Abstract
Staphylococcus epidermidis, which is a causative pathogen of nosocomial infection, expresses its virulent traits such as biofilm and autolysis regulated by two-component signal transduction system SaeRS. In this study, we performed a proteomic analysis of differences in expression between the S. epidermidis 1457 wild-type and saeRS mutant to identify candidates regulated by saeRS using two-dimensional gel electrophoresis (2-DE) combined with matrix-assisted laser desorption/lonization mass spectrometry (MALDI-TOF-MS). Of 55 identified proteins that significantly differed in expression between the two strains, 15 were upregulated and 40 were downregulated. The downregulated proteins included enzymes related to glycolysis and TCA cycle, suggesting that glucose is not properly utilized in S. epidermidis when saeRS was deleted. The study will be helpful for treatment of S. epidermidis infection from the viewpoint of metabolic modulation dependent on two-component signal transduction system SaeRS.
1. Background
With all kinds of artificial medical materials commonly used, the opportunistic pathogen Staphylococcus epidermidis has become a major pathogen of nosocomial infection [1, 2]. S. epidermidis pathogenesis is associated with its ability to form biofilms on the surface of medical indwelling materials [3]. The biofilms have the ability of resistance to antibiotics and the host immune system.
Biofilm formation proceeds in two distinct developmental phases: primary attachment of staphylococcal cells to a polystyrene surface followed by bacterial accumulation in multiple layers. During the initial stage of biofilm formation, extracellular DNA (eDNA) is critical for bacterial attachment [4]. Extracellular DNA release from S. epidermidis is related to AtlE-mediated bacterial autolysis. During the bacterial accumulation phase in S. epidermidis, biofilm formation is mediated by polysaccharide intercellular adhesin (PIA) synthesized by icaADBC operon-encoded enzymes [5].
Glucose can induce the S. aureus biofilm formation by inducing ica gene. The addition of glucose to growth media triggered biofilm formation in S. aureus strain SA113 [6], increased the expression of genes and enzymes of the glycolytic pathway, while genes and proteins of the tricarboxylic acid (TCA) cycle, required for the complete oxidation of glucose, were repressed via catabolite control protein A (CcpA) [7]. Deletion of CcpA, which regulates gene expression in response to the carbon source, abolished the capacity of SA113 to form a biofilm. Biofilm formation is closely related to bacteria energy metabolism and cell wall synthesis [5]. Disintegration of S. epidermidis biofilms under glucose-limiting conditions depends on the activity of the alternative sigma factor sigmaB [8].
Glycolysis that is the principal pathway of glucose metabolism occurs in the bacterial cytoplasm where glucose is oxidized to pyruvate (in aerobic condition) or lactate (in anaerobic condition) and generates energy in the form of ATP. The accumulation of organic acids in the culture medium occurred coincident with a decreased pH. The pH change coincided with changes in cell viability, cell ATP, and the ability to produce acidity by glucose fermentation [9]. Glyceraldehyde 3-phosphate dehydrogenase is an essential enzyme that catalyzes the sixth step of glycolysis and thus serves to break down glucose for energy and carbon molecules [10]. Glucose metabolism of Escherichia coli strain NP315 fda (encoding fructose-1, 6-diphosphate aldolase) and gnd (encoding 6-phosphogluconate dehydrogenase) mutant was decreased [11].
Staphylococcal biofilm formation is a complicated process that is regulated by two-component signal transduction systems (TCSs) [12, 13]. Prokaryotic TCSs modulate gene expression in response to different environmental signals [14]. SaeRS TCS was originally identified in Staphylococcus aureus as a transposon mutant system deficient in the synthesis of several exoproteins, including α- and β-hemolysin and coagulase [15]. In S. aureus COL, proteomic approach showed that extracellular proteins were influenced by a mutation in saeS [16]. In a previous study of the S. epidermidis SaeRS TCS, a saeR deletion mutant exhibited a lower anaerobic growth rate, a significantly reduced rate of nitrate utilization, and a slightly higher biofilm-forming ability compared to the parental strain [17]. In S. epidermidis, the TCS SaeRS is known to be involved in biofilm formation. Furthermore, SaeRS was found to be involved in S. epidermidis autolysis, resulting from altered bacterial eDNA release [12].
In the present study, the influence of the TCS SaeRS on global gene expression in S. epidermidis was investigated by proteomic approach. SaeRS positively regulates the proteins expression involved in glucose metabolism. Furthermore, we show that glucose metabolism by decreasing the pH value affects biofilm-forming ability and bacterial cell viability.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Media
The bacterial strains used in this study are S. epidermidis wild-type (WT), S. epidermidis saeRS mutant (SAE), and S. epidermidis saeRS complemental strain (SAEC). S. epidermidis cells were grown at 37°C in BM medium (Tryptone 10 g, Yeast extract 5 g, NaCl 5 g, K2HPO4 1 g, and Glucose 1 g/L medium) or tryptic soy broth (TSB) (Oxiod, Basingstoke, Hampshire, England) supplemented with antibiotics when necessary. Spectinomycin (spc) was used at 300 μg/mL for S. epidermidis.
2.2. Sample Preparation for Two-Dimensional Gel Electrophoresis (2-DE)
SAE and WT were grown until the exponential phase (OD600 = 1.6) in TSB medium and the bacteria (7.56 × 109 cells of each) were harvested by centrifugation at 7,000 g for 10 min at 4°C. The bacterial pellets were solubilized with lysis buffer (7 M urea, 2 M thiourea, 2% (w/v) CHAPS, and 50 mM dithiothreitol (DTT), 2% (v/v) pH 4–7 immobilized pH gradient (IPG) buffer (Amersham Biosciences) containing 1% protease inhibitor cocktail (Roche) and 1 mM PMSF (Sigma)) and then sonicated on ice for 12 cycles, each consisting of 5 s pulse and 10 s pause. After centrifugation at 7,000 g for 1 h at 4°C, the supernatants of lysates were divided into aliquots and the protein concentrations were determined by the Bradford assay. Then, aliquots were stored at −80°C for further analysis.
2.3. Two-Dimensional Gel Electrophoresis and Image Analysis
The 2-DE gels were performed using 24 cm IPG strips (pH 4–7, GE Healthcare) in Ettan IPGphor isoelectric focusing system (Amersham Biosciences) plus Ettan-Dalt Six system (Amersham Biosciences) according to the manufacturer's instructions. To compensate the variability of gel electrophoresis, at least three replicate gels were performed for each group. In the first dimensional isoelectric focusing (IEF), 120 μg proteins of each sample were diluted to 450 μL with rehydration buffer containing 8 M urea, 2% (w/v) CHAPS, 50 mM DTT, and 0.5% (v/v) ampholyte (pH 4–7, Amersham Biosciences), and IPG strips were allowed to rehydrate in the above solution under mineral oil. IEF was performed as follows: 30 V for 6 h (active rehydration), 60 V for 6 h (active rehydration), 500 V for 2 h, rapid, 1,000 V for 2 h, rapid, 4,000 V for 2 h, linear, linear ramping to 8,000 V for 2 h, and finally 8,000 V for about 7 h with a total of 64 KVh at 20°C. Then the IPG strips were incubated in equilibration buffer (75 mM Tris-HCl (pH 8.8), 6 M urea, 29.3% (v/v) glycerol, 2% (w/v) SDS, and 0.002% (w/v) bromophenol blue) containing 2% (w/v) DTT for 15 min with gentle agitation, followed by incubation in the same equilibration buffer supplemented with 2.5% (w/v) iodoacetamide for 15 min at room temperature. The second dimension SDS-PAGE was performed on 1 mm thick 12.5% polyacrylamide vertical gels in Ettan-Dalt Six system using 5 W/gel for 30 min and followed by 12 W/gel at 10°C until the bromophenol blue dye front reached the end of the gels. The gels were stained by a modified silver staining method compatible with MS analysis and scanned at 300 dpi (dots/inch) using ImageScanner (UMAX, Amersham Biosciences). Images were captured and analyzed by ImageMaster 2D platinum 6.0 software (Amersham Biosciences). The percentage of the volume of the spots representing a certain protein was determined in comparison with the total proteins present in the 2-DE gel. To select differentially expressed protein spots, quantitative analysis was performed using the Student's t-test to compare the percentage volumes of spots between SAE and WT groups. The differentially expressed protein spots with P values less than 0.05 were considered significant differences, and at least 1.5-fold difference in percentage of the volume for each spot was set as a threshold. These protein spots were selected and subjected to in-gel tryptic digestion and identification by MS.
2.4. In-Gel Tryptic Digestion
The differentially expressed protein spots were manually excised from the sliver-stained gels (each gel of 120 μg protein) and placed into a 96-well microplate. The gel pieces were destained with a solution of 15 mM potassium ferricyanide and 50 mM sodium thiosulfate (1 : 1) at room temperature for 10 min, then washed twice with deionized water, each for 30 min, and dehydrated in 80 μL of acetonitrile (ACN) twice. Then the samples were swollen in a digestion buffer containing 25 mM NH4HCO3 and 12.5 ng/μL trypsin (Promega) at 4°C after 30 min incubation and incubated at 37°C for more than 12 h. The peptide mixtures from the gel were extracted twice using 0.1% trifluoroacetic/50% ACN at room temperature, resuspended with 0.7 μL matrix solution (α-cyano-4-hydroxycinnamic acid (Sigma) in 0.1% trifluoroacetic, 50% ACN), and allowed to dry in air under the protection of N2.
2.4.1. Mass Spectrometric Analysis and Database Searching
The peptide mixtures from samples were analyzed by 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems). The UV laser was operated at a 200 Hz repetition rate with wavelength of 355 nm. The accelerated voltage was operated at 20 kV. Myoglobin digested by trypsin was used to calibrate the mass instrument with internal calibration mode. All acquired spectra of samples were processed using 4700 series Explore software (Applied Biosystems) in a default mode. The parent mass peaks with mass range 700–3200 Da and minimum S/N 20 were picked out for tandem TOF/TOF analysis. Combined MS and MS/MS spectra were submitted to MASCOT (V2.1, Matrix Science) by GPS Explorer software (V3.6, Applied Biosystems) and searched with the following parameters: NCBInr database (release date: November 2009), taxonomy of bony vertebrates or viruses, trypsin digest with one missing cleavage, none fixed modifications, MS tolerance of 100 ppm, MS/MS tolerance of 0.6 Da, and possible oxidation of methionine. Known contaminant ions (human keratin, tryptic autodigest peptides, etc.) were excluded. MASCOT protein scores (based on MS and MS/MS spectra) with greater than 72 were considered statistically significant (P < 0.05). The individual MS/MS spectrum with statistically significant (confidence interval >95%) and best ion score (based on MS/MS spectra) was accepted.
2.5. Quantitative Real-Time PCR Analysis
Total RNA was extracted from the SAE and WT after incubation for 12 h. For each RNA sample, duplicate reverse transcription reactions were performed using reverse transcriptase MMLV (Promega, Madison, USA), with a control without reverse transcriptase. RT real-time sequence-specific detection was performed using SYBR Premix Ex Taq (TaKaRa) and relative quantification of gene expression was calculated using the comparative cycle threshold method as described for the ABI 7500 real-time PCR system (Applied Biosystems, California, USA) by the manufacturer's instructions. The housekeeping gene gyrB was used as an endogenous control. Gene-specific primers were designed according to the gene sequences at GenBank (Accession no. CP000029) (Table 1). All samples were analyzed in triplicate and normalized against gyrB gene expression.
Table 1.
Oligonucleotide primers used for RT real-time PCR.
| Target gene | Gene description | GenBank accession no. | Primer* | Primer sequence | Location | Function |
|---|---|---|---|---|---|---|
| gyrB | DNA gyrase, B subunit | 57636585 | gyrBF gyrBR |
CTTATATGAGAATCCATCTGTAGG AGAACAATCTGCCAATTTACC |
1110–1263 | House keeping gene |
|
| ||||||
| pta | Phosphate acetyltransferase | 57866177 |
ptaF
ptaR |
AGGTTCTGCTAAATCGGATGATG GGTGCTTTCTTCTCTGCTACG |
767–922 | Catalyzes the conversion of acetyl-CoA and phosphate to CoA and acetyl phosphate |
|
| ||||||
| gapA-1 | Glyceraldehyde 3-phosphate dehydrogenase | 57866427 |
gapA-1F
gapA-1R |
ACACAAGACGCACCTCAC TCTGGAATAACTTTACCGATAGC |
709–821 | Catalyzes the formation of 1,3-bisphosphoglycerate from glyceraldehyde-3-P in glycolysis |
|
| ||||||
| eno | Enolase; phosphopyruvate hydratase | 57866440 |
enoF
enoR |
GCCAATTATTACAGATGTTTACG CAGTAGAAGCACCAGAAGG |
200–327 | Catalyzes the formation of phosphoenolpyruvate from 2-phospho-D-glycerate in glycolysis |
|
| ||||||
| sucD | Succinyl-CoA synthetase alpha subunit | 57866714 |
sucDF
sucDR |
AGAAGAAGAAGCGGCACAATG GCAGTTTCAACACCACAATCG |
779–958 | Catalyzes the only substrate-level phosphorylation in the TCA cycle |
|
| ||||||
| icd | Isocitrate dehydrogenase | 57867183 |
icdF
icdR |
ACTATATCTCTGATGCTCTTG TTCTGATGACGGATTAACC |
1118–1272 | Converts isocitrate to alpha ketoglutarate |
|
| ||||||
| pyk | Pyruvate kinase | 57867187 |
pykF
pykR |
CGAGTTTCAGTGTTAGGTC CGTAGAGGTTAATTGATTATTCC |
4–159 | Catalyzes the formation of pyruvate from phosphoenolpyruvate in glycolysis |
|
| ||||||
| fbaA | Fructose-bisphosphate aldolase | 57867608 |
fbaAF
fbaAR |
TGCTGATGGCGTTATCTATG CCGTGTAATACTAAAGGTAATCC |
575–757 | Catalyzes the formation of glycerone phosphate and glyceraldehyde 3-phosphate from fructose 1,6-bisphosphate |
*F: forward primer; R: reverse prime.
2.6. Biofilm and PIA Assay
The biofilm-forming ability of S. epidermidis strains was determined by the microtiter-plate test as described by Christensen. Briefly, overnight cultures of S. epidermidis were diluted 1 : 200 in BM medium and inoculated into wells of polystyrene microtiter plates (200 μL per well) at 37°C for 24 h. Glucose (G8270, Sigma) was added into BM medium at 0.125%, 0.25%, 0.5%, and 1% (g/mL), respectively. After incubation, the wells were gently washed three times with 200 μL PBS and stained with 2% crystal violet for 5 min. Absorbance was determined at 570 nm.
S. epidermidis cells for PIA immunoblot assays were grown in 6-well plates (Nunc, DK-4000 Roskitde, Denmark) under static conditions at 37°C for 24 h. The cells were scraped off and resuspended in 0.5 M EDTA (pH 8.0). The supernatant was treated with proteinase K (final concentration 4 mg/mL; Roche, MERCK, Darmstadt, Germany) for 3 h (37°C). Serial dilutions of the PIA extract were then transferred to a nitrocellulose membrane (Millipore, Billerica, MA) using a 96-well dot blot vacuum manifold (Gibco). The air-dried membrane was blocked with 3% (wt/vol) bovine serum albumin and subsequently incubated with 3.2 μg/mL wheat germ agglutinin coupled to horseradish peroxidase (WGA-HRP conjugate; Lectinotest Laboratory, Lviv, Ukraine) for 1 h. Horseradish peroxidase (HRP) activity was visualized via chromogenic detection. The gray scale of the spots corresponding to PIA was quantified using the Quantity One software.
2.7. CFU Count and Acidity Determination
Overnight cultures of S. epidermidis were diluted 1 : 200 in BM medium with or without 0.125%, 0.25%, 0.5%, and 1% (g/mL) glucose and incubated at 37°C for 24 h. Colony forming units (CFU) on TSA plates were further counted with serial dilutions of each sample plated on 6 agar plates. Then, the cultures were centrifuged. The pH of the supernatant was determined by using a pH meter (FE20, Mettler-Toledo, Shanghai, China). Then, five drops of methyl red were added to the supernatant and then gently rolled to disperse the methyl red. The supernatant will turn yellow at neutral pH but turn red at pH < 4.0.
2.8. Statistical Analysis
Experimental data were analyzed with the SPSS software and compared using the Student's t-test. Differences with a P value of <0.05 were considered statistically significant.
3. Results
3.1. Protein Expression Profiles of S. epidermidis Affected by Two-Component Signal Transduction System SaeRS
Two-dimensional gel electrophoresis (2-DE) was performed to analyze the expression of S. epidermidis proteins regulated by two-component signal transduction system SaeRS. The gel images were compared and analyzed. The patterns of protein expression in SAE and WT appeared largely similar but some clear differences in protein expression levels of certain protein spots were evident (Figure 1). Fifty-five protein spots were excised for further analysis by the LC-MS/MS. Of these, 15 spots were up-regulated, and 40 were downregulated (Table 2).
Figure 1.
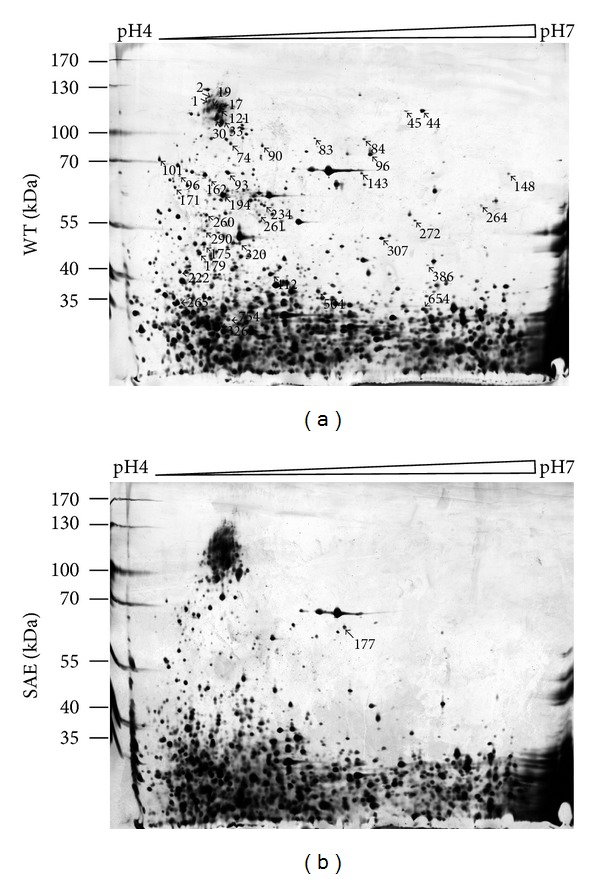
Identified spots on the 2D proteome map of SAE and WT. SAE and WT were grown in TSB medium at 37°C until the postexponential growth phase; the bacteria were then separated by centrifugation. Bacteria cell pellets were dissolved in lysis buffer and sonicated on ice. The 2-DE gels were performed using 24 cm immobilized dry strips (IPG, nonlinear, pH 4–7, GE Healthcare) and analyzed by ImageMaster 2D platinum 6.0 software (Amersham Biosciences). Differentially expressed protein spots on 2-DE gels were identified by MS/MS analysis and shown in gels with unique protein spot numbers. Protein spots were identified using a 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems, California, USA).
Table 2.
Protein expression regulated by saeRS in S. epidermidis.
| Spot numbera | Predicted MW (Da) | Predicted pI | Protein hit | Mowse Scoreb/number of match peptides | Accession numberc | Fold change (2D: WT/SAE) |
|---|---|---|---|---|---|---|
| Upregulated protein spots | ||||||
| 179 | 18124 | 4.6 | PTS system glucose-specific enzyme II A component [Staphylococcus epidermidis ATCC 12228] | 215/13 | 27468033 | 1/4 |
| 175 | 18124 | 4.6 | PTS system glucose-specific enzyme II A component [Staphylococcus epidermidis ATCC 12228] | 156/11 | 27468033 | 1/4 |
| 2 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 88/6 | 7242804 | 1/10 |
| 121 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 190/16 | 7242804 | 1/10 |
| 17 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 72/6 | 7242804 | 1/10 |
| 19 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 124/9 | 7242804 | 1/10 |
| 30 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 187/11 | 7242804 | 1/10 |
| 33 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 127/7 | 7242804 | 1/10 |
| 1 | 131887 | 4.73 | Accumulation-associated protein [Staphylococcus epidermidis RP62A] | 83/9 | 7242804 | 1/10 |
| 74 | 35066 | 4.72 | Phosphate acetyltransferase [Staphylococcus epidermidis ATCC 12228] | 360/19 | 27467277 | 1/5 |
| 177 | 33131 | 5.18 | Cysteine synthase [Staphylococcus epidermidis ATCC 12228] | 110/15 | 27469188 | 1/2 |
| 93 | 33200 | 4.7 | 5-Oxo-1,2,5-tricarboxylic-3-penten acid decarboxylase [Staphylococcus epidermidis ATCC 12228] | 83/9 | 27467583 | 1/10 |
| 222 | 19971 | 4.4 | Heat shock protein 60 [Staphylococcus muscae] | 81/6 | 17224365 | 1/1.5 |
| 326 | 15342 | 4.74 | Hypothetical protein SE0591 [Staphylococcus epidermidis ATCC 12228] | 174/10 | 27467509 | 1/2 |
| 265 | 18685 | 4.5 | Hypothetical protein SE1560 [Staphylococcus epidermidis ATCC 12228] | 89/5 | 27468478 | 1.5/1 |
|
| ||||||
| Downregulated protein spots | ||||||
| 96 | 31679 | 5.35 | Succinyl-CoA synthetase alpha subunit [Staphylococcus epidermidis ATCC 12228] | 113/9 | 27467842 | A/Nd |
| 412 | 41998 | 4.83 | Succinyl-CoA synthetase beta subunit [Staphylococcus epidermidis ATCC 12228] | 80/11 | 27467841 | A/N |
| 272 | 41998 | 4.83 | Succinyl-CoA synthetase beta subunit [Staphylococcus epidermidis ATCC 12228] | 74/14 | 27467841 | A/N |
| 143 | 31679 | 5.35 | Succinyl-CoA synthetase alpha subunit [Staphylococcus epidermidis ATCC 12228] | 99/10 | 27467842 | 7/1 |
| 307 | 41998 | 4.83 | Succinyl-CoA synthetase beta subunit [Staphylococcus epidermidis ATCC 12228] | 134/19 | 27467841 | A/N |
| 293 | 66196 | 4.99 | D-Fructose-6-phosphate amidotransferase [Staphylococcus epidermidis ATCC 12228] | 130/16 | 27468669 | A/N |
| 290 | 34981 | 4.66 | Oxidoreductase ion channel [Staphylococcus epidermidis ATCC 12228] | 74/9 | 27467285 | 6/1 |
| 504 | 32490 | 5.17 | Elongation factor Ts [Staphylococcus epidermidis ATCC 12228] | 159/17 | 27467851 | A/N |
| 234 | 18748 | 6.04 | Putative vls recombination cassette Vls15 [Borrelia burgdorferi] | 77/7 | 2039294 | A/N |
| 264 | 36442 | 5.29 | Catabolite control protein A [Staphylococcus epidermidis RP62A] | 75/9 | 57867258 | A/N |
| 84 | 52589 | 5.63 | Inositol-monophosphate dehydrogenase [Staphylococcus epidermidis ATCC 12228] | 92/16 | 27469266 | A/N |
| 260 | 22329 | 4.59 | Hexulose-6-phosphate synthase, putative [Staphylococcus epidermidis RP62A] | 124/11 | 57866142 | A/N |
| 386 | 34713 | 5.04 | Glycerate dehydrogenase [Staphylococcus epidermidis RP62A] | 101/13 | 57867831 | A/N |
| 754 | 47217 | 4.58 | Enolase [Staphylococcus epidermidis ATCC 12228] | 84/14 | 27467479 | 6/1 |
| 194 | 34185 | 4.61 | Putative manganese-dependent inorganic pyrophosphatase [Staphylococcus epidermidis RP62A] | 190/19 | 57867405 | A/N |
| 163 | 33131 | 5.18 | Cysteine synthase [Staphylococcus epidermidis ATCC 12228] | 88/13 | 27469188 | 1.5/1 |
| 424 | 66196 | 4.99 | D-Fructose-6-phosphate Amidotransferase [Staphylococcus epidermidis ATCC 12228] | 77/10 | 27468669 | A/N |
| 171 | 26556 | 4.51 | 30S ribosomal protein S1 [Staphylococcus epidermidis ATCC 12228] | 137/12 | 27468083 | 10/1 |
| 162 | 42075 | 4.62 | DNA polymerase III Subunit beta [Staphylococcus epidermidis ATCC 12228] | 79/11 | 27466920 | 1.5/1 |
| 94 | 62994 | 5.24 | Pyruvate kinase [Staphylococcus epidermidis ATCC 12228] | 230/20 | 27468291 | A/N |
| 415 | 46590 | 5.01 | Isocitrate dehydrogenase [Staphylococcus epidermidis ATCC 12228] | 72/7 | 27468288 | 7/1 |
| 82 | 66105 | 4.57 | DnaK protein [Staphylococcus epidermidis RP62A] | 257/17 | 57867037 | 5/1 |
| 96 | 66105 | 4.57 | DnaK protein [Staphylococcus epidermidis RP62A] | 77/7 | 57867037 | 5/1 |
| 72 | 66105 | 4.57 | DnaK protein [Staphylococcus epidermidis RP62A] | 84/10 | 57867037 | 5/1 |
| 21 | 66105 | 4.57 | DnaK protein [Staphylococcus epidermidis RP62A] | 157/12 | 57867037 | 5/1 |
| 74 | 66105 | 4.57 | DnaK protein [Staphylococcus epidermidis RP62A] | 88/14 | 57867037 | A/N |
| 142 | 33031 | 4.82 | Fructose-bisphosphate aldolase [Staphylococcus epidermidis ATCC 12228] | 261/24 | 27469074 | 3/1 |
| 481 | 14906 | 5.96 | Urease beta subunit [Staphylococcus epidermidis ATCC 12228] | 218/18 | 27468780 | 4/1 |
| 90 | 36168 | 4.83 | Glyceraldehyde-3-phosphate dehydrogenase [Staphylococcus epidermidis ATCC 12228] | 123/13 | 27467475 | A/N |
| 83 | 52589 | 5.63 | Inositol-monophosphate dehydrogenase [Staphylococcus epidermidis ATCC 12228] | 86/13 | 27469266 | A/N |
| 45 | 52589 | 5.63 | Inositol-monophosphate dehydrogenase [Staphylococcus epidermidis ATCC 12228] | 106/16 | 27469266 | A/N |
| 44 | 52589 | 5.63 | Inositol-monophosphate dehydrogenase [Staphylococcus epidermidis ATCC 12228] | 205/27 | 27469266 | A/N |
| 101 | 48702 | 4.28 | Trigger factor [Staphylococcus epidermidis ATCC 12228] | 89/13 | 27468268 | 3/1 |
| 148 | 51524 | 4.62 | ATP synthase subunit B [Staphylococcus epidermidis ATCC 12228] | 96/6 | 27468618 | 4/1 |
| 148 | 51524 | 4.62 | ATP synthase subunit B [Staphylococcus epidermidis ATCC 12228] | 107/7 | 27468618 | 4/1 |
| 227 | 20203 | 5.71 | Immunodominant antigen B [Staphylococcus epidermidis ATCC 12228] | 81/8 | 27468493 | 1.5/1 |
| 237 | 35151 | 5.42 | Hypothetical protein SE1260 [Staphylococcus epidermidis ATCC 12228] | 112/16 | 27468178 | 4/1 |
| 261 | 35151 | 5.42 | Hypothetical protein SE1260 [Staphylococcus epidermidis ATCC 12228] | 111/16 | 27468178 | 1.5/1 |
| 654 | 17753 | 5.55 | Hypothetical protein SE2391 [Staphylococcus epidermidis ATCC 12228] | 126/10 | 27469309 | 35/1 |
| 320 | 19182 | 4.69 | Hypothetical protein SE0264 [Staphylococcus epidermidis ATCC 12228] | 113/15 | 27467182 | 10/1 |
aSpots numbers correspond to the numbers in Figure 2.
bProtein score was from MALDI-TOF/TOF identification. The proteins that had a score great than 72 (P < 0.05) were considered statistically significant.
cAccession no. is the MASCOT result of MALDI-TOF/TOF searched from the NCBInr database.
dA/N indicates the spot detected only in the protein extracts of S. epidermidis 1457 wild-type strain.
The 55 identified proteins that varied in expression levels showed diverse annotated functions (the identified proteins were functionally classified into the category of metabolic enzyme, biofilm formation, transcriptional regulator, chaperone, energy metabolism, or unknown function), including the enzymes involved in glycolysis (fructose-bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, enolase and pyruvate kinase) and TCA cycle (succinyl-CoA synthetase alpha and beta subunits, and isocitrate dehydrogenase), biofilm forming-related proteins (accumulation-associated protein), stress response protein (heat shock protein 60 and dnaK protein), serine proteinase inhibitor (alpha-2-macroglobulin), cytoskeletal proteins (actin, tubulin, and twinstar), enzymes involved in energy metabolism (ATP synthase subunit B) and proteins involved in glucose transport (PTS system glucose-specific enzyme II A), and regulator of carbon metabolism (catabolite control protein A).
3.2. Enzymes
Approximately 40% of the identified proteins were defined as enzymes. Most of the identified enzymes were upregulated at the postexponential phase and categorized as involved in glucose metabolism. Figure 2 showed that enzymes related to glycolysis (fructose-bisphosphate aldolase (GI: 27469074), glyceraldehyde 3-phosphate dehydrogenase (GI: 27467475), enolase (GI: 27467479), pyruvate kinase (GI: 27468291)), the TCA cycle (succinyl-CoA synthase (GI: 27467842), and isocitrate dehydrogenase (GI: 27468288)) were downregulated at the post-exponential phase. This is the first report that the expression of enzymes related to carbohydrate metabolism in Staphylococcus species is down-regulated by deletion of TCS SaeRS.
Figure 2.
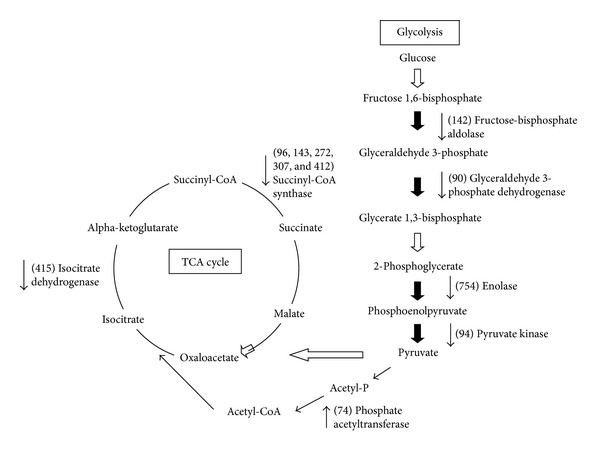
Carbohydrate metabolic pathways in the saeRS mutant under aerobic conditions. The following enzymes (identified in the 2-DE experiment) were found at lower levels in the saeRS mutant: fructose bisphosphate aldolase, glyceraldehyde 3-phosphate dehydrogenase, enolase, pyruvate kinase, succinyl-CoA synthase, and isocitrate dehydrogenase. Phosphate acetyltransferase was found at higher level in the saeRS mutant.
3.3. Real-Time RT-PCR of the Differentially Expressed Genes
To verify the differentially expressed proteins regulated by SaeRS at the transcriptional level, glucose metabolism-related genes were chosen for the quantitative real-time RT-PCR analysis. The gyrB gene was used as an internal control. The transcription of glyceraldehyde 3-phosphate dehydrogenase, enolase, succinyl-CoA synthase alpha subunit, isocitrate dehydrogenase, pyruvate kinase, and fructose-bisphosphate aldolase was significantly decreased in SAE, approximately 0.8-, 0.8-, 0.6-, 0.75-, 0.72-, and 0.7-fold lower than that in WT, respectively (Figure 3). The phosphate acetyltransferase transcription expression levels were significantly upregulated 1.72-fold over than those of the control.
Figure 3.
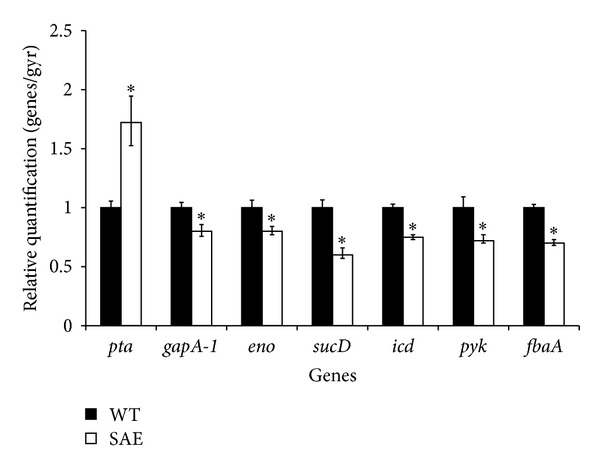
Quantitative transcript analysis of glucose metabolism-related gene transcript levels. Total RNA was extracted from SAE and WT grown in TSB medium at 37°C until the postexponential growth phase and the cDNA were then synthesized. The mRNA expression levels of glyceraldehyde 3-phosphate dehydrogenase (gapA-1), enolase (eno), succinyl-CoA synthase alpha subunit (sucD), isocitrate dehydrogenase (icd), pyruvate kinase (pyk), fructose-bisphosphate aldolase (fbaA), and phosphate acetyltransferase (pta) were determined using the beta-actin gene as an internal control. Data are shown as the mean ± S.D. Means with different letters are significantly different (P < 0.05).
3.4. Effect of saeRS Deletion on S. epidermidis Biofilm Formation in Response to Glucose
As TCS SaeRS regulated glucose metabolism and biofilm formation-related genes expression, the effect of saeRS deletion on S. epidermidis biofilm formation in response to various concentrations of glucose was further examined. Upon the addition of glucose to the BM medium, the biofilm forming ability of SAE was significantly increased compared to the parental strain and reached the peak level at 0.5% (g/mL) glucose (Student's t-test, P < 0.05) (Figure 4). However, the biofilm forming ability of WT showed no obvious difference upon the stimulation of glucose. Although it did not reach the level of the wild-type strain, complementation of saeRS resulted in decreased biofilm formation.
Figure 4.
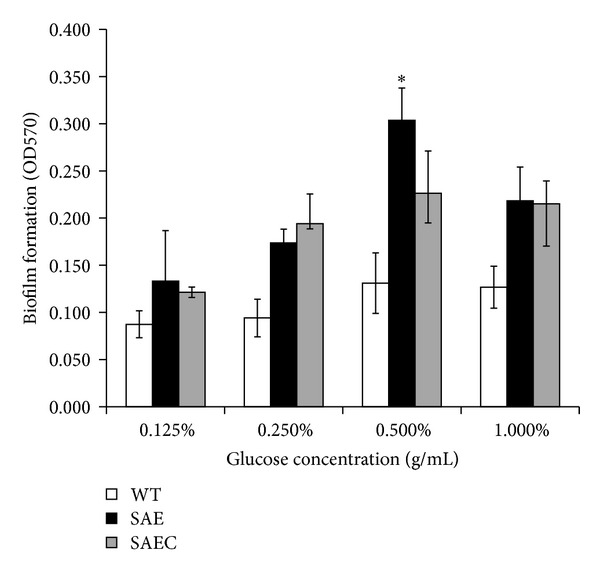
Effect of glucose on SAE, WT, and SAEC biofilm formation. SAE, WT, and SAEC biofilms were formed in the absence (black bars) or presence of glucose (0.125%, 0.25%, 0.5%, and 1%); then the biofilms were washed and then stained with crystal violet. Their retained biomass was quantified by measuring the absorbance of each well at 570 nm. Mean values and standard deviations from three independent experiments are shown, *P < 0.05.
3.5. Effect of saeRS Deletion on S. epidermidis Viability in Response to Glucose
In our previous study, saeRS deletion in S. epidermidis decreased the bacterial cell viability. To investigate whether the decreased cell viability that resulted from saeRS deletion was affected by glucose metabolism, colony-forming unit (CFU) counts of SAE, WT, and SAEC in BM medium with or without glucose (0.125%, 0.25%, 0.5%, and 1%) were determined. Cultures were inoculated with approximately 104 CFU/mL of each strain and incubated for 24 h. When cultured in medium containing 0.25% glucose, SAE cultures had the highest CFU count (4.23 × 1010 CFU/mL) compared to WT (7 × 109 CFU/mL) and SAEC (2 × 1010 CFU/mL) (Figure 5); SAE showed the highest viability when grown in medium containing 0.25% glucose, compared with the other growth conditions.
Figure 5.
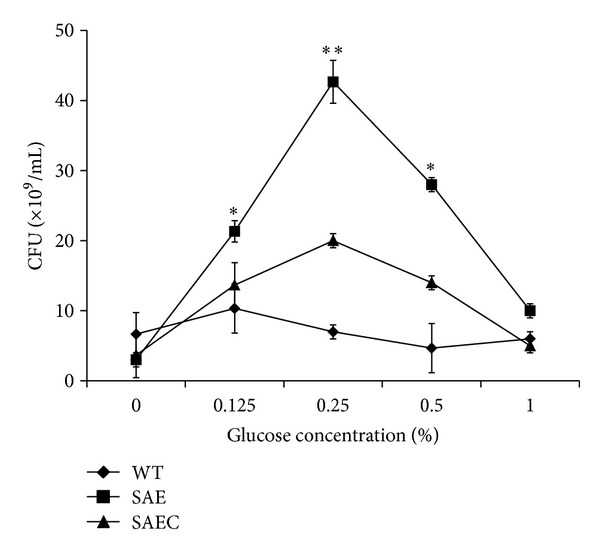
Viability of S. epidermidis 1457 in BM medium containing various concentrations of glucose. Overnight cultures of S. epidermidis were diluted 1 : 200 in BM medium with or without 0.125%, 0.25%, 0.5%, and 1% (g/mL) glucose and incubated at 37°C for 24 h. Colony forming units (CFU) on TSA plates were further counted with serial dilutions of each sample plated on 6 agar plates.
3.6. Effect of saeRS Deletion on Acidity in Response to Glucose
Glucose is oxidized to pyruvate (in aerobic condition) or lactate (in anaerobic condition) in S. epidermidis, the pH value and the pH indicator methyl red may reflect the utilization efficiency of glucose. After 24 h, 0.25% glucose decreased the pH value of WT growth medium to 7.0 (lower than 7.4) and maintained the pH value of SAE and SAEC to 8.07 and 7.5 (higher than 7.4), respectively (Figure 6). For the methyl red test, SAE showed the lowest degree of red when cultured in BM medium containing 0.25% glucose, compared to WT and SAEC, indicating that SAE possesses the low utilization efficiency of glucose (Figure 7).
Figure 6.
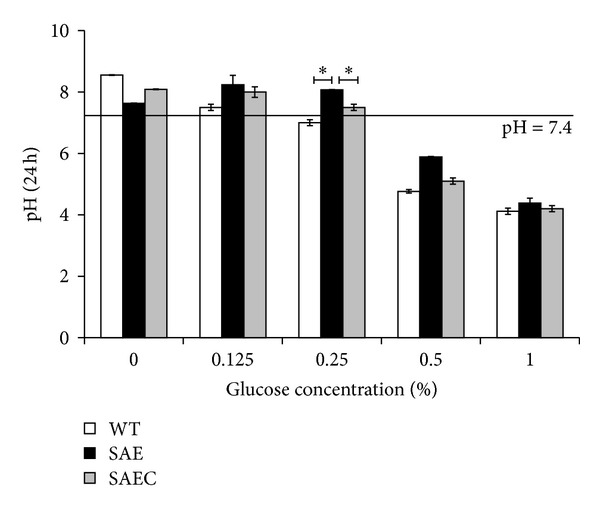
pH value of S. epidermidis 1457 in BM medium containing various concentrations of glucose. SAE, WT and SAEC were grown with or without 0.125%, 0.25%, 0.5%, and 1% (g/mL) glucose for 24 h. Then, the cultures were centrifuged. The pH of the supernatant was determined by using a pH meter (FE20, Mettler-Toledo, Shanghai, China).
Figure 7.
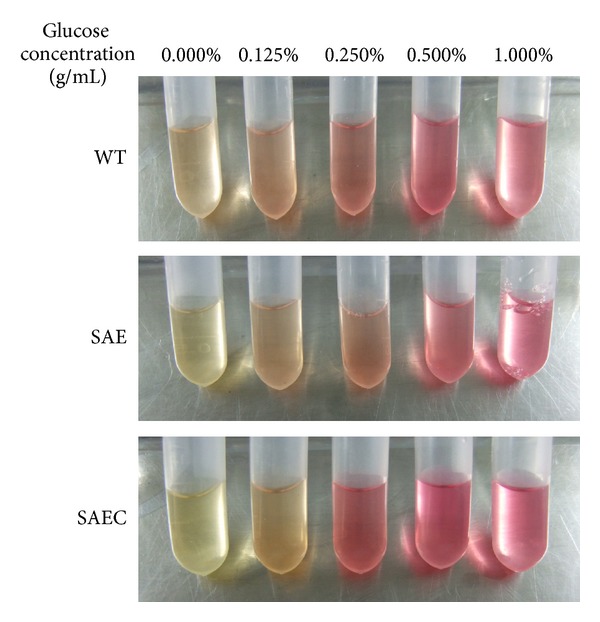
Methyl red test. SAE, WT, and SAEC were grown with or without 0.125%, 0.25%, 0.5%, and 1% (g/mL) glucose for 24 h. Five drops of methyl red were added to the culture supernatant and then gently rolled to disperse the methyl red. The supernatant will turn yellow at neutral pH but turn red at pH < 4.0.
4. Discussion
Staphylococci biofilm formation and autolysis are influenced by a variety of different factors such as glucose, NaCl, pH, temperature, and growth phase, suggesting the existence of a mechanism that can sense environmental conditions [18]. In S. epidermidis, the two-component signal transduction system (TCS) SaeRS has been proven to negatively regulate biofilm formation and autolysis [12].
To further illustrate the regulation mechanism of SaeRS on biofilm formation and autolysis, two-dimensional gel electrophoresis (2-DE) was performed to analyze the expression of S. epidermidis proteins regulated by TCS SaeRS. In preliminarily work, the appropriate pH range for the first dimension isoelectric focusing (IEF) was determined using a 24 cm IPG strip of pH 3–10. Since most of the protein spots were present in the pH range of 4–7 (data not shown), subsequent 2-DE utilised 24 cm IPG strips of pH 4–7 for better separation and resolution. The amounts of protein and staining procedure used in the experiment were also empirically optimised. It was found that 120 μg protein samples were required for the optimal silver staining.
Protein profile analysis of the saeRS mutant and the wild-type strain found 55 differentially expressed proteins in the present study. The transcription of four genes taking part in glycolysis was downregulated—gapA1, eno, fbaA, and pyk—indicating decreased glycolytic activity of SAE. We also found that the expression of catabolite control protein A (CcpA) was down-regulated in SAE. CcpA inhibits the genes expression involved in tricarboxylic acid cycle (TCA), such as acnA gene (encoding Aconitase) in S. epidermidis [19]. The formation of S. epidermidis biofilms is enhanced by conditions that repress tricarboxylic acid (TCA) cycle activity [19], indicating that there are other mechanisms involved in SaeRS-mediated biofilm formation.
S. epidermidis saeRS deletion mutant showed increased biofilm-forming ability in our previous study, and the mechanism includes increased extracellular DNA which may be caused by enhanced autolytic activity [12]. In the present study, appropriate concentration of glucose promotes the biofilm-forming ability of S. epidermidis, while excess glucose inhibits its biofilm-forming ability. In S. epidermidis, the effect of excess glucose on the metabolome was essentially identical to the effect of TCA cycle inactivation [20]. The formation of S. epidermidis biofilms may be enhanced by conditions that repress tricarboxylic acid (TCA) cycle activity. SAE showed strongest survival ability in BM medium containing 0.25% (g/mL) glucose; this may be caused by the low acidity of the growth medium. SAE showed lower pH value than WT grown in medium without glucose and higher pH value in medium with glucose, indicating the low ability of utilization of glucose in SAE.
PIA synthesis is important for biofilm formation [21–23]. In our previous study, no obvious difference in PIA production was observed between SAE and WT. PIA may not be the main cause of enhanced biofilm forming ability of SAE. The enhanced expression of accumulation-associated protein (identified in the 2-DE experiment) may play important role in SAE biofilm formation. In the present study, we have shown that glucose significantly promotes the biofilm formation of SAE, while having little effect on that of WT. The higher pH value that resulted from the low glucose utilization efficiency of SAE may also contribute to the biofilm formation. In addition, we have detected that high viability of SAE incubated in glucose excess medium, which may be caused by the high pH value, may also contribute to the biofilm formation.
5. Conclusions
TCS SaeRS positively regulates glucose metabolism in S. epidermidis. The repression of glucose utilization in SAE leads to decreased acidity of growth medium which may contribue to the higher viability and the enhanced biofilm forming ability. The present study suggests that in S. epidermidis, the external factor glucose plays an important role in the biofilm formation and viability regulated by SaeRS. The study will be helpful for treatment of S. epidermidis infection from the viewpoint of metabolic modulation dependent on two-component signal transduction system SaeRS.
Acknowledgment
This work was supported by Henan University Research Funding (no. 2011YBZR004) and the Foundation of He'nan Educational Committee (no. 14B310015).
Abbreviations
- WT:
S. epidermidis wild-type
- SAE:
S. epidermidis saeRS mutant
- SAEC:
S. epidermidis saeRS complemental strain
- 2-DE:
Two-dimensional gel electrophoresis
- TCA:
Tricarboxylic acid
- PIA:
Polysaccharide intercellular adhesion
- TCS:
Two-component signal transduction system
- CFU:
Colony-forming unit.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Otto M. Staphylococcus epidermidis—the “accidental” pathogen. Nature Reviews Microbiology. 2009;7(8):555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. The Lancet Infectious Diseases. 2002;2(11):677–685. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 3.Mohammed M, Aboki M, Saidu H, Victor O, Tawakalitu A, Maikano S. Phytochemical and some antmicrobial activity of Cassia Occidentalis L.(Caesalpiniaceae) International Journal of Science and Technology. 2012;2(4) [Google Scholar]
- 4.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):p. 1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 5.Vuong C, Kocianova S, Voyich JM, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. Journal of Biological Chemistry. 2004;279(52):54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 6.Seidl K, Goerke C, Wolz C, Mack D, Berger-Bächi B, Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infection and Immunity. 2008;76(5):2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seidl K, Müller S, François P, et al. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus . BMC Microbiology. 2009;9, article 95 doi: 10.1186/1471-2180-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jäger S, Mack D, Rohde H, Horstkotte MA, Knobloch JK-M. Disintegration of Staphylococcus epidermidis biofilms under glucose-limiting conditions depends on the activity of the alternative sigma factor σB. Applied and Environmental Microbiology. 2005;71(9):5577–5581. doi: 10.1128/AEM.71.9.5577-5581.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opekarova M, Sigler K. Acidification power: indicator of metabolic activity and autolytic changes in Saccharomyces cerevisiae. Folia Microbiologica. 1982;27(6):395–403. doi: 10.1007/BF02876450. [DOI] [PubMed] [Google Scholar]
- 10.Purves J, Cockayne A, Moody PCE, Morrissey JA. Comparison of the regulation, metabolic functions, and roles in virulence of the glyceraldehyde-3-phosphate dehydrogenase homologues gapA and gapB in Staphylococcus aureus . Infection and Immunity. 2010;78(12):5223–5232. doi: 10.1128/IAI.00762-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreyer R, Bock A. Phenotypic suppression of a fructose 1,6 diphosphate aldolase mutation in Escherichia coli . Journal of Bacteriology. 1973;115(1):268–276. doi: 10.1128/jb.115.1.268-276.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lou Q, Zhu T, Hu J, et al. Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiology. 2011;11, article 146 doi: 10.1186/1471-2180-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Wang J, Xu T, et al. The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica-dependent manner. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0040041.e40041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norvick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals agr quorum sensing. Microbiology. 2003;149(10):2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 15.Giraudo AT, Cheung AL, Nagel R. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Archives of Microbiology. 1997;168(1):53–58. doi: 10.1007/s002030050469. [DOI] [PubMed] [Google Scholar]
- 16.Rogasch K, Rühmling V, Pané-Farré J, et al. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. Journal of Bacteriology. 2006;188(22):7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handke LD, Rogers KL, Olson ME, et al. Staphylococcus epidermidis saeR is an effector of anaerobic growth and a mediator of acute inflammation. Infection and Immunity. 2008;76(1):141–152. doi: 10.1128/IAI.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilpin RW, Chatterjee AN, Young FE. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus . Journal of Bacteriology. 1972;111(1):272–283. doi: 10.1128/jb.111.1.272-283.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadykov MR, Hartmann T, Mattes TA, et al. CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis . Microbiology. 2011;157(12):3458–3468. doi: 10.1099/mic.0.051243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Halouska S, Schiaffo CE, Sadykov MR, Somerville GA, Powers R. NMR analysis of a stress response metabolic signaling network. Journal of Proteome Research. 2011;10(8):3743–3754. doi: 10.1021/pr200360w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluckiger U, Ulrich M, Steinhuber A, et al. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infection and Immunity. 2005;73(3):1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schommer NN, Christner M, Hentschke M, Ruckdeschel K, Aepfelbacher M, Rohde H. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infection and Immunity. 2011;79(6):2267–2276. doi: 10.1128/IAI.01142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerca F, França Â, Guimarães R, et al. Modulation of poly-N-acetylglucosamine accumulation within mature Staphylococcus epidermidis biofilms grown in excess glucose. Microbiology and Immunology. 2011;55(10):673–682. doi: 10.1111/j.1348-0421.2011.00368.x. [DOI] [PubMed] [Google Scholar]


