase inhibitor I and II (Sigma). After centrifugation, the protein sample (100 μg in 20 μl) was mixed with 20 μl immobilized NeutrAvidin Gel in 500 μl RIPA buffer and placed on a rocker overnight at 4°C. The bead fraction was harvested, mixed with 2× sample buffer, boiled for 5 min, and resolved by electrophoresis. The biotin-labeled ROMK1 was detected with ROMK antibody or GFP antibody. The surface expression of ROMK1 among different groups was normalized by calculating the ratio between the surface and the total expression of ROMK1.
Electrophysiology experiments.
Within 24 h after transfection, the cells were treated with trypsin-containing medium (Tryple Ecpresscare; GIBCO) for 10 min to detach the cells. The cell suspension (0.2 ml in volume) was carefully removed to a 5 × 5-mm cover glass coated with polylysin followed by additional incubation for 30 min to allow the cells to adhere to the cover glass. The cover glass was transferred to a chamber (1 ml) mounted on the stage of a Nikon inverted microscope. We carried out the perforated whole cell patch-clamp experiments at room temperature. The cells were incubated with a bath solution containing 140 mM KCl, 1.8 mM MgCl2, 1.8 mM CaCl2, and 10 mM HEPES (pH 7.4). Fluorescence signal (an indication of positive transfection) was detected with an intensified video imaging system including SIT 68 camera (Long Island Industries). Borosilicate glass (1.7-mm OD) was used to make the patch-clamp pipettes that were pulled with a Narishege electrode puller. The pipette had a resistance of 2–4 MΩ when filled with 140 mM KCl. The tip of the pipette was filled with pipette solution containing 140 mM KCl, 1.8 mM MgCl2, 1 mM CaCl2, 1 mM EGTA, and 5 mM HEPES (pH 7.4). The pipette was then back-filled with amphotericin B (2 μg/0.1 ml) containing pipette solution. After a high-resistance seal (>2 GΩ) was formed, the membrane capacitance was monitored until the whole cell patch configuration was formed. The cell membrane capacitance was measured and compensated. The K currents were measured by an Axon 200A patch-clamp amplifier. The currents were low-pass filtered at 1 kHz and digitized by an Axon interface (Digidata 1200) and were analyzed using the pClamp software system 9 (Axon). The cell membrane capacitance was measured and compensated and K currents were presented as pA/25 pF.
Experimental materials and statistics.
Actin antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) while Flag and GFP antibodies were purchased from Sigma (St. Louis, MO) and Clontech Lab (Mountain View, CA), respectively. Antibody for ITSN1 was a gift from Dr. J. O'Bryan. We purchased miR-194 mimic/inhibitor and corresponding control oligo from Qiagen (Foster City, CA). GFP-ROMK1 or flag-tagged ROMK1 was subcloned into pcDNA3 (Invitrogen, San Diego, CA) as described previously (29). The data are presented as means ± SE. We used the paired Student's t-test or one-way ANOVA to determine the statistical significance.
RESULTS
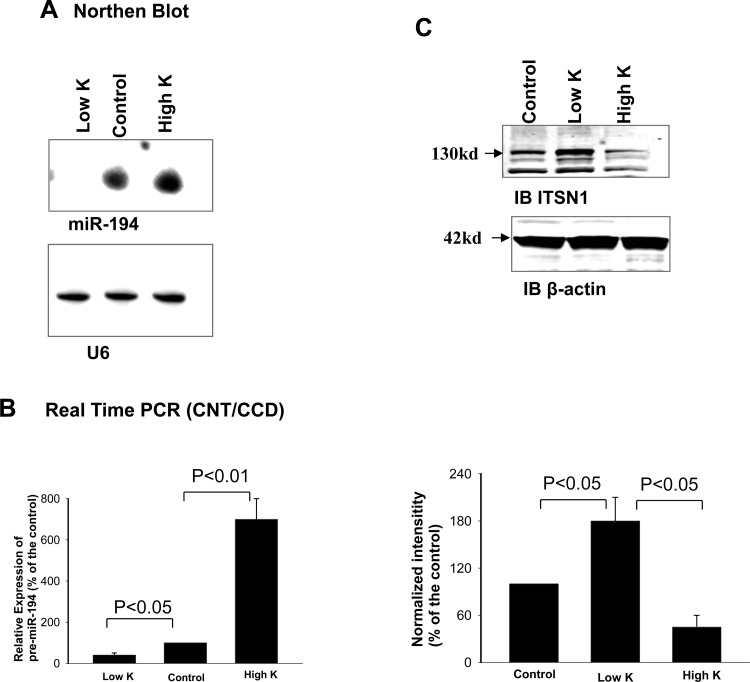
The microRNA array assay performed with renal tissue of mice on a HK, an NK (control), and a LK diet for 7 days showed that HK increased the expression of miR-802, miR-665 and miR-21 while LK intake increased the expression of miR-142, miR-144, miR-146a, miR-223, miR-451, and miR-455 (data not shown). Since miR-194 has been reported to be expressed highly in the distal nephrons (30), we carried out Northern blot experiments to examine whether dietary K intake regulated the expression of miR-194 in the mouse kidney using a 32P-labeled probe containing oligo-nucleotides (TCCACATGGAGTTGCTGTTACA) complementary to mmu-miR194. Figure 1A is a representative Northern blot from four such experiments demonstrating that HK intake increased the expression of the mature form of miR-194 by 80 ± 20%, whereas a LK intake decreased the expression of miR-194 by 60 ± 20% in the mouse kidney. In contrast, changes in dietary K intake had no significant effect on U6 expression. Because an alteration of dietary K intake has been shown to affect K transport in the CNT and CCD and miR-194 is highly expressed in the distal nephron (30), we then used the real-time PCR technique to examine whether changes in dietary K intake had an effect on miR-194 expression in the CNT and CCD. We harvested RNA from the isolated CNT and CCD of mice on control, HK, or LK diet for 7 days and conducted qRT-PCR assay. We confirmed the previous finding that HK intake significantly and specifically increased the expression of miR-802 and decreased the expression of miR-192 in the CNT/CCD (4, 20). Moreover, a HK intake increased miR-194 expression in the CNT/CCD. Figure 1B summarizes the results from four such experiments demonstrating that HK intake significantly increased (600 ± 100%) while LK intake significantly decreased pre-miR-194 expression (by 60 ± 10%) compared with those on control diet. Thus, our data suggest that dietary K intake regulates the expression of miR-194 in the CNT/CCD.
Fig. 1.
High-K intake increases miR-194 transcription and decreases intersectin 1 (ITSN1) expression. A: Northern blot shows the effect of dietary K intake on the mature form of miR-194 and U6 in the mouse kidney. The sequence of oligo probe (mmu-miR-194) used for the Northern blot was TCCACATGGAGTTGCTGTTACA and probe sequence of U6 was ATATGGAACGCTTCACGAATT. The probes were labeled with [γ-32P]. The RNA isolated from the kidney was hybridized with the probe, and the miR-194 was detected by X-ray film. Mice were kept on low-K (<0.001%), control (1% K), or high-K (5%) diets for 7 days. B: bar graph summarizes results of real-time PCR experiments showing the relative expression of pre-miR-194 in the isolated connecting tubule (CNT) and cortical collecting duct (CCD) of mice on low-K, control, or high-K diet for 7 days. C: Western blot shows the effect of dietary K intake on the expression of ITSN1 in the mouse cortex tissue. A bar graph summarizes results showing normalized changes in ITSN1 expression (bottom).
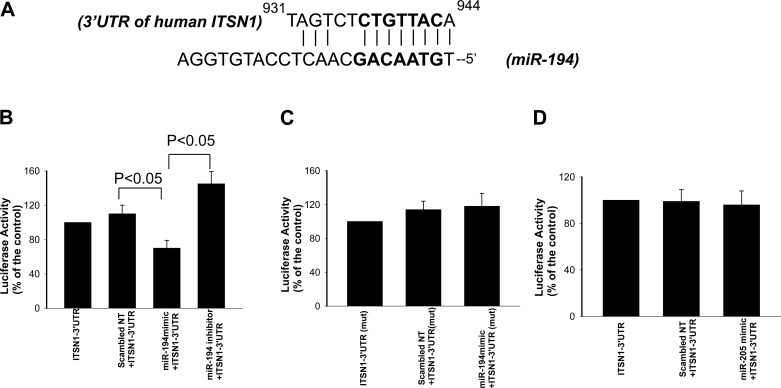
We then searched the potential target protein for miR-194 through databases (Target-scan, Miranda, and miRDB) and identified that human ITSN1 to 3′UTR contains a putative binding site of miR-194 at nt931 to 944 (Fig. 2A). This sequence is also conserved in mouse and rats. If dietary K intake regulates the expression of ITSN1 through miR-194, we speculate that changes in dietary K intake should alter the expression of ITSN1 in an opposite way as it affects miR-194 expression. Thus, we examined the effect of dietary K intake on the expression of ITSN1 in the mouse kidney. Figure 1C is a Western blot showing that LK intake increased (80 ± 20%) while HK intake decreased (55 ± 15%) the expression of ITSN1 in the kidney. This suggests the possibility that miR-194 might indeed regulate the expression of ITSN1. To test whether miR-194 regulates ITSN1 translation, we cloned a full-length 3′UTR (1,478 bp) of the human ITSN1 gene into the downstream of a constitutively active luciferase cassette. HEK293T cells were transfected with the vector containing the ITSN1 to 3′UTR (ITSN1–3′UTR) and the effect of mir-194 on luciferase activity was examined 24 h after the transfection. The experimental results (n = 5) are summarized in Fig. 2B showing that cotransfection of miR-194 mimics significantly decreased activity of the luciferase (30 ± 7%) while suppression of miR-194 increased the activity (45 ± 6%) in cells transfected with vector containing human ITSN1–3′UTR. The effect of miR-194 on luciferase activity was specifically related to the presence of the seed sequence of ITSN1-3′UTR because miR-194 did not affect luciferase activity in the cells transfected with mutant ITSN1-3′UTR without seed sequence (Fig. 2C). Finally, we examined the effect of miR-205 expression on the luciferase activity in HEK293T cells transfected with ITSN1-3′-UTR. Because miR-205 is not detected in the CCD and is also not predicted to bind with ITSN1-3′UTR (data not shown), the experiments would be served as a negative control. As demonstrated in Fig. 2D, expression of miR-205 had no effect on the luciferase activity in the cells transfected with ITSN1-3′UTR. These data strongly suggest that miR-194 plays a role in regulating the expression of ITSN1.
Fig. 2.
Mir-194 interacts with 3′UTR of ITSN1. A: putative anneal site of mature human miR-194 to the human ITSN1 to 3′UTR. The bold fronts indicate the seed sequence. B: expression of miR-194 mimic decreases while expression of miR-194 inhibitor increases the activity of luciferase report gene in HEK293T cells transfected with a construct containing the ITSN-3′UTR. Scrambled nucleotide sequence (nt-control) was used as a negative control. C: bar graph showing that miR-194 mimic did not affect the activity of luciferase report gene in the cells transfected with mutant ITSN1-3′UTR (mut 3′UTR) in which the seed sequence for mir-194 was deleted. D: bar graph showing that miR-205 mimic did not affect the activity of luciferase reporter gene in the cells transfected with ITSN1-3′UTR.
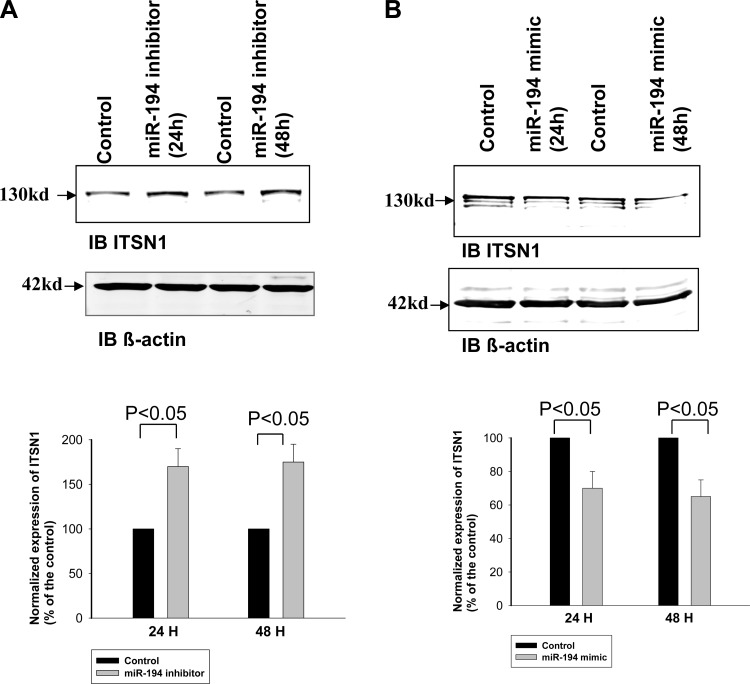
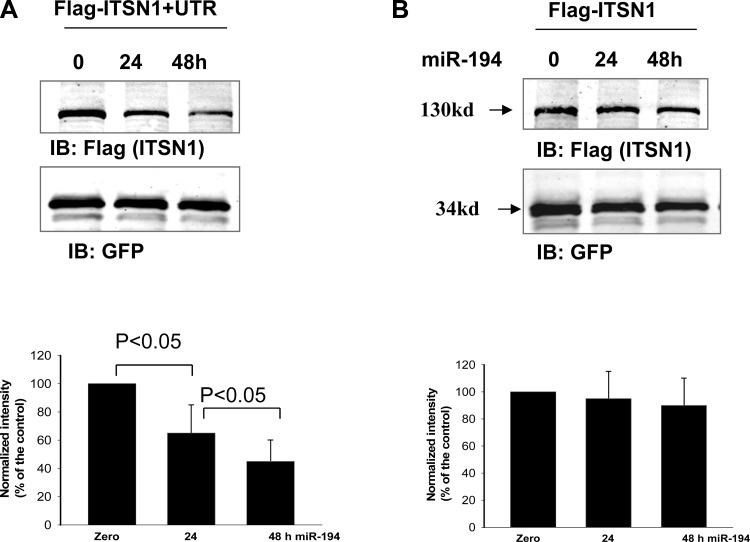
We next examined whether inhibiting endogenous miR-194 expression could increase the translation of endogenous ITSN1 in HEK293T cells. Figure 3A is a Western blot showing the effect of inhibiting endogenous miR-194 on ITSN1 expression in HEK293T cells. Results from four such experiments are summarized in a bar graph (bottom) showing that transfection of the cells with commercially available miR-194 inhibitor for 24 or 48 h increased the expression of endogenous ITSN1 by 70 ± 20 and 75 ± 20%, respectively, whereas the application of control oligonucleotide had no significant effect on ITSN1 expression (data not shown). We also examined the effect of expression of miR-194 mimic on ITSN1 expression in HEK cells. Figure 3B is a Western blot showing that transfection of the cells with commercially available miR-194 mimic for 24 or 48 h decreased the expression of endogenous ITSN1 by 30 ± 8 and 35 ± 10% (n = 4), respectively. To confirm that the effect of miR-194 on ITSN1 expression depends on its 3′UTR, we examined the effect of miR-194 on ITSN1 expression in HEK293T cells transfected with flag-tagged human ITSN1 with or without 3′UTR. The flag-tagged ITSN1 was cloned into pTracerCMV vector which contained GFP driven by a different promoter. Figure 4A is a Western blot demonstrating the expression of flag-tagged ITSN1 in HEK293T cells 24 or 48 h after transfection of miR-194 mimic. From the inspection of Fig. 4A, it is apparent that expression of miR-194 significantly decreased ITSN1 expression by 35 ± 15% (24 h) and 60 ± 20% (48 h; n = 5). We also repeated the experiments in HEK293T cells transfected with flag-tagged human ITSN1 without its 3′UTR. Figure 4B is a Western blot showing the expression of flag-tagged ITSN1 24 and 48 h after transfection of miR-194 mimic. Results (n = 4) are summarized in a bar graph (bottom) demonstrating that miR-194 had no significant effect on ITSN1 expression in the cells transfected with 3′UTR-free ITSN1. This suggests that the effect of miR-194 on ITSN1 expression is achieved by modulating the function of ITSN1–3′UTR.
Fig. 3.
Inhibition of miR-194 increases the expression of endogenous ITSN1. A: Western blot shows the effect of miR-194 inhibitor on the expression of endogenous ITSN1 in HEK293T cells. A bar graph demonstrates the normalized expression of ITSN1 at 24 and 48 h after the transfection (bottom). B: Western blot shows the effect of miR-194 mimic on the expression of endogenous ITSN1 in HEK293T cells. A bar graph demonstrates the normalized expression of ITSN1 at 24 and 48 h after the transfection (bottom).
Fig. 4.
miR-194 regulates the expression of ITSN1. A: expression of miR-194 decreases the expression of ITSN1 expression but not the expression of green fluorescent protein (GFP) in HEK293T cells transfected with flag-tagged ITSN1-3′UTR cloned into pTracerCMV vector which contains GFP driven by a different promoter. A bar graph demonstrates the normalized expression of ITSN1 at 0, 24, and 48 h after the transfection (bottom). B: mir-194 failed to inhibit 3′UTR-free ITSN1 expression. A Western blot shows the effect of miR-194 on the expression of ITSN1 in the cells transfected with flag-tagged and 3′UTR-free-ITSN1 cloned into pTracerCMV vector. The results are summarized in a bar graph (bottom).
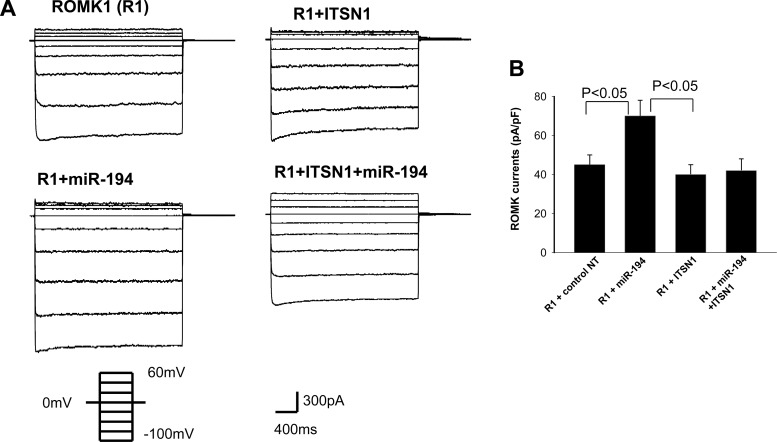
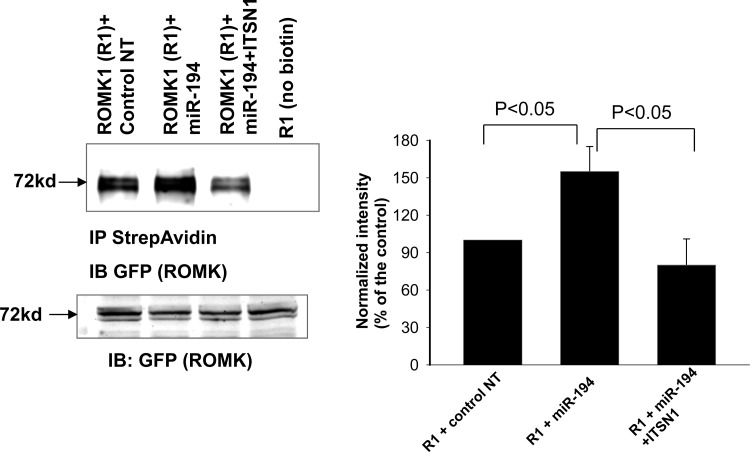
ITSN1 has been shown to regulate ROMK channel activity by stimulating WNK-induced endocytosis of ROMK channels (8). Thus, it is conceivable that miR-194 should have an effect on ROMK channel activity through regulating ITSN1 expression. To test this notion, we used the perforated whole cell recording to examine the effect of miR-194 on ROMK channel activity in HEK293T cells transfected with GFP-tagged ROMK1 channel. Twenty-four hours after the transfection, the patch-clamp experiments were performed in GFP-positive cells, an indication of successful expression of ROMK. Figure 5A is a set of whole cell recording showing the Ba2+-sensitive K currents measured from −100 to 60 mV at 20-mV step. Expression of pre-miR-194 significantly increased K currents from 45 ± 5 to 70 ± 8 pA/pF (n = 9). The stimulatory effect of miR-194 on ROMK channels was completely abolished in the cells cotransfected with human ITSN1 (K currents, 42 ± 6 pA/pF), a value that was not significantly different from that measured in cells transfected with human ITSN1 alone (40 ± 5pA/pF). The reason that expression of ITSN1 failed to further decrease ROMK channel currents was due to the possibility that ITSN1 was not a rate-limiting factor for mediating WNK-dependent inhibition of ROMK under control conditions. Since the endogenous ITSN1 level is high enough to interact with WNK for inhibiting ROMK channels, expression of exogenous ITSN1 could not further enhance ITSN1-WNK interaction and would not decrease ROMK channels. To examine whether miR-194 expression-induced increase in ROMK channel activity was the result of enhancing ROMK channel surface expression, we used biotin-labeling technique to measure the surface expression of ROMK channels. ROMK1-transfected HEK293T cells were cotransfected with control oligonucleotide, pre-miR-194 or pre-miR-194+ITSN1. Figure 6 is a Western blot showing that expression of pre-miR-194 significantly increased ROMK surface expression by 55 ± 15% (n = 4), an effect that was blocked by cotransfected ITSN1 (80 ± 22% of the control). Figure 6 also showed that the total ROMK expression was similar among four groups. Therefore, the expression of miR-194 increased ROMK channel activity by enhancing surface expression of the K channels. This effect was most likely achieved by inhibiting ITSN1-WNK-induced endocytosis of ROMK because coexpression of ITSN1 reversed the stimulatory effect of miR-194 on ROMK channels.
Fig. 5.
Expression of miR-194 increases ROMK channel activity. A: whole cell recordings show the effect of expression of ITSN1 and miR-194 on the Ba2+-sensitive K currents. The experiments were performed 24 h after transfection with the perforated whole cell recording at the voltage from −100 to 60 mV at a 20-mV step. B: bar graph summarizes the above data from 6 such experiments at −100 mV. The bath and pipette solutions contained 140 mM KCl, 1.8 mM MgCl2, 1.8 mM CaCl2, and 10 mM HEPES (pH 7.4). The HEK293 T cells were transfected with GFP-tagged ROMK1 and pre-miR-194 which was cloned into pSilencerCMV vector. The HEK293T cells were transfected with GFP-ROMK1 (1 μg) + 1 μg negative control nucleotides (nt), 1 μg GFP-ROMK1 + 0.5 μg miR-194 mimic + 0.5 μg nt, 1 μg GFP-ROMK1 + 0.5 μg miR-194 + 0.5 μg ITSN1 for 24 h.
Fig. 6.
Mir-194 increases ROMK expression in the plasma membrane. HEK293T cells were transfected with GFP-tagged ROMK1 with control nt, pre-miR-194, or pre-miR-194+ITSN1 for 24 h. The biotin-labeled ROMK1 was immunoprecipitated with neutravidin beads and detected with GFP (ROMK) antibody. The surface expression of ROMK1 was normalized by calculating the ratio between the surface and total expression of ROMK1. The HEK293T cells were transfected with GFP-ROMK1 (1 μg) + 1 μg negative control nucleotides (nt), 1 μg GFP-ROMK1 + 0.5 μg miR-194 mimic + 0.5 μg nt, 1 μg GFP-ROMK1 + 0.5 μg miR-194 + 0.5 μg ITSN1 for 24 h.
DISCUSSION
MicroRNAs play a role not only in regulating renal function under physiological conditions but also in modulating the pathological process in the kidney. For example, miR-15a has been shown to regulate the expression of the cell division cycle 25 homolog A (Cdc25A). Moreover, a decrease in miR-15a expression was associated with accelerating cell proliferation and increasing cyst growth (15). Alteration in miR-192 expression is associated with renal fibrosis through modulating TGF-β-dependent signaling (3, 12). Also, miR-192 is highly expressed in the proximal tubule and regulates the expression of Na-K-ATPase β-subunit (25). Our previous and the present studies strongly suggested that microRNA plays a role in the regulation of renal K secretion and, specifically, in mediating the effect of dietary K intake on ROMK channels (20).
The main finding of the present study is that dietary K intake regulates miR-194 expression such that HK intake increased while LK intake decreased miR-194 transcription in the kidney. MicroRNA-194 has two loci in chromosome 1 and 11, defined as miR-194-1 and miR-194-2. Moreover, miR-194-2 is clustered with miR-192 in 11q13.1. Interestingly, a HK intake decreased miR-192 expression but increased miR-194 expression in the CNT/CCD, suggesting the possibility that HK may regulate miR-194 by a mechanism different from that of miR-192. However, further study is required to explore how a HK intake stimulates miR-194 expression. Three lines of evidence strongly suggest that miR-194 regulates the expression of ITSN1 through modulating its 3′UTR. First, inhibition of miR-194 increased while stimulation of miR-194 decreased luciferase activity in the cells transfected with 3′UTR of ITSN1 but it had no effect in those without 3′UTR. Second, inhibition of miR-194 increased the expression of endogenous ITSN1 in HEK293T cells. Third, expression of pre-miR-194 decreased the translation of exogenous ITSN1 with its 3′UTR but it had no effect on ITSN1 in the cells transfected with 3′UTR-free constructor.
The possibility that miR-194 modulates the expression of ITSN1 in the kidney is also suggested by the observation that dietary K intake regulated the expression of ITSN1 such that a HK intake decreased while a LK intake increased the expression of ITSN1. ITSN1 is ubiquitously expressed in a variety of tissues including the kidney and it has two EH (Eps15 homology) domains, a coiled-coil region, and five SH3 (Src-homology 3) domains (27). Moreover, it has been demonstrated that NH2-terminal proline-rich motifs of WNK1 interacted with the 3rd SH3 domain of ITSN1. ITSN1 has been shown to be expressed in the aldosterone-sensitive distal nephron and plays a role in regulation of ROMK channel expression (8). Therefore, it has been suggested that ITSN1 links WNK kinases to endocytosis of ROMK channels.
ROMK channels are expressed in the apical membrane of aldosterone-sensitive distal nephron including the late distal convoluted tubule, CNT, and CCD (9). It is well-established that ROMK channels are mainly responsible for K secretion under control conditions (NK diet) and also are involved in K secretion during increasing dietary K intake (32). Several protein kinases including WNK, Src family protein tyrosine kinase (SFK), serum-glucocorticoid-induced kinase (SGK) have been shown to play a role in regulating ROMK channel activity (11, 28, 36). For instance, phosphorylation of the ROMK1 channel by SGK1 stimulates the export of ROMK1 (26, 35), whereas WNK and SFK enhance ROMK endocytosis (8, 10, 17, 21, 23, 31). Moreover, WNK1 and WNK4 have been demonstrated to inhibit ROMK channels by stimulation of endocytosis (13, 28, 31).
A large body of evidence demonstrates that dietary K intake plays a key role in the regulation of ROMK channel activity (1, 6, 16, 20, 32, 34): an increase in dietary K intake augments apical ROMK channel expression in the CNT/CCD, whereas a decrease in dietary K intake has an opposite effect (2, 18). The effect of dietary K intake on ROMK channel activity is at least partially achieved by modulating the effect of WNK on ROMK channels. For instance, a LK intake has been shown to enhance the inhibitory effect of full-length WNK1 by suppressing the expression of the kidney-specific WNK1 (KS-WNK1) which inhibits the effect of full-length WNK1 (13, 31). Conversely, an HK intake diminished the inhibitory effect of full-length WNK1 on ROMK by increasing KS-WNK1 expression. Also, an HK intake promotes SGK1-induced WNK4 phosphorylation which stimulates ROMK channel activity (36). The role of microRNA in mediating the effect of dietary K intake on ROMK channels has been suggested by our previous experiments (20). We demonstrated that HK intake stimulated miR-802 expression in the collecting duct and that the upregulation of miR-802 stimulated the ROMK channel activity by targeting calveolin-1. The present study further demonstrated that an HK intake also increased the expression of miR-194 and that miR-194 is involved in stimulating ROMK channel activity. We speculate that miR-802 and miR-194 should have a synergistic effect on ROMK channel activity rather than two independent actions. Moreover, it is very likely that a mechanism other than miR-194 could also be responsible for the regulation of ITSN1 expression in the CNT/CCD.
The physiological significance of the present study is to illustrate the role of miR-194 in mediating the effect of dietary K intake on ROMK channels. Moreover, two lines of evidence suggest that miR-194-induced regulation of ROMK channels is the result of suppressing the ITSN1 expression: 1) miR-194-induced increase in ROMK channel currents was abolished by coexpression of ITSN1; 2) expression of exogenous ITSN1 reversed the stimulatory effect of miR-194 on ROMK channel surface expression. Previous studies demonstrated that HK intake decreased while LK intake enhanced the WNK-induced endocytosis of ROMK channels (23, 31). Since ITSN1 is involved in mediating the WNK-induced endocytosis of ROMK channels, a decrease in ITSN1 induced by HK intake is expected to inhibit the endocytosis and to increase the surface expression of ROMK channels in the distal nephron. However, an increase in ITSN1 induced by LK intake should enhance WNK-induced endocytosis of ROMK. Although we showed the role of miR-194 in regulating ITSN1 expression, miR-194-induced downregulation of ITSN1 may play a role in the regulation of ROMK channel only during changing dietary K intake. Other microRNAs may also be involved in the regulation of ITSN1 in response to different physiological stimuli. We conclude that miR-194 plays a role in mediating the effect of dietary K intake on ROMK channels and that the effect of miR-194 is achieved by regulating the expression of ITSN1 in the aldosterone-sensitive distal nephron.
GRANTS
The work is supported by National Institutes of Health Grant DK54983 and D.-H. Lin is supported by American Heart Association Scientific Development Award.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: D.-H.L. and W.-H.W. conception and design of research; D.-H.L., P.Y., and C.Z. performed experiments; D.-H.L., P.Y., C.Z., and W.-H.W. analyzed data; D.-H.L. and W.-H.W. interpreted results of experiments; D.-H.L. and W.-H.W. prepared figures; D.-H.L. and W.-H.W. drafted manuscript; D.-H.L. and W.-H.W. edited and revised manuscript; D.-H.L. and W.-H.W. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. John O'Bryan for ITSN1 antibody.
REFERENCES
- 1.Babilonia E, Wei Y, Sterling H, Kaminski P, Wolin MS, Wang WH. Superoxide anions are involved in mediating the effect of low K intake on c-Src expression and renal K secretion in the cortical collecting duct. J Biol Chem 280: 10790–10796, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu PY, Quigley R, Babich V, Huang CL. Dietary potassium restriction stimulates endocytosis of ROMK channel in rat cortical collecting duct. Am J Physiol Renal Physiol 285: F1179–F1187, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Chung ACK, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-β/Smad3-driven renal fibrosis. J Am Soc Nephrol 21: 1317–1325, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elvira-Matelot E, Zhou XO, Farman N, Beaurain GV, Henrion-Caude A, Hadchouel J, Jeunemaitre X. Regulation of WNK1 expression by miR-192 and aldosterone. J Am Soc Nephrol 21: 1724–1731, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flunt AS, Lai EC. Biological principals of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genetics 9: 831–842, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frindt G, Shah A, Edvinsson J, Palmer LG. Dietary K regulates ROMK channels in connecting tubule and cortical collecting duct of rat kidney. Am J Physiol Renal Physiol 296: F347–F354, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci USA 107: 14339–14344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He G, Wang HR, Huang SK, Huang CL. Intersectin links WNK kinase to endocytosis of ROMK1. J Clin Invest 117: 1078–1087, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebert SC, Desir G, Giebisch G, Wang W. Molecular diversity and regulation of renal potassium channels. Physiol Rev 85: 319–371, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahle KT, Wilson FH, Leng Q, Lalioti MD, O'Connell AD, Dong K, Rapson AK, MacGregor GG, Giebisch G, Hebert SC, Lifton RP. WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet 35: 372–376, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Kahle KT, Ring AM, Lifton RP. Molecular physiology of the WNK kinases. Annu Rev Physiol 70: 329–355, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 21: 438–447, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazrak A, Liu Z, Huang CL. Antagonistic regulation of ROMK by long and kidney-specific WNK1 isoforms. Proc Natl Acad Sci USA 103: 1615–1620, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SO, Masyuk T, Splinter P, Banales JSM, Masyuk A, Stroope A, LaRusso N. MicroRNA15a modulates expression of the cell cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest 118: 3714–3724, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin DH, Sterling H, Lerea KM, Welling P, Jin L, Giebisch G, Wang WH. K depletion increases the protein tyrosine-mediated phosphorylation of ROMK. Am J Physiol Renal Physiol 283: F671–F677, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin DH, Sterling H, Wang WH. The protein tyrosine kinase-dependent pathway mediates the effect of K intake on renal K secretion. Physiology (Bethesda) 20: 140–146, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Lin DH, Sterling H, Yang B, Hebert SC, Giebisch G, Wang WH. Protein tyrosine kinase is expressed and regulates ROMK1 location in the cortical collecting duct. Am J Physiol Renal Physiol 286: F881–F892, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin DH, Yue P, Pan CY, Sun P, Zhang X, Han Z, Roos M, Caplan M, Giebisch G, Wang WH. POSH stimulates the ubiquitination and the clathrin-independent endocytosis of ROMK1 channels. J Biol Chem 284: 29614–29624, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol 22: 1087–1098, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin D, Kamsteeg EJ, Zhang Y, Jin Y, Sterling H, Yue P, Roos M, Duffield A, Spencer J, Caplan M, Wang WH. Expression of tetraspan protein CD63 activates protein-tyrosine kinase (PTK) and enhances the PTK-induced inhibition of ROMK channels. J Biol Chem 283: 7674–7681, 2008 [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 55: 974–982, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Wang HR, Huang CL. Regulation of ROMK channel and K+ homeostasis by kidney-specific WNK1 kinase. J Biol Chem 284: 12198–12206, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macconi D, Tomasoni S, Romagnani P, Trionfini P, Sangalli F, Mazzinghi B, Rizzo P, Lazzeri E, Abbate M, Remuzzi G, Benigni A. MicroRNA-324–3p promotes renal fibrosis and is a target of ACE inhibition. J Am Soc Nephrol 23: 1496–1505, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mladinov D, Liu Y, Mattson DL, Liang M. MicroRNAs contribute to the maintenance of cell type-specific physiological characteristics: miR-192 targets Na+-K+-ATPase +1. Nucleic Acids Res 41: 1273–1283, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connell AD, Leng Q, Dong K, MacGregor GG, Giebisch G, Hebert SC. Phosphorylation-regulated endoplasmic reticulum retention signal in the renal outer medullary K+ channel (ROMK). Proc Natl Acad Sci USA 102: 9954–9959, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pucharcos C, Casas C, Nadal M, Estivill X, de la Luna S. The human intersectin genes and their spliced variants are differentially expressed. Biochim Biophys Acta 1521: 1–11, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Ring AM, Leng Q, Rinehart J, Wilson FH, Kahle KT, Hebert SC, Lifton RP. An SGK1 site in WNK4 regulates Na+ channel and K+ channel activity and has implications for aldosterone signaling and K+ homeostasis. Proc Natl Acad Sci USA 104: 4025–4029, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sterling H, Lin DH, Gu RM, Dong K, Hebert SC, Wang WH. Inhibition of protein-tyrosine phosphatase stimulates the dynamin-dependent endocytosis of ROMK1. J Biol Chem 277: 4317–4323, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Z, Creene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 18: 404–411, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wade JB, Fang L, Liu J, Li D, Yang CL, Subramanya AR, Maouyo D, Mason A, Ellison DH, Welling PA. WNK1 kinase isoform switch regulates renal potassium excretion. Proc Natl Acad Sci USA 103: 8558–8563, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W. Regulation of renal K transport by dietary K intake. Annu Rev Med 66: 547–569, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol 21: 756–761, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Y, Bloom P, Lin DH, Gu RM, Wang WH. Effect of dietary K intake on the apical small-conductance K channel in the CCD: role of protein tyrosine kinase. Am J Physiol Renal Physiol 281: F206–F212, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Yoo D, Kim BY, Campo C, Nance L, King A, Maouyo D, Welling PA. Cell surface expression of the ROMK (Kir 1.1) channel is regulated by the aldosterone-induced kinase, SGK-1, and protein kinase A. J Biol Chem 278: 23066–23075, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Yue P, Lin DH, Pan CY, Leng Q, Giebisch G, Lifton RP, Wang WH. Src family protein tyrosine kinase (PTK) modulates the effect of SGK1 and WNK4 on ROMK channels. Proc Natl Acad Sci USA 106: 15061–15066, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]