Abstract
Most DNA transposons move from one genomic location to another by a cut-and-paste mechanism and are useful tools for genomic manipulations. Short inverted repeat (IR) DNA sequences marking each end of the transposon are recognized by a DNA transposase (encoded by the transposon itself). This enzyme cleaves the transposon ends and integrates them at a new genomic location. We report here a comparison of the biophysical and biochemical properties of two closely related and active mariner/Tc1 family DNA transposases: Mboumar-9 and Mos1. We compared the in vitro cleavage activities of the enzymes on their own IR sequences, as well as cross-recognition of their inverted repeat sequences. We found that, like Mos1, untagged recombinant Mboumar-9 transposase is a dimer and forms a stable complex with inverted repeat DNA in the presence of Mg2+ ions. Mboumar-9 transposase cleaves its inverted repeat DNA in the manner observed for Mos1 transposase. There was minimal cross-recognition of IR sequences between Mos1 and Mboumar-9 transposases, despite these enzymes having 68% identical amino acid sequences. Transposases sharing common biophysical and biochemical properties, but retaining recognition specificity toward their own IR, are a promising platform for the design of chimeric transposases with predicted and improved sequence recognition.
Transposable elements (TEs) are genomic units that can move from their original location to a new place in the genome. They are an important source of genome evolution and diversity and are useful tools for manipulating genomes.1,2 Members of the mariner/Tc1 family of DNA transposons are particularly useful in this regard, as they move by a simple cut-and-paste mechanism and are present in a broad range of living organisms from protozoa and fungi to humans.3
To date, five naturally active eukaryotic mariner/Tc1 family elements have been described: Mos1 from Drosophila mauritiana,4 Famar1 from the earwig Forficula auriculata,5 Mboumar-9 from the ant Messor bouvieri,6 Minos from the fly Drosophila hydei,7 and Passport from the plaice Pleuronectes platessa.8 In addition, four active mariner/Tc1 elements have been reconstructed from the sequences of inactive elements, e.g., Sleeping Beauty from the fish Danio rerio,9 Frog Prince from the frog Rana pipiens,10 Himar1 from the horn fly Haematibia irritans,11 and Hsmar1 from humans12 (reviewed in ref (2)).
mariner/Tc1 transposons have terminal inverted repeats (IR) and encode a single protein, transposase, required for transposition. The transposase has an N-terminal DNA-binding domain, which recognizes the IR in a sequence-specific manner, and a C-terminal catalytic domain. A transposase dimer brings the ends together in a paired-end complex (PEC).13 After DNA excision, the transposon integrates at a TA dinucleotide in a new genomic location. The resulting duplication of the TA target site either side of the transposon is a signature of mariner/Tc1 transposition.
Sleeping Beauty, Frog Prince, Passport, and Minos transposases were shown to be active when expressed from a helper plasmid inside transfected or, in the case of Minos, microinjected cells,8−10,14 and recombinant Mos1, Mboumar-9, Himar1, and Hsmar1 transposases are able to transpose their elements in vitro.6,11,12,15 In those experiments, transposases were purified and refolded from inclusion bodies11,16 or expressed as soluble protein fusions with maltose binding protein at the N-terminus.6,17−20 Previously, we expressed a soluble mutant of Mos1 transposase, without a tag, in Escherichia coli.21 The purified protein has activity similar to that of refolded Mos1 transposase and was amenable to structural analysis, providing insight into the mechanism of Mos1 IR DNA recognition and cleavage.13,22 Unlike DNA transposons from other families,23,24 excision of Mos1 proceeds without formation of a hairpin intermediate.16 First, the nontransferred strand (NTS) is cleaved three bases within the IR, and then a second hydrolysis reaction cleaves the transferred strand (TS) precisely at the junction of the IR and the flanking DNA.16
We have compared the biochemical activity and biophysical properties of two closely related active mariner transposases, Mos1 and Mboumar-9, and tested the cross-reactivity of the enzymes on each other’s inverted repeats. Like Mos1,25,26 purified Mboumar-9 transposase forms a dimer in the absence of DNA, but the protein is less thermally stable. Mboumar-9 transposase can bind to an IR DNA substrate and divalent metal ions, increasing its thermal stability. Mboumar-9 and Mos1 transposases specifically cleave their own IR sequence to produce staggered ends but have minimal cross-reactivity on the other’s IR. Our results suggest that these closely related enzymes could be used to design chimeric proteins with predictable DNA recognition properties and enhanced efficiencies as genomic manipulation tools.
Materials and Methods
Sequence Alignments
The transposase sequences were obtained from http://www.ncbi.nlm.nih.gov, and the GenBank entries are CAH03740 for Mboumar-9 and AAC16614.1 for Mos1. The amino acid sequences were aligned using the T-Coffee web server (EMBL-EBI). The integration site sequences were aligned using the WebLogo 2.8.2 server (http://weblogo.berkeley.edu).
Mos1 Cloning, Expression, and Purification
An artificially synthesized (GeneArt) codon-optimized Mos1 gene of 1035 bp was cloned into the pET30a expression vector by NdeI and XhoI restriction enzymes sites. Protein expression was induced in E. coli BL21(DE3) strain with 0.5 mM IPTG for 24 h at 25 °C with 250 rpm agitation. Purification was conducted as described previously.21
Mboumar-9 Cloning, Expression, and Purification
A codon-optimized Mboumar-9 gene (1035 bp) was artificially synthesized (GeneArt) and cloned into the pET30a expression vector by NdeI and XhoI restriction enzyme sites. Protein expression was induced in E. coli BL21(DE3) strain with 0.5 mM IPTG for 24 h at 18 °C with 250 rpm agitation. Cells were pelleted for 1 h at 8000g and 4 °C. The cell pellet was resuspended to a concentration of 10% (w/v) in 20 mM PIPES (pH 6.8), 400 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1.3 Kunitz of DNase, 0.25 mg/mL lysozyme, and protease inhibitor cocktail (Roche) and incubated for 1–2 h (rocking) at 4 °C. The cell suspension was passed through a 0.8 mm needle prior to cell disintegration in a cell disruptor. Cell debris was pelleted for 1 h at 50000g and 4 °C. The supernatant was filtered through a 5 μm filter followed by a 0.45 μm filter before being loaded onto a POROS 20HS cation exchange column (PerSeptive Biosystems). Mboumar-9 transposase was eluted using a gradient from 400 to 1000 mM NaCl in 20 mM PIPES (pH 6.8) and 1 mM DTT. Mboumar-9 transposase eluted at 46 mS/cm conductivity, equivalent to ∼620 mM NaCl. Peak fractions were pooled and concentrated in a Vivaspin 6 30000 molecular weight cutoff PES (GE Healthcare) at 4 °C to a volume of <500 μL. Size-exclusion chromatography was conducted on a Superdex 200 30/100 GL column (GE Healthcare) connected to an ÄKTA purification system at 4 °C. Mboumar-9 transposase was eluted [5.7 μM in 50 μL of 20 mM PIPES (pH 6.8), 500 mM NaCl, 5 mM MgCl2, and 1 mM DTT] at a flow rate of 0.5 mL/min. Fractions containing Mboumar-9 were pooled, concentrated, snap-frozen, and then stored at −80 °C. The protein purity was estimated using ImageLab (Bio-Rad).
Thermal Denaturation Assay
Mboumar-9 transposase was in a buffer containing 50 mM PIPES (pH 6.8), 500 mM NaCl, and 1 mM DTT. Mos1 transposase was in a buffer containing 50 mM PIPES (pH 7.5), 250 mM KCl, and 1 mM DTT. Mboumar-9 and Mos1 IR DNA substrates had staggered ends mimicking the products of DNA cleavage. These were prepared by annealing the 32- or 28-nucleotide TS sequences of the Mboumar-9 or Mos1 IR, respectively (shown in Figure 1b) with the complementary 29- or 25-nucleotide NTS. The two strands were cooled from 95 °C to room temperature overnight in a buffer containing 100 mM Tris-HCl (pH 8.0), 10 mM EDTA (pH 8.0), and 100 mM NaCl. In the reaction mixtures, the final transposase concentration was 5 μM and, where added, the DNA concentration was 7.5 μM and the MgCl2 concentration 5 mM. Reaction mixtures were incubated on ice for 30 min before the fluorescent dye Sypro Orange (Sigma) was added to a final concentration of 5×. Any aggregates were removed by centrifugation for 10 min at 13000 rpm. Each experiment was performed in 45 μL in a 96-well plate and repeated three times. The temperature was increased in an iQ5 thermo cycler (Bio-Rad) from 4 to 95 °C, in 1 °C steps with 30 s between steps. The point of the most rapid change in fluorescence (excitation at 485 nm and emission at 575 nm) corresponds to the melting temperature (Tm) of the sample.
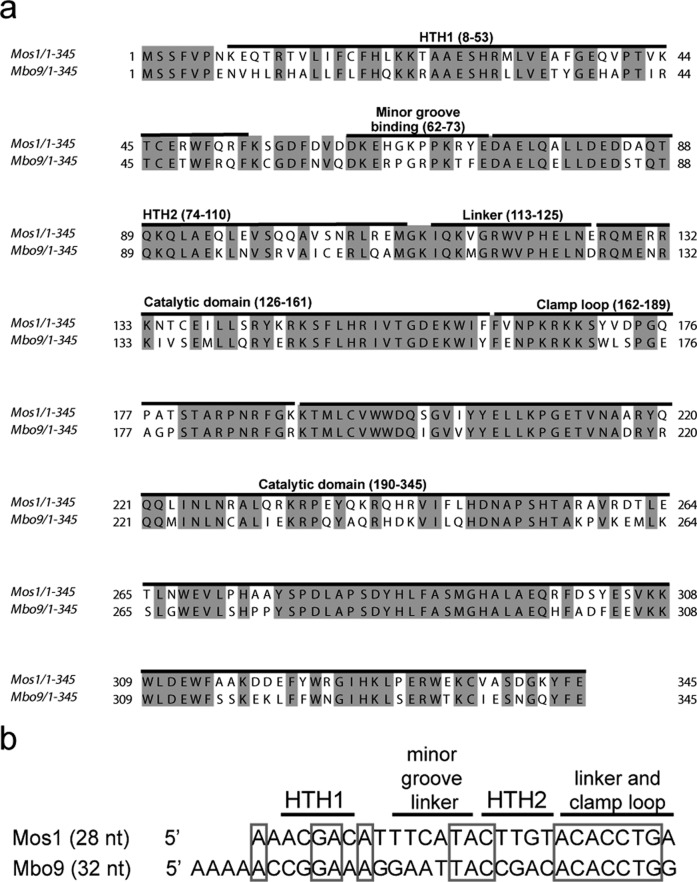
Figure 1.
Mos1 and Mboumar-9 are closely related active mariner transposases. (a) Alignment of the amino acid sequences of Mos1 and Mboumar-9 (Mbo9) transposases. Identical residues are shown with a shaded background. The domain names and amino acid ranges are indicated above the sequence. (b) Alignment of the Mos1 and Mboumar-9 inverted repeats, with identical bases boxed. Regions of the Mos1 transposase that interact with the Mos1 IR in the PEC structure are indicated above the Mos1 TS.
First- and Second-Strand Cleavage Assays
First- and second-strand cleavages were performed in a final volume of 20 μL. Reaction mixtures contained 25 mM HEPES (pH 7.5), 50 mM CH3COOK, 10% (v/v) glycerol, 0.25 mM EDTA, 1 mM DTT, 10 mM MgCl2, 50 μg/mL BSA, 20% (v/v) DMSO, 15 nM DNA, and transposase at 50, 100, or 200 nM. Reaction mixtures were incubated for 1 h at 30 °C, and reactions were stopped by the addition of 20 μL of loading buffer (90% (v/v) formamide and 20 mM EDTA). Samples were incubated at 95 °C for 5 min and transferred to ice, and 10 μL of each sample was loaded into each well of an 8% polyacrylamide denaturing gel containing 7.5 M urea and 1× TTE buffer (89 mM Tris base, 29 mM taurine, and 0.5 mM EDTA).
Substrate sequences for the first-strand cleavage assay were labeled with IRDye 700 on the 5′ end of the NTS. Mboumar-9 substrates (90 bp) are TS (5′-cacaaaatttaacgtgttttttgatttaAAAAACCGGAAAGGAATTACCGACACACCTGGtagtttctatattcaccgactggagcccgt-3′) and NTS (3′-gagttttaaattgcacaaaaaactaaatTTTTTGGCCTTTCCTTAATGGCTGTGTGGACCatcaaagatataagtggctgacctcgggca-5′), where the IR sequence is in uppercase and flanking DNA is in lowercase. Mos1 substrates (100 bp) are NTS (5′-tttctttttccacaaaatttaacgtgttttttgatttaaaaaAAACGACATTTCATACTTGTACACCTGAtagtttctatattcaccgactggagcccgt-3′) and TS (3′-aaagaaaaaggtgttttaaattgcacaaaaaactaaatttttTTTGCTGTAAAGTATGAACATGTGGACTatcaaagatataagtggctgacctcgggca-5′). DNA markers, 28 and 33 nucleotides in length, were used in these experiments and had the sequence of the Mos1 IR NTS.
Substrates for second-strand cleavage were labeled with IRDye 700 on the 5′ end of the TS (with the sequences described above) and mimicked the product of first-strand cleavage. These “prenicked” substrates were obtained by annealing the labeled TS with two NTS oligonucleotides. For Mboumar-9, the NTS strands had the sequences 3′-gagttttaaattgcacaaaaaactaaatTTTTTGGCCTTTCCTTAATGGCTGTGTGG-5′ and 3′-ACCatcaaagatataagtggctgacctcgggca-5′. For Mos1, the NTS oligonucleotides had the sequences 3′-aaagaaaaaggtgttttaaattgcacaaaaaactaaatttttTTTGCTGTAAAGTATGAACATGTGG-5′ and 3′-ACTatcaaagatataagtggctgacctcgggca-5′. The products were visualized after excitation of the IRDye 700 at 680 nm and detection on a LI-COR Odyssey scanner. The 70-nucleotide DNA marker used in these experiments had the sequence of the Mos1 IR TS.
In Vitro Transposon Cleavage Assay
Donor transposon plasmid (5.6 kb, 500 ng, 7.24 nM) was incubated with Mos1 or Mboumar-9 transposase (25, 50, 100, or 200 nM) in a final volume of 20 μL for 90 min at 30 °C in a buffer containing 25 mM HEPES (pH 7.5), 12.5 μg/mL BSA, 2 mM DTT, 100 mM NaCl, 10% (v/v) glycerol, and 10 mM MnCl2. To stop the reaction, 0.5 μL of 500 mM EDTA was added and the products were analyzed by agarose gel electrophoresis.
In Vitro Transposition Assay
Transposition was performed as described previously.27 The transposon donor plasmid contained a kanamycin resistance cassette (1.3 kb) flanked by IR sequences within a pEP185.2 plasmid backbone (4.3 kb), carrying the conditional origin of replication oriR6K. Transposon donor plasmid (5.6 kb, 500 ng, 7.24 nM) was incubated with pBSKS+ recipient plasmid (3 kb, 300 ng) and 72.4 nM transposase for 1 h at 30 °C in a final volume of 20 μL in buffer containing 25 mM HEPES (pH 7.5), 100 mM NaCl, 10% (v/v) glycerol, 2 mM DTT, 200 μg/mL acetylated BSA, and 10 mM MnCl2 or MgCl2. The buffer and DNA were mixed first, and transposase was added just before incubation. After 1 h, the reaction was stopped by addition of 80 μL of buffer containing 50 mM Tris (pH 7.5), 500 μg/mL proteinase K, 10 mM EDTA, and 6.25 μg/mL yeast tRNA and incubated for 1 h at 37 °C. DNA was phenol extracted and ethanol precipitated usually overnight. The DNA pellet was gently resuspended in 10 μL of dH2O at 70 °C. Competent cells were transfected with 10 μL of DNA and plated out on LB agar with 50 μg/mL kanamycin, and in dilutions on LB agar with 100 μg/mL carbenicillin to establish the competency of the cells. The transposition efficiency was calculated as the number of colonies resistant to kanamycin divided by the competency of the cells. Under the optimal conditions, approximately 4000 kanamycin resistant colonies per reaction were observed.
Results
Sequence Comparisons of Mboumar-9 and Mos1 Transposases and Inverted Repeats
Sequence comparisons reveal that Mos1 and Mboumar-9 transposases are the most closely related active mariner transposases described to date, and they share 68% identical amino acid sequences6,28 (Figure 1a). However, the transposon terminal inverted repeats are only 50% identical (Figure 1b), with the seven bases near the 3′ end of the TS, recognized by linker and clamp loop in the Mos1 PEC crystal structure, being the most highly conserved nucleotides.
Recombinant Mboumar-9 Transposase Is a Dimer in Solution
Mboumar-9 transposase, containing the same mutation (T216A) that rendered the recombinant Mos1 protein soluble, was expressed in E. coli and purified by cation exchange and size-exclusion chromatography (panels a and b of Figure 2, respectively), as described in Materials and Methods. The purity of the resulting protein was estimated to be 91% by SDS–PAGE (Figure 2b). We analyzed the oligomeric state of the DNA-free Mboumar-9 transposase in solution using analytical size-exclusion chromatography (Figure 2c). Mboumar-9 transposase eluted at a volume similar to that of Mos1 transposase from the same column (data not shown). The elution volume (13.8 mL) corresponds to an approximate globular mass of 98.4 kDa. Because the mass of the Mboumar-9 transposase monomer is 40.7 kDa, we infer that, like Mos1,26 Mboumar-9 transposase exists as a dimer with an elongated shape in the absence of DNA.
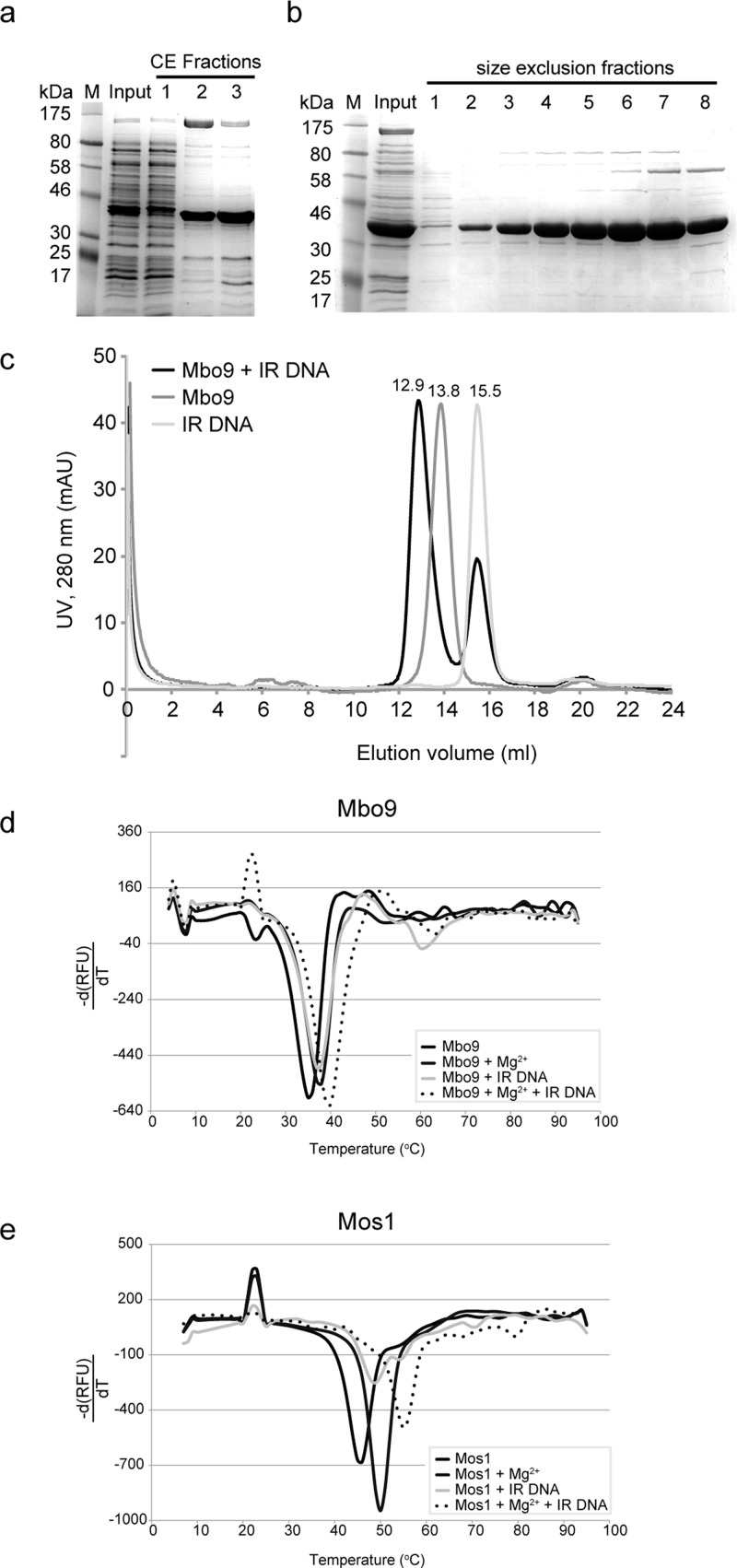
Figure 2.
Purification and DNA binding of Mboumar-9 transposase. (a) SDS–PAGE of fractions from the cation exchange (CE) purification step. The input is crude, soluble cell extract; fraction 1 is the flow-through, and fractions 2 and 3 are from the Mboumar-9 peak. (b) SDS–PAGE of fractions from the second purification step by size-exclusion chromatography. The input is the fraction from CE; fractions 1–8 are across the Mboumar-9 transposase peak. (c) Elution profiles from analytical size-exclusion chromatography of Mboumar-9 (Mbo9) IR DNA (light gray), Mboumar-9 transposase (dark gray), and a complex (black). The complex eluted at 12.9 mL, earlier than the DNA-free transposase (at 13.8 mL) and the IR DNA (at 15.5 mL). (d) Thermal denaturation of Mboumar-9 transposase in the absence or presence of Mg2+ or IR DNA, and with both. The rate of change of fluorescence, measured in relative fluorescence units (RFU), is plotted vs temperature. (e) Thermal denaturation of Mos1 transposase under equivalent conditions.
Mboumar-9 Transposase Forms a Stable Complex with IR DNA
To test if Mboumar-9 transposase could bind IR DNA, we incubated (10 μM) transposase with the DNA substrate (10 μM) containing the Mboumar-9 IR sequence that mimics the product of staggered Mboumar-9 excision (see below). While we did not attempt to detect the binding of the transposase to DNA by an electrophoretic mobility shift assay, we analyzed complex formation by analytical size-exclusion chromatography. We observed a peak indicative of a protein–DNA complex eluting at 12.9 mL (Figure 2c), earlier than the DNA-free transposase and the DNA substrate (elution volume of 15.5 mL).
Mboumar-9 Transposase Is Stabilized by IR DNA and Mg2+ Ions
Next we used thermal denaturation assays to analyze the thermal stability of Mboumar-9 transposase and its complex with IR DNA. We found that Mboumar-9 transposase was less stable than Mos1 transposase, because the melting temperature (Tm) of Mboumar-9 transposase was 35.0 °C (Figure 2d) compared to 46.0 °C for Mos1 transposase (Figure 2e). mariner transposases have a characteristic DDD motif involved in binding the divalent metal ions necessary for DNA cleavage and integration.3,22 We found that both Mboumar-9 and Mos1 transposases were stabilized in the presence of 5 mM MgCl2; the Tm of Mboumar-9 increased by 2.7 °C (Figure 2d) and by 4.0 °C for Mos1 (Figure 2e) under this condition.
Upon addition of IR DNA (in the absence of Mg2+ ions), the Tm increased to 37.0 °C for Mboumar-9 and 48.7 °C for Mos1, consistent with formation of a complex in each case. The thermal stability of the transposase–DNA complex increased further when 5 mM MgCl2 was added, which we observed by an increase in the Tm to 39.7 °C for Mboumar-9 and 55.0 °C for Mos1.
In Vitro DNA Cleavage of Mboumar-9 Inverted Repeats
To assay the first- and second-strand cleavage activity of Mboumar-9 transposase, we incubated linear, fluorescently labeled DNA substrates containing the Mboumar-9 IR sequence with increasing concentrations of Mboumar-9 transposase (as shown schematically in panels a and b of Figure 3). We compared this with the activity of Mos1 transposase on DNA substrates containing the Mos1 IR (Figure 3c,d). The cleavage products were visualized on 8% polyacrylamide denaturing gels. To establish if there is any cross-recognition between mariner transposases, we also compared the cleavage activity of Mos1 transposase on the Mboumar-9 IR substrates and vice versa (Figure 3).
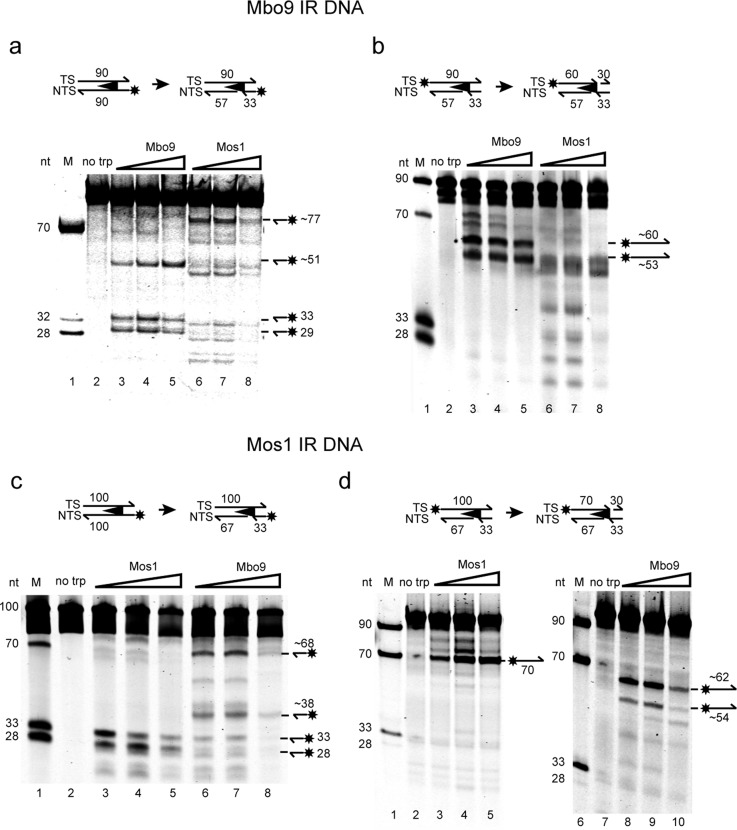
Figure 3.
In vitro first- and second-strand cleavage by Mboumar-9 and Mos1 transposases. (a) Schematic for first-strand cleavage of the Mboumar-9 IR. The star denotes the fluorescent label at the 5′ end of the NTS. Denaturing polyacrylamide gel of reactions performed with no transposase (lane 2), Mboumar-9 (Mbo9) transposase (lanes 3–5), or Mos1 transposase (lanes 6–8). Lane 1 contained fluorescently labeled DNA markers of 70, 32, and 28 nucleotides. (b) Second-strand cleavage of the Mboumar-9 IR, prenicked to bypass the first-strand cleavage reaction. The denaturing PAGE of reactions, with lanes as described for panel a except a 33-nucleotide marker, was used in place of the 32-nucleotide marker. (c) Schematic of first-strand cleavage of the Mos1 IR and denaturing PAGE of the reaction products. Lane 1 contained markers as in panel b. Reaction mixtures in lane 2 had no transposase; those in lanes 3–5 contained Mos1 transposase, and those in lanes 6–8 contained Mboumar-9 transposase. (d) Second-strand cleavage of Mos1 IR substrate, prenicked at the site of first-strand cleavage: lanes 1 and 6, DNA markers; lanes 2 and 7, no transposase controls; lanes 3–5, reaction mixtures with Mos1 transposase; lanes 8–10, reaction mixtures with Mboumar-9 transposase.
First-strand cleavage of the 90 bp Mboumar-9 IR (labeled at the 5′ end of the NTS) by Mboumar-9 transposase produced three fluorescent products 33, 29, and ∼51 nucleotides in length (Figure 3a). The 33- and 29-nucleotide products correspond to excision three bases within the IR and one base outside the IR, respectively. The 51-nucleotide product is most likely due to cleavage 21 nucleotides within the inverted repeat. Incubation of this Mboumar-9 IR substrate with Mos1 transposase resulted in nonspecific cleavage (Figure 3a).
Second-strand cleavage reactions were performed using Mboumar-9 DNA substrates with a prenicked NTS (mimicking the product of first-strand cleavage) and a fluorescent label on the 5′ end of the TS. Incubation of the Mboumar-9 IR with Mboumar-9 transposase resulted in one product of 60 nucleotides, corresponding to cleavage precisely at the junction of the IR and flanking DNA, and a second product of ∼53 nucleotides (Figure 3b). Incubation of the Mboumar-9 IR substrate with Mos1 transposase resulted in nonspecific cleavage of the TS, indicating that Mos1 transposase does not recognize the Mboumar-9 IR as a substrate for transposition.
We also performed the reciprocal experiments using DNA substrates containing the Mos1 IR sequence (Figure 3c,d). As observed previously,13 Mos1 transposase cleaved the Mos1 NTS to produce two products of 33 and 28 nucleotides. Mboumar-9 transposase cleaved this substrate nonspecifically, although one of the minor products was 33 nucleotides in length, corresponding to cleavage 3 bp inside the Mos1 IR, as observed for cleavage with Mos1 transposase.
In the second-strand cleavage assay, Mos1 transposase cleaved the Mos1 TS precisely at the junction of the IR and flanking DNA sequence, to give a 70-nucleotide product, as observed previously.13 Mboumar-9 transposase also cleaved the Mos1 TS to produce a major band of ∼62 nucleotides and a minor band at ∼54 nucleotides, neither of which corresponds to cleavage allowing normal transposition of Mos1.
Cross-Recognition and Activity of mariner Transposases on Plasmid DNA
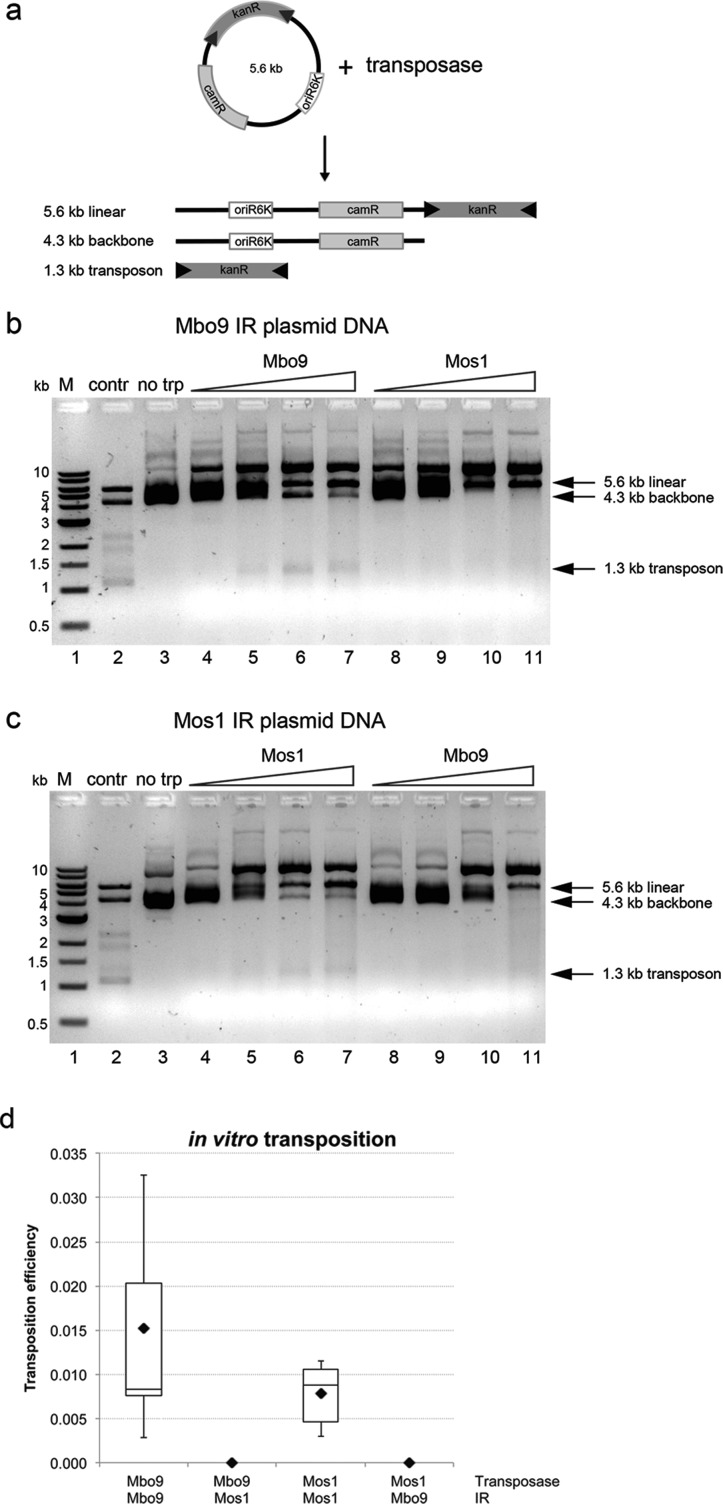
To test transposase activity on plasmid DNA, we first assayed excision of a kanamycin gene flanked by either Mboumar-9 or Mos1 inverted repeats, as shown schematically in Figure 4a. Incubation of the plasmid bearing Mboumar-9 inverted repeats with Mboumar-9 transposase resulted in excision of the 1.3 kb transposon (Figure 4b). However, with Mos1 transposase, we observed only the relaxed and linear plasmid, indicating nonspecific cleavage activity. Similar results were obtained in the reciprocal experiment: Mos1 transposase excised the 1.3 kb transposon flanked with Mos1 inverted repeats, whereas Mboumar-9 transposase cleaved this plasmid nonspecifically (Figure 4c). Thus, the cleavage activities of the enzymes on both plasmid DNA and linear DNA substrates are consistent, with no cross-recognition of inverted repeat sequences evident in either case.
Figure 4.
Transposon excision and in vitro transposition. (a) Schematic of the plasmid-based cleavage assays. The transposon donor plasmids (5.6 kb) contain a kanamycin resistance gene flanked by the inverted repeats (black triangles) of either Mos1 or Mboumar-9 (Mbo9). The expected products and their size (in kilobases) are indicated. (b) Agarose gel of the products of cleavage of the Mboumar-9 plasmid by Mboumar-9 (lanes 4–7) and Mos1 transposase (lanes 8–11). (c) Agarose gel of the products of cleavage of the Mos1 plasmid by Mos1 (lanes 4–7) and Mboumar-9 transposase (lanes 8–11). The control (lane 2) contained Mboumar-9 plasmid linearized with XbaI and digested with SacI, which cleaves outside the IRs. (d) Efficiencies of in vitro transposition reactions performed using Mboumar-9 or Mos1 transposase and donor plasmids containing either the Mboumar-9 or Mos1 inverted repeat (IR). Nine repeats were performed for experiments in which transposase acted on their own repeats, and cross-reactivity experiments were performed in triplicate.
Next, we tested if Mboumar-9 and Mos1 transposases could catalyze in vitro transposition of noncognate sites, using bacterial donor and target plasmids. Transposition of the kanamycin resistance gene flanked by Mboumar-9 inverted repeats occurred with Mboumar-9 transposase but not with Mos1 transposase (Figure 4d). Similarly, when the gene was flanked by Mos1 IRs, we observed transposition only with Mos1 transposase. Thus, the enzymes catalyze in vitro transposition from their cognate inverted repeat sequences only.
Target Site Selection Is Random and Depends Only on the Presence of TA Dinucleotides
To analyze the transposon insertion sites, we sequenced a total of 31 insertion sites for Mboumar-9 transposition and 30 for Mos1 transposition. For reactions performed in the presence of Mg2+, all the insertions occurred at TA target sites, the hallmark of transposons of the mariner/Tc1 family (Figure 5 and Figure 1 of the Supporting Information). As observed previously,6,27 the preference for insertion at TA is weakened in the presence of Mn2+, but this effect is less dramatic for Mboumar-9 than for Mos1 (Figure 5b,d).
Figure 5.
Sequence logos of Mboumar-9 and Mos1 transposition insertion sites. Graphical representation of the alignment of in vitro transposition integration sites. The 19 nucleotides of target plasmid on either side of the central TA dinucleotide were aligned. Results are from (a) 16 in vitro Mboumar-9 (Mbo9) transposition reactions performed with Mg2+ and (b) 15 reactions with Mn2+ and (c) 14 Mos1 transposition reactions with Mg2+ or (d) 16 with Mn2+. This figure was prepared using the WebLogo server.33
Within the target plasmid, 94 TA sites are available for integration in nonessential regions of the plasmid (i.e., out with the antibiotic resistance gene and the origin of replication). We did not observe any sequence preference around these TA sites for either of the transposons, consistent with similar experiments on Hsmar129 and Mboumar-9.6 We found that of the 61 insertion sites sequenced from both transposons, nine common sites were chosen by the two transposases. This is close to the mean number of integration events expected by chance, suggesting that the nine observed common sites were selected randomly by both transposases.
Discussion
We have expressed and purified untagged full-length Mboumar-9 transposase, a mariner/Tc1 family DNA transposase closely related to Mos1 transposase. Like Mos1, the enzyme forms a dimer in solution, and this was recently established as a prerequisite for autoregulation in mariner transposition.30 The enzyme is stabilized by binding of Mg2+ ions, which are required for the DNA cleavage and integration reactions. Purified Mboumar-9 transposase forms a stable complex with DNA substrates representing precleaved Mboumar-9 inverted repeats.
Transposon excision and in vitro transposition occurred only when the transposase acted on its own IR sequence; we observed minimal cross-recognition of the transposon ends in cleavage assays using linear DNA substrates or plasmid substrates. In second-strand cleavage assays, two aberrant products were observed for cleavage of the Mos1 TS by Mboumar-9 transposase. Similar results were observed for Himar1 transposase, which aberrantly cleaved the Mos1 IR in first-strand cleavage assays.19 Cleavage of the Mos1 IR by both Mboumar-9 and Himar1 transposases could reflect the lower specificity of these transposases for the DNA substrate. The inability of Mos1 transposase to cleave the IR DNA of Mboumar-9 or Himar1 indicates the higher specificity of this enzyme.
We confirmed previous observations31,32 that Mos1 shows no target site selection in vitro except for integration at TA nucleotides. Our work shows that Mboumar-9 transposase behaves similarly; no integration site specificity was noticed for Mboumar-9 transposition in vitro. Furthermore, we noticed the loss of TA specificity for Mboumar-9 transposition in the presence of Mn2+, as found previously for Mos1.27
This study showed that two closely related and active mariner transposases, Mos1 and Mboumar-9, have similar biophysical properties. They also have similar DNA cleavage and integration activities. This raises the possibility of using these transposases to create chimeric transposases with designed IR DNA recognition specificity, for example, by swapping DNA-binding motifs between the two enzymes or by mutating Mboumar-9 helix–turn–helix motif residues to mimic sequence-specific interactions observed in the Mos1 PEC crystal structure. These may also lead to improvement of the efficiencies of mariner transposition systems as genome manipulation tools.
Acknowledgments
We thank Dr. Sean Colloms (University of Glasgow, Glasgow, Scotland) for useful discussions and Dr. Alastair Kerr (University of Edinburgh, Edinburgh, Scotland) for help with the statistical analysis of transposon insertion sites.
Glossary
Abbreviations
- TE
transposable element
- IR
inverted repeat
- PEC
paired-end complex
- NTS
nontransferred strand
- TS
transferred strand
- nt
nucleotide
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- EDTA
ethylenediaminetetraacetic acid
- PIPES
1,4-piperazinediethanesulfonic acid
- DTT
dithiothreitol
- Dnase
deoxyribonuclease
- tRNA
transfer ribonucleic acid
- PAGE
polyacrylamide gel electrophoresis
- Mbo9
Mboumar-9
- SDS
sodium dodecyl sulfate.
Supporting Information Available
Supplementary Figure 1. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by a Wellcome Trust grant (085176/Z/08/Z) to J.M.R. and a Ph.D. studentship from the Darwin Trust of Edinburgh to M.T.
Supplementary Material
References
- Delauriere L.; Chenais B.; Hardivillier Y.; Gauvry L.; Casse N. (2009) Mariner transposons as genetic tools in vertebrate cells. Genetica 137, 9–17. [DOI] [PubMed] [Google Scholar]
- Ivics Z.; Li M. A.; Mates L.; Boeke J. D.; Nagy A.; Bradley A.; Izsvak Z. (2009) Transposon-mediated genome manipulation in vertebrates. Nat. Methods 6, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H.; Izsvak Z.; Ivics Z. (1999) Resident aliens: The Tc1/mariner superfamily of transposable elements. Trends Genet. 15, 326–332. [DOI] [PubMed] [Google Scholar]
- Medhora M.; Maruyama K.; Hartl D. L. (1991) Molecular and functional analysis of the mariner mutator element Mos1 in Drosophila. Genetics 128, 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry E. G.; Witherspoon D. J.; Lampe D. J. (2004) A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics 166, 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Lopez M.; Siddique A.; Bischerour J.; Lorite P.; Chalmers R.; Palomeque T. (2008) Transposition of Mboumar-9: Identification of a new naturally active mariner-family transposon. J. Mol. Biol. 382, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukeris T. G.; Arca B.; Livadaras I.; Dialektaki G.; Savakis C. (1995) Introduction of the transposable element Minos into the germ line of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 92, 9485–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. J.; Carlson D. F.; Leaver M. J.; Foster L. K.; Fahrenkrug S. C. (2009) Passport, a native Tc1 transposon from flatfish, is functionally active in vertebrate cells. Nucleic Acids Res. 37, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z.; Hackett P. B.; Plasterk R. H.; Izsvak Z. (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510. [DOI] [PubMed] [Google Scholar]
- Miskey C.; Izsvak Z.; Plasterk R. H.; Ivics Z. (2003) The Frog Prince: A reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31, 6873–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe D. J.; Churchill M. E.; Robertson H. M. (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15, 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- Miskey C.; Papp B.; Mates L.; Sinzelle L.; Keller H.; Izsvak Z.; Ivics Z. (2007) The ancient mariner sails again: Transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell. Biol. 27, 4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. M.; Colloms S. D.; Finnegan D. J.; Walkinshaw M. D. (2009) Molecular architecture of the Mos1 paired-end complex: The structural basis of DNA transposition in a eukaryote. Cell 138, 1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A.; Berghammer A. J.; Averof M.; Klingler M. (2004) Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: Quantitative and qualitative analysis of genomic integration events. Genetics 167, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates C. J.; Jasinskiene N.; Morgan D.; Tosi L. R.; Beverley S. M.; James A. A. (2000) Purified mariner (Mos1) transposase catalyzes the integration of marked elements into the germ-line of the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 30, 1003–1008. [DOI] [PubMed] [Google Scholar]
- Dawson A.; Finnegan D. J. (2003) Excision of the Drosophila mariner transposon Mos1. Comparison with bacterial transposition and V(D)J recombination. Mol. Cell 11, 225–235. [DOI] [PubMed] [Google Scholar]
- Auge-Gouillou C.; Hamelin M. H.; Demattei M. V.; Periquet G.; Bigot Y. (2001) The ITR binding domain of the Mariner Mos-1 transposase. Mol. Genet. Genomics 265, 58–65. [DOI] [PubMed] [Google Scholar]
- Claeys B. C.; Chalmers R. (2010) Transposition of the human Hsmar1 transposon: Rate-limiting steps and the importance of the flanking TA dinucleotide in second strand cleavage. Nucleic Acids Res. 38, 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan N. (2007) Comparison of the mariner transposons Mos1 and Himar1. M.Sc. Research Thesis, University of Edinburgh, Edinburgh, Scotland. [Google Scholar]
- Germon S.; Bouchet N.; Casteret S.; Carpentier G.; Adet J.; Bigot Y.; Auge-Gouillou C. (2009) Mariner Mos1 transposase optimization by rational mutagenesis. Genetica 137, 265–276. [DOI] [PubMed] [Google Scholar]
- Richardson J. M.; Zhang L.; Marcos S.; Finnegan D. J.; Harding M. M.; Taylor P.; Walkinshaw M. D. (2004) Expression, purification and preliminary crystallographic studies of a single-point mutant of Mos1 mariner transposase. Acta Crystallogr. D60, 962–964. [DOI] [PubMed] [Google Scholar]
- Richardson J. M.; Dawson A.; O’Hagan N.; Taylor P.; Finnegan D. J.; Walkinshaw M. D. (2006) Mechanism of Mos1 transposition: Insights from structural analysis. EMBO J. 25, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A.; Goryshin I. Y.; Reznikoff W. S. (1999) Hairpin formation in Tn5 transposition. J. Biol. Chem. 274, 37021–37029. [DOI] [PubMed] [Google Scholar]
- Hickman A. B.; Perez Z. N.; Zhou L.; Musingarimi P.; Ghirlando R.; Hinshaw J. E.; Craig N. L.; Dyda F. (2005) Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 12, 715–721. [DOI] [PubMed] [Google Scholar]
- Auge-Gouillou C.; Brillet B.; Germon S.; Hamelin M. H.; Bigot Y. (2005) Mariner Mos1 transposase dimerizes prior to ITR binding. J. Mol. Biol. 351, 117–130. [DOI] [PubMed] [Google Scholar]
- Cuypers M. G.; Trubitsyna M.; Callow P.; Forsyth V. T.; Richardson J. M. (2013) Solution conformations of early intermediates in Mos1 transposition. Nucleic Acids Res. 41, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Dawson A.; Finnegan D. J. (2001) DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 29, 3566–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomeque T.; Antonio C. J.; Munoz-Lopez M.; Lorite P. (2006) Detection of a mariner-like element and a miniature inverted-repeat transposable element (MITE) associated with the heterochromatin from ants of the genus Messor and their possible involvement for satellite DNA evolution. Gene 371, 194–205. [DOI] [PubMed] [Google Scholar]
- Claeys Bouuaert C.; Chalmers R. (2013) Hsmar1 Transposition Is Sensitive to the Topology of the Transposon Donor and the Target. PLoS One 8, e53690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys B. C.; Lipkow K.; Andrews S. S.; Liu D.; Chalmers R. (2013) The autoregulation of a eukaryotic DNA transposon. Elife 2, e00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi L. R.; Beverley S. M. (2000) cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res. 28, 784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenes G.; Moundras C.; Demattei M. V.; Bigot Y.; Petit A.; Renault S. (2010) Target site selection by the mariner-like element, Mos1. Genetica 138, 509–517. [DOI] [PubMed] [Google Scholar]
- Crooks G. E.; Hon G.; Chandonia J. M.; Brenner S. E. (2004) WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.