Abstract
In this issue of Molecular Cell, Conaway and colleagues (Yao et al., 2008) provide a glimpse into an interesting mechanism to control a deubiquitylating enzyme via interaction of two complexes—the 19S proteasome regulatory particle and an ATP-dependent nucleosome remodeling complex.
Enzyme activity must be tightly regulated spatially and temporally. The deubiquitylases (DUBs) are an increasingly large class of enzymes whose diverse roles in biological processes include genomic regulation. A vexing question has been the basis of tight regulation of these enzymes, which can occur in large, modular protein complexes linked to transcription and to DNA repair. In this issue of Molecular Cell, a novel regulatory interaction is revealed, which involves 19S proteasome regulatory particle (RP) activation of latent DUB Uch37 residing within a nucleosome remodeling complex and provides a model for timely enzyme activation (Yao et al., 2008).
There have been tantalizing hints over many years that the ubiquitylation system is directly linked to genomic regulation and to transcription (Baker and Grant, 2005; Collins and Tansey, 2006). This evidence includes observations that the 19S RP is recruited to genes to regulate transcriptional elongation and that conjugation of ubiquitin to transcriptional activators and ubiquitin-mediated proteolytic turnover are actually required for activation. More recently, histones have been shown to be dynamically ubiquitylated and deubiquitylated to activate transcription. Indeed, the SAGA histone acetylation complex is both recruited by the 19S RP and, itself, contains a histone deubiquitylation enzyme. Moreover, recent evidence indicates that histones are also dynamically ubiquitylated at sites of DNA damage. These observations indicate an intricate cycle of ubiquitylation/deubiquitylation during transcriptional activation and DNA repair and implicate the proteasome, or the proteasome 19S RP, in this regulation. The findings further beg the question of just how the activity of deubiquitylation enzymes is held in check until the right time and the right place.
The DUB Uch37 is a component of the 19S RP, where it deubiquitylates substrates prior to their proteolysis. It associates with the 19S RP via the Rpn13 subunit, the proteasomal ubiquitin receptor that activates Uch37 deubiquitylating activity (Husnjak et al., 2008; Yao et al., 2006). Recently, Uch37 has also been suggested to be a subunit of the human INO80 complex. INO80 has been of intense interest in recent years—it is an ATP-dependent chromatin remodeling complex that alters nucleosome positioning on DNA during both transcription and DNA repair (Cai et al., 2007). Conaway and colleagues now provide a detailed mechanistic analysis of Uch37; their results show that INO80 and Rpn13/19S proteasome have opposing effects on Uch37 activity and that the complexes work together to regulate deubiquitylation by Uch37 (Yao et al., 2008).
These researchers purified an epitope-tagged Uch37 from mammalian cells and used rigorous quantitative mass spectroscopy to identify Uch37 complexes in cytoplasmic or nuclear fractions. They found that INO80-associated Uch37 is exclusively nuclear. Further analysis revealed that the association of Uch37 is mediated by the INO80 subunit NFRKB, a DNA-binding transcriptional activator (see below for comments on this finding). Uch37 is also associated with the 26S proteasome in the nucleus and the cytoplasm.
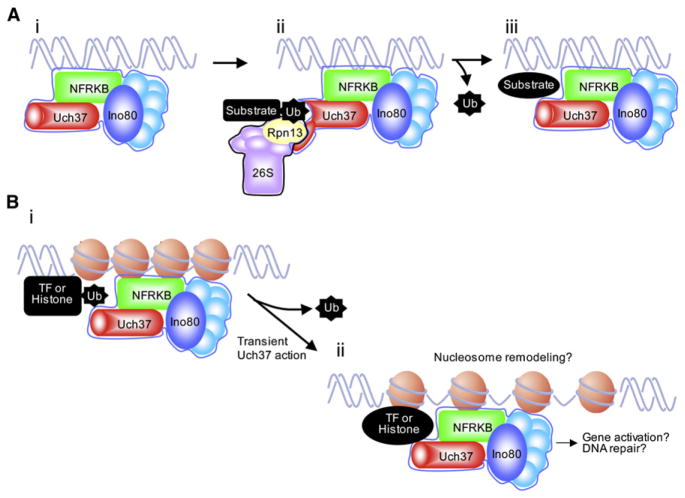
One of the key observations in the study is that, in association with INO80, Uch37 could neither bind nor hydrolyze its ubiquitin substrate. However, when the INO80 complex was exposed to Rpn13, Uch37 was able to hydrolyze ubiquitin. This occurred while Uch37 was still complexed to INO80, as Rpn13 addition did not remove Uch37 from the INO80 complex. Furthermore, Rpn13 did not stably bind Uch37 when Uch37 was complexed with INO80. Therefore, it appears that Rpn13 can transiently activate Uch37 in a “hit-and-run” manner without disrupting its association with INO80. This mechanism of transient Uch37 activation allows tight control over its activity when it is part of the INO80 complex. These findings are summarized in the model shown in Figure 1A.
Figure 1. Uch37 Regulation and Activation.
(A) Mechansim of Uch37 regulation. (Ai) Uch37 associates with the INO80 complex through the DNA-binding NFRKB subunit and is in an inactive state. (Aii) Rpn13 binds ubiquitin and may deliver ubiquitylated substrates to Uch37. It interacts with the autoinhibitory tail of Uch37 to activate it, leading to substrate deubiquitylation. The identity of the substrate is unknown. Rpn13 is depicted here as a part of the 26S proteasome, but it can also perform these functions alone. (Aiii) The interaction between Rpn13 and Uch37 is transient and does not disrupt the Uch37-INO80 complex.
(B) Possible biological outcomes of Uch37 activation. (Bi) Rpn13-mediated Uch37 activation at chromatin might function to deubiquitylate histones or transcription factors, which could alter their conformation or activity. (Bii) This could be involved in INO80-mediated transcriptional activation or DNA repair. It is also possible that Uch37 activity affects nucleosome remodeling by INO80.
Whether the proteasome itself has a role in the Uch37/INO80 pathway remains unclear. As described above, proteasomal subunits have established roles in regulating gene transcription and DNA-damage repair, but only some of these rely on proteolytic activity. For example, termination of transcription after DNA damage is achieved through proteasome-mediated degradation of RNA polymerase. Other gene regulatory functions of proteasomal subunits are nonproteolytic but are dependent on proteasomal ATPases. These include targeting SAGA, promoting histone modifications, and promoting transcriptional elongation (Baker and Grant, 2005). Stimulation of deubiquitylating activity can now be added to this list of proteasomal functions, but neither ATPase nor proteolytic function of the proteasome is required, as Rpn13 alone can activate Uch37. However, it remains to be seen whether Uch37-mediated biological outcomes rely on linking INO80-Uch37-mediated deubiquitylation to proteolysis. Perhaps Rpn13, the proteasomal ubiquitin receptor, delivers substrates to INO80-Uch37 for deubiquitylation prior to feeding them into the 20S proteolytic barrel for degradation. For example, as mentioned above, certain transcriptional activators are ubiquitylated to regulate their stability or activity (Collins and Tansey, 2006) and INO80-bound Uch37 could regulate transcription through these mediators.
An alternative scenario is that Uch37-mediated deubiquitylation occurs in the absence of the 20S subunit, most likely to alter substrate function in transcription or DNA repair. Because INO80 is involved in transcriptional regulation, possible substrates for Uch37 are the histones H2A and H2B and transcriptional activators. When histones H2A and H2B are mono-ubiquitylated, transcription is stalled. Histone deubiquitylation is required for assembly of protein complexes at the promoter and movement through the nucleosome (Weake and Workman, 2008), which apparently does not involve proteolysis. Coupling deubiquitylating activity with the chromatin remodeling function of INO80 would promote gene expression. A general model encompassing these speculations is shown in Figure 1B.
A host of questions emerge from this work. First, does proteasome-activated Uch37 affect INO80-mediated nucleosome remodeling activity? This question can be addressed in vitro in classic nucleosome remodeling assays, using INO80 bearing inactive or proteasomally activated Uch37 and nucleosomes bearing ubiquitylated H2A/H2B. Second, is Uch37 directly involved in gene regulation or DNA repair, two processes that INO80 has been linked to in vivo? And finally, what genes are targeted by Uch37-containing INO80? The association of Uch37 with the DNA-binding NFRKB subunit of INO80 suggests that Uch37’s role in gene regulation could be restricted to specific genes that contain NFRKB binding sites. This may be an emerging theme for gene-regulatory deubiquitylases, as another ubiquitin protease, Ubp8, is targeted to gene loci through association with SAGA and the activators that recruit the coactivator complex to promoters. Identification of Uch37 substrates and NFRKB-dependent genes will shed light on exactly what biological pathways are mediated by this enzyme.
References
- Baker SP, Grant PA. Cell. 2005;123:361–363. doi: 10.1016/j.cell.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]