Abstract
Highly pathogenic H5N1 influenza viruses have been isolated from a number of avian and mammalian species. Despite intensive control measures the number of human and animal cases continues to increase. A more complete understanding of susceptible species and of contributing environmental and molecular factors is crucial if we are to slow the rate of new cases. H5N1 is currently endemic in domestic poultry in only a handful of countries with sporadic and unpredictable spread to other countries. Close contact of terrestrial bird or mammalian species with infected poultry/waterfowl or their biological products is the major route for interspecies transmission. Intra-species transmission of H5N1 in mammals, including humans, has taken place on a limited scale though it remains to be seen if this will change; recent laboratory studies suggest that it is indeed possible. Here we review the avian and mammalian species that are naturally susceptible to H5N1 infection and the molecular factors associated with its expanded host range.
Keywords: H5N1, Host range, Mammals, Avian influenza
1. Introduction
Influenza A viruses belong to the family Orthomyxoviridae with the majority of viruses maintained in wild aquatic bird reservoirs, predominantly those of the anseriform species. Classification of these viruses is determined by the two surface glycoproteins, the hemagglutinin (HA) and neuraminidase (NA) of which 16 HA and 9 NA subtypes have been isolated from aquatic birds (Kim et al., 2009). The migratory nature of the aquatic fowl reservoir results in the wide geographic spread and distribution of most circulating subtypes. A consequence of this geographic distribution in migratory birds is the is the potential contact of infected bird with domestic avian and mammalian species, including humans, with novel influenza A subtypes that can cross the host range barrier and initiate local epidemics or widespread influenza pandemics (Webster et al., 1992).
Since their detection in Asia in 1996, the highly pathogenic avian influenza (HPAI) H5N1 viruses have spread to many countries in the eastern hemisphere becoming endemic in populations of domestic birds in a more restricted number of countries and transiently infecting a number of other hosts along the way. The mechanism of virus spread throughout the hemisphere is not entirely clear, but probably as a result of both wild birds and poultry trade. While wild and domestic species may have contributed to spread, it is at the level of the domestic waterfowl and poultry interface that many mammalian species, humans included, are likely at the highest risk of acquiring H5N1 infection (Fig. 1). Because of the high potential for H5N1 to negatively impact veterinary and human health and subsequently the global economy, understanding the host range of the virus is of utmost importance. This review will focus on detailing the avian species susceptible to infection with HPAI H5N1 influenza viruses and the factors (and species) that influence the onward transmission to mammalian hosts (Fig. 2).
Fig. 1. Routes of H5N1 species distribution.
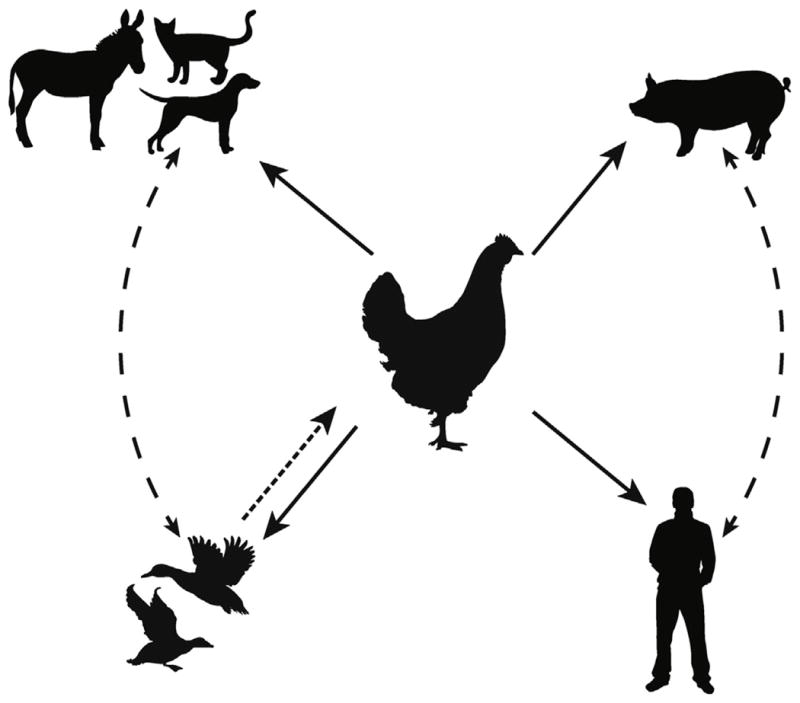
H5N1 influenza viruses cross the species barrier through close contact with infected birds, in particular domestic poultry. Contact with domestic poultry (chickens, ducks, etc.) has resulted in the inter-species transmission of H5N1 to non-avian hosts. Infrequently documented is the transmission of H5N1 from wild migratory birds to domestic and/or captive mammals. Though not documented, swine to human transmission of influenza subtypes H1N1, H2N2, and H3N2 does occur in nature.
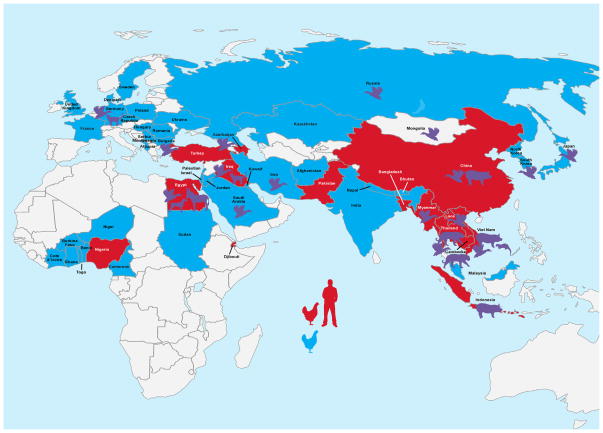
Fig. 2. Global distribution of H5N1 infection.
Countries shaded in red have concurrent H5N1 outbreak in domestic poultry and humans. Blue countries signify outbreaks only in wild avian species. Additional cases of mammalian H5N1 are depicted as shaded figures (http://www.fao.org/avianflu, http://www.oie.int/wahis2/public/wahid.php/Diseaseinformation/Diseasedistributionmap, http://www.who.int/influenza/humananimalinterface/ENGIP20120810CumulaCumulativeN5N1cases.pdf).
Influenza viruses encoding HA of subtype H5 and paired with varying NA subtypes are maintained as low pathogenic avian influenza (LPAI) forms in many anseriform species (Sharp et al., 1997, 1993; Süss et al., 1994). In 1996 the antecedent HPAI H5N1 virus A/Goose/Guangdong/1/96 was detected on a goose farm in Southern China causing an outbreak in the summer and early fall with 40% morbidity in birds (Xu et al., 1999). Reassortants of this virus were next detected in domestic poultry, ducks, and geese in Hong Kong in 1997 (Shortridge et al., 1998) with associated human cases resulting from direct contact with infected birds {Claas:1998jy}. Genetic analysis of all 8 gene segments from the 1997 viruses revealed substantial homology to other concomitantly circulating avian influenza viruses; with the internal genes being donated by either A/Quail/Hong Kong/G1/1997 (H9N2) or A/Teal/Hong Kong/W312/1997 (H6N1) viruses of domestic poultry (Guan et al., 1999; Hoffmann et al., 2000). While the 1997 outbreak was controlled following depopulation, H5N1 viruses continued to circulate in the wider region in at least ducks and geese (Guan et al., 2002a) generating a number of reassortants. While the reassortment partners of the H5N1 viruses were considered to be viruses of aquatic bird origin, there was no evidence of infection of wild birds with the highly pathogenic virus. This changed in 2002 when H5N1 viruses were detected in resident waterfowl and flamingos as well as wild little egrets and gray herons and other wild migratory birds; of particular note was the fact that the infection was lethal in some of the aquatic birds, an atypical presentation in these hosts {Sturm-Ramirez:2004dq}. While it is unclear what hosts were critical in maintaining the virus during the intervening time, 2004 saw the emergence of H5N1 in an extended number of countries {Li:2004ez}. While the details of this geographic expansion and associated genetic diversification are detailed elsewhere in this publication, it was associated with an expansion of hosts in which the virus has been found.
2. H5N1 influenza in wild bird species
All influenza virus subtypes circulate in wild bird populations taxonomically classified in the orders Anseriformes and Charadriiformes; the order Anseriformes is composed of aquatic species (ducks, geese, swans) whereas shore birds (gulls, sandpipers, surf-birds, terns) are grouped into order Charadriiformes. Though many avian species are susceptible to infection with H5N1 influenza viruses, it is the anseriform species that have been implicated in their global spread (Kim et al., 2009). While it is generally accepted that transition to the highly pathogenic form of influenza virus occurs in gallinaceous poultry species and the majority of prior HPAI outbreaks have been confined to these hosts, the HPAI H5N1 virus has subsequently transmitted back to wild birds on a number of occasions with good evidence of maintenance in the new host for at least limited periods of time. The large global population of anseriform species annually undergo high population turnover allowing for the potential maintenance of H5N1 viruses in wild bird populations (Munster and Fouchier, 2009). The following section focuses on the prevalence of H5N1 in anseriformes and factors dictating viral persistence in host species.
2.1. HPAI H5N1 infection of anseriformes
The main reservoirs for all subtypes of Influenza A viruses are ducks in the Order Anseriformes, family Anatidae (Kim et al., 2009). HPAI H5N1 viruses have been isolated from 36 species of ducks, the most frequent being Mallards (Anas platyrhynchos), Northern Pintail (Anas acuta), and Blue-winged Teal (Anas discors) while common goose species known to be naturally infected with H5N1 influenza virus include Bar-headed geese (Anser indicus), Cackling geese (Branta hutchinsii), Canada geese (Branta Canadensis), and Domestic geese (Anser anser domesticus) (Neufeld et al., 2009; Olsen et al., 2006). It is hypothesized that the feeding behavior of waterfowl determines the prevalence of low pathogenic influenza virus infection and there is no reason to suspect that it would be different for highly pathogenic viruses. Dabbling ducks subsist primarily on food floating on the surface of water where avian influenza virions accumulate, whereas diving ducks obtain sustenance well below the water surface (Olsen et al., 2006) potentially avoiding ingestion and respiratory contact with contaminated water. Though there may be a paucity of genetic determinants, the behavior of avian species may be sufficient to exclude diving duck species, yet preclude dabbling duck species for infection with influenza virus.
In ducks and geese, the primary site of most influenza virus replication is the intestine resulting in the shedding of virus upon defecation (Webster et al., 1978). The HPAI H5N1 viruses are somewhat of an exception and can replicate in and shed from the respiratory tract of infected birds {Go:2012dv}. While there is considerable heterogeneity in the clinical signs in ducks after experimental HPAI H5N1 infection, unlike in chickens where the infection is almost always lethal, the respiratory shedding phenotype is consistent. For example, in an experimental infection of 5 duck species with two H5N1 viruses, Brown et al. detailed consistently higher viral viral titers in oropharyngeal swabs than in cloacal swabs (Brown et al., 2007). Geese experimentally infected with the 1997 H5N1 isolates showed replication to moderate titers in the respiratory and intestinal tract (1–4 log 10/ml) though with low morbidity (3/6) and mortality (1/6) (Shortridge et al., 1998). The high levels of respiratory shedding seen with post 2004 H5N1 viruses along with the absence of clinical signs has led to the postulation that domestic ducks are the “Trojan horses” of H5N1 spread {Anonymous: AgkOs+sT}. As discussed below, HPAI H5N1 infection of gallinaceous birds most often results in substantial mortality therefore is a relatively easy disease to identify which enhances control strategies. The often mild or asymptomatic nature of H5N1 infection in ducks in contrast can aid in undetected spread of the virus.
One of the more controversial aspects of the H5N1 outbreak has been the role of wild birds in its dissemination. During the first few years of its spread there was limited evidence to support such a link, but the temporal association of a large wild bird die-off in Qinghai Lake, China, in 2005 {Chen: 2006iu} with the dissemination of the virus outside of Southeast Asia provided at least circumstantial evidence {Gilbert: 2006kg}. Related to the spread by wild birds has been the question of whether the different genetic clades of circulating H5N1 viruses have different infectivity’s for different species of birds. Of note only 2 clades, clade 2.2 and clade 2.3.2.1, of H5N1 virus have spread outside of Southeast Asia. Each of these clades has also been associated with infection of wild birds, the clade 2.2 viruses in the 2005 Qinghai Lake outbreak and 2.3.2.1 viruses in wild birds in Hong Kong and elsewhere. Unfortunately there have been no large scale comparative experimental studies in wild birds to determine if various genetic backgrounds have enhanced ability to infect different avian hosts. Surveillance studies examining Goose/Guangdong/1/96-like viruses isolated from geese and ducks in Hong Kong during the late 1990s–early 2000s did suggest a distinct association of virus genotypes to either duck or goose species with reassortant viruses being isolated from both hosts (Guan et al., 2002b). Further studies are very much needed.
2.2. H5N1 infection in other wild bird species
In addition to ducks and geese, a wide variety of wild bird species have been shown either experimentally susceptible or naturally infected with HPAI H5N1 virus. The remains of migratory bird species are often found in the vicinity of H5N1 outbreaks near water sources along migratory flyways. The Qinghai Lake outbreak involved a number of other bird species in addition to ducks and geese (Chen et al., 2005; Liu et al., 2005). Among the species infected included brown-headed gulls (Larus brunnicephalus), great black-headed gulls (Larus ichthyaetus), and great cormorants (Phalocrocorax carbo) (Chen et al., 2005). Following the Qinghai lake outbreak an increasing number of terrestrial bird species have tested positive for H5N1 influenza raising concerns that HPAI can circulate amongst these species of birds, many of which come into close contact with humans and other mammalian species.
Passerines are another group of birds that have been associated with HPAI H5N1 infection. For example, tree sparrows (Passer montanus) are wild, peri-domestic birds that have sporadically tested positive for H5N1 influenza. The first report of H5N1 positive tree sparrows came from the outbreaks in Penfold and Kowloon parks in Hong Kong in 2002 (Ellis et al., 2004). More extensive surveillance of tree sparrows in China by Kou et al. (2005) found a high proportion (25/38) of birds sampled were positive. Genotypic analysis of these tree sparrow isolates found them to be closely related to H5N1 strains circulating contemporaneously in ducks and chickens. Experimental infection of sparrows have shown that the viruses replicate in respiratory and gastrointestinal tracts for durations of 4–7 days often causing lethal infections {Boon: 2007bt}; while some differences in susceptibility of different passerines such as finches and starlings have been reported, the unifying feature is the lack of transmission suggesting that they may be incidental hosts and not maintenance reservoir hosts. This does not of course rule out a role for these birds in local dissemination of virus to other bird or mammalian species.
Birds of prey, including raptors and carrion birds, are highly susceptible to H5N1 infection. Carnivorous avian species testing positive for H5N1 include Hodgsons’s Hawk-Eagles (Spizaetus nipalensis), Peregrine falcons (Falco peregrinus), Saker falcons (Falco cherrug), and common buzzards (Buteo buteo) (Marinova-Petkova et al., 2012; Marjuki et al., 2009; Van Borm et al., 2005). Molecular characterization of the raptor-isolated viruses often indicates that they are closely related to chicken strains circulating in domestic poultry near where the birds of prey were found indicating a likely infection route (predation on infected poultry carcasses). In at least one instance, however, a Bulgarian buzzard isolate (A/common buzzard/Bulgaria/38WB/2010) was more similar to exotic strains circulating in Chinese waterfowl than to those circulating in Bulgaria (Marinova-Petkova et al., 2012). Molecular analysis of a Falcon strain showed a single amino acid substitution, G323R, in the multi-basic cleavage site of the HA in addition to E627K mutation in the PB2; both known to confer increased pathogenicity in mice. Conversely, A515T and H436Y mutations in PA and PB1, mutations known to attenuate H5N1 virus in ducks though not in mammals were detected in the falcon strains (Marjuki et al., 2009). Interestingly a related buzzard strain lacked mutations corresponding to increased virulence in mammals. Further study is required to determine if the presence of these mutations are the result of host adaptive pressure within these birds or merely a consequence of species-specific feeding behaviors.
Clinically, H5N1 infection of raptors causes a highly lethal disease with intense CNS involvement. Experimental infection of American Kestrels (Falco sparverius) by Hall et al. (2009) resulted in severe clinical signs (feather fluffing, ataxia, anorexia, tremors, and weight loss) and euthanasia within 4–5 days of inoculation. These findings are consistent with results obtain from the experimental infection of Gyr-Saker hybrid falcons (Lierz et al., 2007). Oropharyngeal swabs from both Kestrels and hybrid falcons reached peak RNA titers one day post-infection and remained positive for the remainder of both studies while cloacal swabs showed a one day delay, peaking at 3–4 days post-infection (Hall et al., 2009; Lierz et al., 2007). Together these studies show raptors are highly susceptible to H5N1 influenza though the natural route of infection and the role of predatory birds in influenza ecology remain unclear.
3. H5N1 in gallinaceous birds
The first detected HPAI H5N1 infections of gallinaceous poultry occurred during the 1997 Hong Kong outbreak causing high mortality (~75%) on three chicken farms in Hong Kong, SAR, China (Shortridge et al., 1998). Surveillance of poultry markets found approximately 20% of chickens and 2% of ducks to be positive for HPAI H5N1. Characterization of representative isolates from chicken, ducks, and geese showed them all to be highly pathogenic in chickens killing all infected study animals within 3 days post-infection; as expected for a virus with a multibasic cleavage site (Shortridge et al., 1998). Although many countries have experienced HPAI H5N1 in the ensuing years, the virus has remained endemic in Bangladesh, China, Egypt, India, Indonesia, and Vietnam (“CDC – Seasonal Influenza (Flu) – H5N1 in Birds and Other Animals,” 2012).
HPAI H5N1 infection manifests intense pathology in domestic chickens. Other gallinaceous bird such as pheasants, quail and guinea fowl are similarly sensitive to H5N1 infection. Outward signs of infection include depression, anorexia, and respiratory congestion in addition to edematous comb and wattles, and hemorrhage of the conjunctiva and bursa of Fabricius (Bahgat et al., 2009; Jeong et al., 2009; Nakamura et al., 2008; Suarez et al., 1998). In studies of chickens experimentally infected with HPAI H5N1, immunohistochemical and histopathological analysis detected viral antigens in vascular epithelial cells, alveolar macrophages and the parenchyma of all major internal organs: lung, colon, brain, spleen, liver, kidney and heart. The presence of viral antigen staining in organ tissue sections was accompanied by severe inflammation and apoptosis (Isoda et al., 2006; Jeong et al., 2009; Karpala et al., 2011; Kwon et al., 2011; Suarez et al., 1998; Suzuki et al., 2009; Wasilenko et al., 2009). Identification of the molecular determinants of H5N1 transmission and virulence in poultry is critical for future influenza surveillance and therapeutic development.
4. Molecular markers of H5N1 replication in different avian hosts
Comparative genetic characterization studies of HPAI and LPAI H5N1 strains have identified molecular markers conferring efficient viral replication and high pathogenicity in domestic poultry. The past decade has seen an increase in our understanding of how point mutations, deletions, and specific gene constellations can influence the initiation of infection in avian hosts and how these genetic alterations contribute to the virulent phenotype of highly pathogenic H5N1.
Single gene reassortant studies highlight the contribution of the HA to host range and virulence of HPAI H5N1 viruses. Genetic characterization of the HA protein showed that all viruses isolated from the initial 1997 Hong Kong out break encoded a multi-basic cleavage site (RERRRKKR) and mutations at amino acid residue 158 of HA1 nullifying a carbohydrate site (Shortridge et al., 1998). HPAI H5 hemagglutinin proteins isolated from chickens continue to encode a multi-basic cleavage site emphasizing a lack of selection for LPAI derivatives. Though the amino acid motif is present, mutations changing the consensus RRRKKR/G sequence, characteristic of A/goose/Guandong/1/97, to RRRKR/G seen in contemporary isolates has been observed (Pfeiffer et al., 2009). More recently, surveillance of Egyptian poultry identified several recurring mutations in the multi-basic cleavage site (K346R, E340K, R341K) highlighting the high level mutation displayed by H5N1 (Kayali et al., 2011). Because of their high susceptibility to H5N1 and close proximity to humans and other mammals, identifying additional factors influencing host range of H5N1 in domestic birds (especially chickens) is of utmost importance.
In addition to the multi-basic HA cleavage site, receptor binding specificity of 1997 H5N1 viruses has been implicated as a determinant of host range. The HA of avian influenza viruses recognizes and binds to Siaα2,3Gal gangliosides. H5 HA binding to Siaα2,3Gal gangliosides differs between strains based on the length of the oligosaccharide moiety (Gambaryan et al., 2003). Duck viruses bind with high affinity to α2,3 sialic acid bound to a type 1 core (Galβ1-4GlcNAc). In contrast, chicken H5 viruses isolated from the 1997 Hong Kong outbreak bind with higher affinity to cells expressing the type 2 core (Galβ1-4GlcNAcβ); a moiety expressed on mammalian cells but lacking on duck cells. The highly glycosylated chicken H5, incorporating the glycosylation site at 158, may influence receptor binding affinity (Matrosovich et al., 1999). Expression of type 2 sialic acid core in both chickens and humans has been hypothesized to promote human infection with chicken origin H5N1 viruses (Gambaryan et al., 2004). Differences in sialic acid core expression between avian species identifies an intrinsic difference between ducks and chickens that directly effects virus host range pointing to a phenotypic change required for duck H5N1 viruses to infect and circulate in domestic chickens. Experiments examining the effect of sialic acid core type on the receptor binding preference of H5N1 from domestic ducks, geese, and mammals are needed to elucidate the full contribution of core binding in H5N1 host range.
Chicken H5N1 isolates from the initial Hong Kong outbreak in 1997 encode a truncated NA protein, characterized by a 19 amino acid deletion in the stalk region. The truncated NA of chicken/Hong Kong/97-like viruses results in the reduced elution efficiency of virions bound to chicken red bloods cells compared to duck isolates (Matrosovich et al., 1999). In addition, the 19 amino acid deletion removed three glycosylation sites though the functional significance of this deletion is unknown (Zhou et al., 1999). Removal of glycosylation sites in the NA globular head reduce the pathogenicity of recombinant viruses by delaying viral clearance in experimentally infected chickens (Hulse et al., 2004), further indicating the NA as a source of phenotypic diversity in H5N1 infected chickens.
The host range of H5N1 viruses can be influenced by mutations in internal gene segments; altering replicative, packaging, and dampening of the innate immune response in infected hosts. Studies with reassortants of a HPAI chicken and a LPAI duck virus show that reassortants encoding the PB2 and NP gene segments from chicken viruses have increased lethality in chickens compared with reassortants encoding duck PB2 and NP gene segments. The chicken virus segments enhanced polymerase activity in chicken embryonic fibroblasts and minigenome replicon systems (Tada et al., 2011).
The Influenza A virus NS1 protein is an antagonist of interferon signaling and inactivating mutations can result in virus attenuation (García-Sastre et al., 1998). NS1 variants of the archetypal strain A/goose/Guandong/1/96 are pathogenic in geese yet are unable to replicate in chickens, indicating that a functional NS1 protein plays a role in adaptation of H5N1 to chickens. Molecular analysis identified the V149A mutation in the NS1 of A/goose/Guandong/2/96 as the specific factor causing the inability of this virus to replicate in chickens (Li et al., 2006). Mutations in neighboring amino acids (E148G) have been shown to reduce virus replication of the more contemporary H5N1 isolate A/chicken/Indonesia/7/2003 in chicken embryonic fibroblasts (Wasilenko et al., 2009). Interestingly, the mutations identified in these studies are immediately adjacent, located in the effector-binding domain near the dimerization site (Bornholdt and Prasad, 2006), suggesting that a deficiency in NS1 dimerzation reduces NS1 function attenuating the virus for chicken hosts.
While there is much to learn, the data clearly show that HPAI H5N1 viruses cannot be treated as a homogeneous viral population and the range of hosts infected is a consequence of a combination of factors including minor differences in the virus itself.
5. HPAI H5N1 infection in mammals
The initial 1997 Hong Kong outbreak of HPAI H5N1 in chickens coincided with the first human cases of H5N1 infection. Influenza viruses with the ability to cross the species barrier, jumping from birds to mammals, have been responsible for previous influenza pandemics, though reassortment contributed to those in 1957 and 1968 (Webster et al., 1992). Surveillance efforts have been stepped up, with increased monitoring of H5N1 infection in mammals. Findings from influenza surveillance show that most mammalian cases of H5N1 occur simultaneously with outbreaks in domestic poultry, indicating contact with poultry as the primary route of interspecies HPAI H5N1 transmission (Fig. 1). Though H5N1 viruses are known to cause infections in mammals there has been no documented evidence of sustained intra-species transmission, possibly associated with the quick identification and response stemming from surveillance efforts. The primary mode of influenza virus transmission in mammals is through respiratory droplet transmission and the inability of H5N1 to display sustained respiratory droplet transmission prohibits the initiation of a pandemic.
5.1. Human infection with H5N1
The first human cases of H5N1 occurred in 1997 coinciding with a simultaneous outbreak in domestic chickens. The initial outbreak resulted in the identification of eighteen individuals causing 6 deaths. Molecular analysis of virus isolates revealed all gene segments to be of avian origin evincing direct avian to human transmission (Subbarao, 1998). As of June 4, 2013, 630 confirmed human cases and 375 deaths from H5N1 infection (“WHO | Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO,” n.d.). Though several instances of human-to-human transmission have been reported (Normile, 2006; Ungchusak et al., 2005), sustained human transmission has not occurred, prohibiting a pandemic.
5.1.1. Molecular markers associated with increased human infection
Animal models, combined with reverse genetics generated mutants, have proved invaluable in deepening our understanding of the molecular factors required for avian H5N1 virus to cross the species barrier and infect mammals. In particular, the mouse and ferret models of H5N1 infection have allowed for the study of virus pathogenicity and transmission. Receptor binding preference has long been known to be a major factor of virus host range. Avian influenza viruses bind to Siaα2,3Gal moieties found on cell surfaces where as human viruses preferentially bind to Siaα2,6Gal moieties (Connor et al., 1994; Gambaryan et al., 2003; Matrosovich et al., 1997). Mutations in receptor binding site at amino acid residues Q226L and N224K of the HPAI H5N1 virus A/Vietnam/1203/04, alter its receptor binding preference from Siaα2,3Gal to Siaα2,6Gal in solid phase binding assays and with the additional N158D mutation allowing for improved replication and transmission in vivo (Imai et al., 2012). Transmission studies in the ferret (Mustela putorius furo) model show that additional mutations outside of the receptor-binding site are required for efficient aerosol transmission. For example, in the study above, the HA mutation T318I served to confer mammalian H1-like fusogenic properties to the H5 virus. In a similar experiment, receptor-binding mutants of A/Indonesia/5/2005 with the Q222L and G224S mutations were used to serially inoculate ferrets for 10 passages. Serial passage resulted in the accumulation of H103Y and T156A mutations in HA of the ferret-adapted virus (Herfst et al., 2012).
Mutations conferring enhanced replication in mammals have also been identified in internals proteins. Known to be a determinant of host range, replacement of glutamate (E) with lysine (K) located at position 627 in the polymerase subunit PB2 alters the permissive temperature of avian influenza viruses, allowing for efficient replication at 33C, the approximate temperature of the mammalian respiratory tract (Massin et al., 2001; Subbarao et al., 1993). Experiments utilizing reassortant virus strains inserting the HA and PB2 gene segments from the reconstructed 1918 pandemic influenza virus on an A/Duck/Alberta/35/76 (H1N1) backbone allowed for efficient aerosol transmission of viruses in the ferret model (van Hoeven et al., 2009). Structural analysis of the PB2 protein found amino acid 627K located on superficial fold with the lysine side chain exposed (Tarendeau et al., 2008). Mechanistically the 627K mutations increases the interaction of PB2 with mammalian Importinα1 resulting in enhanced replication in mammalian cells (Gabriel et al., 2008; Resa-Infante et al., 2008). Currently circulating H5N1 virus strains have the 627K mutation, causing increased pathogenicity in birds and mice, that could further affect the replicative ability of H5N1 in mammalian hosts (Li et al., 2009; Schat et al., 2012).
5.1.2. Epidemiologic factors associated with Human H5N1 Infection
Epidemiology has been useful in identifying risk factors (both medical and behavioral) associated with H5N1 infection in humans. Exposure to infected birds, especially among poultry workers, is associated with a high risk of infection with H5N1 influenza. From 2006 to 2010, 96% of all human H5N1 cases were associated with contact to potentially infected poultry. Human H5N1 case numbers are highest in the northern hemisphere winter and spring (Fiebig et al., 2011), possibly resulting from the introduction of novel H5N1 viruses into domestic flocks from annual bird migration. Analysis of the distribution of confirmed human H5N1 cases from 2004 to 2006 found a correlation with age noting a higher rate of infections in people aged 29 years or younger than in those over 30 years of age. Whether the skewing of human H5N1 infections by age is due to biological or behavioral factors is unknown, though (based on statistical modeling) it has been suggested that persons older than 35 years of age possess immunoglobulins cross-reactive against H5 subtype influenza from exposure to previously circulating strains (Smallman-Raynor and Cliff, 2007).
Extensive epidemiological studies have been conducted in concurrence with H5N1 outbreaks in Egypt. Studies done by Kayali et al. (2011) found that in 5 years (March 2006–December 2010) the 119 human cases of H5N1 influenza in Egypt showed seasonal dependence, predominantly occurring in the winter and spring months. Consistent with previous epidemiological data, a majority (62%) of H5N1 positive individuals were younger than 18 years of age though the highest case-fatality rates (65 and 61%) were observed in the 19–29 and 30–39 years age groups. Additionally, the authors report increased mortality rates amongst women (47%) which they attribute to culture practices though biological effects should not be overlooked.
5.2. HPAI H5N1 in swine
Domestic swine are naturally susceptible to infection with influenza A viruses. Subtypes of influenza viruses naturally circulating in swine are of H1 and H3 and N1 and N2. Influenza in swine is a febrile disease with outward clinical signs including inactivity, nasal discharge, coughing, sneezing, labored breathing, and conjunctivitis. Swine influenza is endemic in most regions of the world with high morbidity (~100%) and low mortality (<1%) seen (Ma et al., 2009; Neumann et al., 2009; Webster et al 1992).
Little is known about the prevalence of H5N1 infection in pigs although the available evidence suggests that they are not a highly susceptible host. The paucity of epidemiologic of HPAI H5N1 in swine is attributed to lack of clinical disease. In swine, H5N1 infection manifests as a mild, non-life threatening disease. HI testing of swine sera from slaughterhouses in Vietnam during 2004 showed a low percentage (0.25%) of pigs had been exposed to H5N1 virus (Choi et al., 2005). Surveillance of swine herds in Fujian, China found no evidence of H5N1 infection among pigs from 2004 to 2007, though a low percentage of the sample population (1% in 2004, 2.6% in 2007) had HI titers against the H9 subtype (Song et al., 2010). Further surveillance efforts in Indonesia from 2005 to 2007 were able to isolate 52 viruses (7.4% success rate) from domestic pig farms. Phylogenetic analysis of the Indonesian swine H5N1 isolates showed clustering of both the HA and NA with those circulating in domestic poultry (Nidom et al., 2010). Together, these studies show that pigs can be directly infected with avian H5N1 viruses, which can co-circulate with other influenza subtypes, though H5N1 display low morbidity in domestic swine.
The extent of porcine H5N1 virus infection has been studied in laboratory settings. Pigs experimentally infected with HPAI H5N1 virus exhibited mild to no changes in food consumption or behavior compared to controls infected with swine H3N2 and H1N1 viruses, which displayed lethargy and listlessness. Despite the lack of outward clinical signs, several study animals showed significant weight loss (5–15% initial body weight) on days 1–4 post-infection, though lost body weight was recovered by day 5 (Choi et al., 2005; Lipatov et al., 2008). In these studies, limited pathology correlated with low levels of virus replication. Experimentally infected swine shed H5N1 influenza virus at titers well below those observed following H5N1 infection in ferrets or mice. Lipatov et al. (2008) report only one piglet out of 12 shed HPAI H5N1 at titers above log 3 EID50/ml after day 3 post-infection with no virus being detected at 7 days post infection. These results are consistent with results from Choi et al. (2005) and Manzoor et al. (2009) showing that wild-type H5N1 replicates to moderate titers and is shed for approximately 6 days in experimentally infected pigs. Together these studies demonstrate the low incidence and pathogenicity of H5N1 viruses in swine.
Limited data exists on swine-to-swine transmission and the molecular factors associated with increased replication in porcine hosts. The best evidence for inter and intra species come from surveillance studies done by Takano et al. (2009) in Indonesia. Phylogenetic analysis of the five viruses isolated from chickens and pigs raised on the same farm were highly related, highlighting repeated incidents of direct transmission to pigs from poultry. Full genome sequencing of chicken and swine isolates belonging to clade 2.1.3 uncovered three mutations present in one of two swine isolates not found in the closely related chicken virus. The mutations identified in PB2 (Q13H and G150V) and NA (G41R) highly attenuate the virus in mice, suggesting multiple passages in swine reduced the virulence of the chicken H5N1 strain (Takano et al., 2009). Further mutations in PB2 allow for increased replication in mammals, i.e. D256G and E627K (Manzoor et al., 2009).
Though circulation of H5N1 viruses in swine is limited, isolation of attenuated virus strains from swine in China show significant divergence from closely related avian strains. Of two H5N1 viruses isolated form infected swine in the Fujian province of China, a 15-nucleotide deletion at position 612–626 is present in the NS gene segment, resulting in a five amino acid deletion in the NS1 protein at positions 191–195 (Zhu et al., 2008). This deletion was shown to reduce weight loss in mice and attenuate the virus in chickens through inefficient dampening of the interferon response. It is unclear what selective advantage is conferred by a reduced ability to control host immune responses, though these data are consistent with the idea that continued circulation of virus in a population can lead to reduced virulence.
5.3. HPAI H5N1 in other mammalian species
HPAI H5N1 viruses have a remarkable ability to infect a number of mammalian species, many being classified in the order Carnivora. Spread of the virus amongst carnivores is likely attributed to predatory habits, as H5N1 has been isolated from the meat of infected animals (Tumpey et al., 2002) and ingestion of infected meat can cause infection (Kuiken, 2004). Captive tigers (Panthera tigris) and leopards (Panthera pardus) succumbed to H5N1 infection at zoos in Thailand following an outbreak of H5N1 influenza in wild-birds in 2003 and 2004 (Amonsin et al., 2006; Keawcharoen et al., 2004; Thanawongnuwech et al., 2005). In addition to tigers and leopards, a 2009 H5N1 outbreak occurring in the Phnom Tamao Wildlife Rescue Centre in Cambodia caused 100% mortality in lions (Panthera leo), Asiatic golden cats (Catopuma temminckii), and clouded leopard (Neofelis nebulosa) (Desvaux et al., 2009). Necropsy of H5N1 infected tigers revealed intense pulmonary pathology and multi-organ spread (Keawcharoen et al., 2004). The tiger H5N1 isolates clustered with genotype Z viruses that were circulating in chickens and detected in humans at that time. Notably, the HA gene contained a multibasic cleavage site and possessed Q222 and G224 at the receptor binding site, both amino acids dictating preferential binding to the avian α2,3 sialic acid receptor. Additionally, the tiger isolates acquired the amino acid lysine at position 627 of PB2, a mutation associate with adaptation to mammals (Amonsin et al., 2006; Keawcharoen et al., 2004). These results indicate no prior adaptation is needed for the infection of Panthera species with avian lineage H5N1 influenza and that big cat species may represent a novel avenue for the adaptation of avian influenza viruses to mammalian species.
Carnivora species from the family Felidae are particularly susceptible to infection with H5N1. Reports of H5N1 infection of domestic cats (Felis catus) began in Thailand in 2004 coinciding with outbreaks in poultry (Kuiken, 2004; Thiry et al., 2007). More recent infections of domestic cats have occurred in Austria and Iraq both coinciding with H5N1 outbreaks in birds (Leschnik et al., 2007; Yingst et al., 2006). Experimentally infected cats show increased body temperature, weight-loss, and decreased activity similar to H5N1 infection in humans. In studies examining different routes of infection (intratracheal and oral), experimentally infected cats displayed acute clinical disease with virus replicating to moderate titers (2–5 log TCID50/ml) at day 7 post-infection from pharyngeal, nasal, and rectal swabs (Kuiken, 2004; Rimmelzwaan et al., 2006). Interestingly, naïve animals placed in direct contact with infected animals became infected and exhibited a similar disease pattern compared to directly infected animals. These findings indicate cats can be infected through multiple routes of infection and shed virus through nasal secretion and feces both of which may be important for horizontal transmission in felids (Rimmelzwaan et al., 2006; Vahlenkamp et al., 2010).
Laboratory confirmed cases of H5N1 influenza have also been identified from other mammalian species in isolated incidents. In 2004, the remains of a domestic dog (Canis familiaris) tested positive by RT-PCR for H5N1 days after feeding on the carcass of ducks from H5N1 infected areas (Songserm et al., 2006). In Egypt, isolates of H5N1 were collected from the nasal swabs of donkeys (Equus africanus asinus) in 2009 following close contact of the animals with infected poultry (Abdel-Moneim et al., 2010). Additionally, a wild stone marten (Martes foina) (Klopfleisch et al., 2007) and Owston’s civets (chrotogale owstani) (Roberton et al., 2006) have tested positive for H5N1 infection in Germany and Vietnam, respectively. Finally, red foxes (Vulpes vulpes) have been shown to be susceptible to H5N1 infection in experimental studies (Reperant et al., 2008).
6. Discussion
HPAI H5N1 viruses have the potential to infect a diverse range of animal species which can contribute to high levels of genetic diversity. Because of this, the potential for the spread of H5N1 to uninfected countries and novel hosts remains high. Increased active surveillance efforts will ensure the movement of H5N1, and other influenza subtypes and viruses, is minimal. The risk of continued human and animal infection with HPAI H5N1 is, at present, not completely appreciated and can only be fully understood through continued experimentation.
In particular, gain-of-function studies will be invaluable in identifying the contributions of molecular mutations to H5N1 host range. Because of the unprecedented diversity of H5N1 and other influenza subtypes in nature, direct experimentation on the agents in question is the most efficient way to maximize our understanding of factors contributing to host range. Studies like those of Imai et al. (2012) and Herfst et al. (2012) serve as prime examples of the value of gain-of-function research. The findings from these papers are immediately applicable to influenza surveillance efforts. Future studies employing ferret adapted virus are necessary to increase our understanding of H5N1 transmission beyond the identification of individual mutations, but to characterize the underlying biochemical and biophysical properties contributing aerosol transmission in mammalian hosts.
Concurrently, fully understanding the dynamic interplay of H5N1 transmission between domestic and wild avian species is necessary for the protection of fragile ecosystems. Massive die offs of wild birds, like those seen at Qinghai Lake (Chen et al., 2005). In addition the susceptibility of endangered mammalian species, such as leopards and tigers (Amonsin et al., 2006; Keawcharoen et al., 2004; Thanawongnuwech et al., 2005; Desvaux et al., 2009), may lead to the continued decline of threatened species. Further work is required to detail the role of wild birds in the spread of H5N1, the role of domestic animals in maintaining H5N1, and the species susceptible to H5N1 infection.
HPAI H5N1 has been at the forefront of public health and surveillance efforts for nearly 15 years. Despite intense surveillance efforts and laboratory studies a full understanding of the ecology and molecular mechanisms dictating host range still eludes us. The consequences of this knowledge gap include a continuing threat to human health, large scale agricultural losses and irreversible changes. Only through further, unrestricted study will the biological, behavioral, and ecological factors contributing to the uncanny promiscuity of H5N1 influenza be elucidated.
References
- Abdel-Moneim AS, Abdel-Ghany AE, Shany SAS. Isolation and characterization of highly pathogenic avian influenza virus subtype H5N1 from donkeys. J Biomed Sci. 2010;17:25. doi: 10.1186/1423-0127-17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amonsin A, Payungporn S, Theamboonlers A, Thanawongnuwech R, Suradhat S, Pariyothorn N, Tantilertcharoen R, Damrongwantanapokin S, Buranathai C, Chaisingh A, Songserm T, Poovorawan Y. Genetic characterization of H5N1 influenza A viruses isolated from zoo tigers in Thailand. Virology. 2006;344:480–491. doi: 10.1016/j.virol.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Bahgat MM, Kutkat MA, Nasraa MH, Mostafa A, Webby R, Bahgat IM, Ali MAA. Characterization of an avian influenza virus H5N1 Egyptian isolate. J Virol Methods. 2009;159:244–250. doi: 10.1016/j.jviromet.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BVV. X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol. 2006;13:559–560. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- Brown JD, Swayne DE, Cooper RJ, Burns RE, Stallknecht DE. Persistence of H5 and H7 avian influenza viruses in water. Avian Dis. 2007;51:285–289. doi: 10.1637/7636-042806R.1. [DOI] [PubMed] [Google Scholar]
- CDC – Seasonal Influenza (Flu) – H5N1 in Birds and Other Animals [WWW Document], 2012. CDC – Seasonal Influenza (Flu) – H5N1 in Birds and Other Animals [WWW Document]. cdc.gov. URL http://www.cdc.gov/flu/avianflu/h5n1-animals.htm
- Chen H, Smith GJD, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JSM, Guan Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P, Buranathal C, Chaisingh A, Auewarakul P, Hanh NTH, Ma SK, Hui PY, Guan Y, Peiris JSM, Webster RG. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol. 2005;79:10821–10825. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Desvaux S, Marx N, Ong S, Gaidet N, Hunt M, Manuguerra JC, Sorn S, Peiris M, van der Werf S, Reynes JM. Highly pathogenic avian influenza virus (H5N1) outbreak in captive wild birds and cats, Cambodia. Emerg Infect Dis. 2009;15:475–478. doi: 10.3201/eid1503.081410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TM, Bousfield RB, Bissett LA, Dyrting KC, Luk GSM, Tsim ST, Sturm-Ramirez K, Webster RG, Guan Y, Peiris JSM. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Path. 2004;33 (5):492–505. doi: 10.1080/03079450400003601. [DOI] [PubMed] [Google Scholar]
- Fiebig L, Soyka J, Buda S, Buchholz U, Dehnert M, Haas W. Avian influenza A(H5N1) in humans: new insights from a line list of World Health Organization confirmed cases, September 2006 to August 2010. Euro Surveill. 2011;16 [PubMed] [Google Scholar]
- Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan AS, Tuzikov AB, Bovin NV, Yamnikova SS, Lvov DK, Webster RG, Matrosovich MN. Differences between influenza virus receptors on target cells of duck and chicken and receptor specificity of the 1997 H5N1 chicken and human influenza viruses from Hong Kong. Avian Dis. 2003;47:1154–1160. doi: 10.1637/0005-2086-47.s3.1154. [DOI] [PubMed] [Google Scholar]
- Gambaryan AS, Tuzikov AB, Pazynina GV, Webster RG, Matrosovich MN, Bovin NV. H5N1 chicken influenza viruses display a high binding affinity for Neu5Acalpha2-3Galbeta1-4(6-HSO3)GlcNAc-containing receptors. Virology. 2004;326:310–316. doi: 10.1016/j.virol.2004.06.002. [DOI] [PubMed] [Google Scholar]
- García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Guan Y, Peiris JSM, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc Natl Acad Sci U S A. 2002a;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Peiris M, Kong KF, Dyrting KC, Ellis TM, Sit T, Zhang LJ, Shortridge KF. H5N1 influenza viruses isolated from geese in Southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology. 2002b;292:16–23. doi: 10.1006/viro.2001.1207. [DOI] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JS, Ip HS, Franson JC, Meteyer C, Nashold S, TeSlaa JL, French J, Redig P, Brand C. Experimental infection of a North American raptor, American Kestrel (Falco sparverius), with highly pathogenic avian influenza virus (H5N1) PLoS ONE. 2009;4:e7555. doi: 10.1371/journal.pone.0007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Leneva I, Krauss S, Scholtissek C, Chin PS, Peiris M, Short-ridge KF, Webster RG. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J Virol. 2000;74:6309–6315. doi: 10.1128/jvi.74.14.6309-6315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse DJ, Webster RG, Russell RJ, Perez DR. Molecular determinants within the surface proteins involved in the pathogenicity of H5N1 influenza viruses in chickens. J Virol. 2004;78:9954–9964. doi: 10.1128/JVI.78.18.9954-9964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda N, Sakoda Y, Kishida N, Bai GR, Matsuda K, Umemura T, Kida H. Pathogenicity of a highly pathogenic avian influenza virus, A/chicken/Yamaguchi/7/04 (H5N1) in different species of birds and mammals. Arch Virol. 2006;151:1267–1279. doi: 10.1007/s00705-005-0723-6. [DOI] [PubMed] [Google Scholar]
- Jeong OM, Kim MC, Kim MJ, Kang HM, Kim HR, Kim YJ, Joh SJ, Kwon JH, Lee YJ. Experimental infection of chickens, ducks and quails with the highly pathogenic H5N1 avian influenza virus. J Vet Sci. 2009;10:53–60. doi: 10.4142/jvs.2009.10.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpala AJ, Bingham J, Schat KA, Chen LM, Donis RO, Lowenthal JW, Bean AGD. Highly pathogenic (H5N1) avian influenza induces an inflammatory T helper type 1 cytokine response in the chicken. J Interferon Cytokine Res. 2011;31:393–400. doi: 10.1089/jir.2010.0069. [DOI] [PubMed] [Google Scholar]
- Kayali G, El-Shesheny R, Kutkat MA, Kandeil AM, Mostafa A, Ducatez MF, McKenzie PM, Govorkova EA, Nasraa MH, Webster RG, Webby RJ, Ali MA. Continuing threat of influenza (H5N1) virus circulation in Egypt. Emerg Infect Dis. 2011a;17:2306–2308. doi: 10.3201/eid1712.110683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayali G, Webby RJ, Ducatez MF, El Shesheny RA, Kandeil AM, Govorkova EA, Mostafa A, Ali MA. The epidemiological and molecular aspects of influenza H5N1 viruses at the human–animal interface in Egypt. PLoS ONE. 2011b;6:e17730. doi: 10.1371/journal.pone.0017730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J, Oraveerakul K, Kuiken T, Fouchier RAM, Amonsin A, Payungporn S, Noppornpanth S, Wattanodorn S, Theambooniers A, Tantilertcharoen R, Pattanarangsan R, Arya N, Ratanakorn P, Osterhaus DME, Poovorawan Y. Avian influenza H5N1 in tigers and leopards. Emerg Infect Dis. 2004;10:2189–2191. doi: 10.3201/eid1012.040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Negovetich NJ, Forrest HL, Webster RG. Ducks: the “Trojan horses” of H5N1 influenza. Influenza Other Respi Viruses. 2009;3:121–128. doi: 10.1111/j.1750-2659.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfleisch R, Wolf PU, Wolf C, Harder T, Starick E, Niebuhr M, Mettenleiter TC, Teifke JP. Encephalitis in a stone marten (Martes foina) after natural infection with highly pathogenic avian influenza virus subtype H5N1. J Comp Pathol. 2007;137:155–159. doi: 10.1016/j.jcpa.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Kuiken T. Avian H5N1 influenza in cats. Science. 2004;306:241. doi: 10.1126/science.1102287. [DOI] [PubMed] [Google Scholar]
- Kwon HI, Song MS, Pascua PNQ, Baek YH, Lee JH, Hong SP, Rho JB, Kim JK, Poo H, Kim CJ, Choi YK. Genetic characterization and pathogenicity assessment of highly pathogenic H5N1 avian influenza viruses isolated from migratory wild birds in 2011, South Korea. Virus Res. 2011;160:305–315. doi: 10.1016/j.virusres.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Leschnik M, Weikel J, Möstl K, Revilla-Fernández S, Wodak E, Bagó Z, Vanek E, Benetka V, Hess M, Thalhammer JG. Subclinical infection with avian influenza A (H5N1) virus in cats. Emerg Infect Dis. 2007;13:243–247. doi: 10.3201/eid1302.060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishaq M, Prudence M, Xi X, Hu T, Liu Q, Guo D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009;144:123–129. doi: 10.1016/j.virusres.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang Y, Jiao P, Wang A, Zhao F, Tian G, Wang X, Yu K, Bu Z, Chen H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol. 2006;80:11115–11123. doi: 10.1128/JVI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lierz M, Hafez HM, Klopfleisch R, Lüschow D, Prusas C, Teifke JP, Rudolf M, Grund C, Kalthoff D, Mettenleiter T, Beer M, Hardert T. Protection and virus shedding of falcons vaccinated against highly pathogenic avian influenza A virus (H5N1) Emerg Infect Dis. 2007;13:1667–1674. doi: 10.3201/eid1311.070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatov AS, Kwon YK, Sarmento LV, Lager KM, Spackman E, Suarez DL, Swayne DE. Domestic pigs have low susceptibility to H5N1 highly pathogenic avian influenza viruses. PLoS Pathog. 2008;4:e1000102. doi: 10.1371/journal.ppat.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang XW, Zhang XL, Zhao D, Wang G, Feng Y, Ma J, Liu W, Wang J, Gao GF. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- Ma W, Lager KM, Vincent AL, Janke BH, Gramer MR, Richt JA. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health. 2009;56:326–337. doi: 10.1111/j.1863-2378.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- Manzoor R, Sakoda Y, Nomura N, Tsuda Y, Ozaki H, Okamatsu M, Kida H. PB2 protein of a highly pathogenic avian influenza virus strain A/chicken/Yamaguchi/7/2004 (H5N1) determines its replication potential in pigs. J Virol. 2009;83:1572–1578. doi: 10.1128/JVI.01879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova-Petkova A, Georgiev G, Seiler P, Darnell D, Franks J, Krauss S, Webby RJ, Webster RG. Spread of influenza virus A (H5N1) clade 2.3.2.1 to Bulgaria in common buzzards. Emerg Infect Dis. 2012;18:1596–1602. doi: 10.3201/eid1810.120357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjuki H, Wernery U, Yen HL, Franks J, Seiler P, Walker D, Krauss S, Webster RG. Isolation of highly pathogenic avian influenza H5N1 virus from Saker falcons (Falco cherrug) in the Middle East. Adv Virol. 2009;2009:1. doi: 10.1155/2009/294520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massin P, van der Werf S, Naffakh N. Residue 627 of PB2 is a determinant of cold sensitivity in RNA replication of avian influenza viruses. J Virol. 2001;75:5398–5404. doi: 10.1128/JVI.75.11.5398-5404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Zhou N, Kawaoka Y, Webster R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- Munster VJ, Fouchier RAM. Avian influenza virus: of virus and bird ecology. Vaccine. 2009;27:6340–6344. doi: 10.1016/j.vaccine.2009.02.082. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Imada T, Imai K, Yamamoto Y, Tanimura N, Yamada M, Mase M, Tsukamoto K, Yamaguchi S. Pathology of specific-pathogen-free chickens inoculated with H5N1 avian influenza viruses isolated in Japan in 2004. Avian Dis. 2008;52:8–13. doi: 10.1637/8027-060607-Reg. [DOI] [PubMed] [Google Scholar]
- Neufeld JL, Embury-Hyatt C, Berhane Y, Manning L, Ganske S, Pasick J. Pathology of highly pathogenic avian influenza virus (H5N1) infection in Canada geese (Branta canadensis): preliminary studies. Vet Pathol. 2009;46:966–970. doi: 10.1354/vp.08-VP-0168-E-FL. [DOI] [PubMed] [Google Scholar]
- Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, Suzuki T, Suzuki Y, Shinya K, Iwatsuki Horimoto K, Muramoto Y, Kawaoka Y. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis. 2010;16:1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Avian influenza. Human transmission but no pandemic in Indonesia. Science. 2006;312:1855. doi: 10.1126/science.312.5782.1855b. [DOI] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Pfeiffer J, Pantin-Jackwood M, To TL, Nguyen T, Suarez DL. Phylogenetic and biological characterization of highly pathogenic H5N1 avian influenza viruses (Vietnam 2005) in chickens and ducks. Virus Res. 2009;142:108–120. doi: 10.1016/j.virusres.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Reperant LA, van Amerongen G, van de Bildt MW, Rimmelzwaan GF, Dobson AP, Osterhaus AD, et al. Highly pathogenic avian influenza virus (H5N1) infection in red foxes fed infected bird carcasses. Emerg Infect Dis. 2008;14:1835. doi: 10.3201/eid1412.080470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Jorba N, Zamarreño N, Fernández Y, Juárez S, Ortín J. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS ONE. 2008;3:e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan GF, van Riel D, Baars M, Bestebroer TM, van Amerongen G, Fouchier RAM, et al. Influenza A virus (H5N1) infection in cats causes systemic disease with potential novel routes of virus spread within and between hosts. Am J Pathol. 2006;168:176–183. doi: 10.2353/ajpath.2006.050466. (quiz 364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberton SI, Bell DJ, Smith GJD, Nicholls JM, Chan KH, Nguyen DT, Tran PQ, Streicher U, Poon LLM, Chen H, Horby P, Guardo M, Guan Y, Peiris JSM. Avian influenza H5N1 in viverrids: implications for wildlife health and conservation. Proc Biol Sci. 2006;273:1729–1732. doi: 10.1098/rspb.2006.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat KA, Bingham J, Butler JM, Chen LM, Lowther S, Crowley TM, Moore RJ, Donis RO, Lowenthal JW. Role of position 627 of PB2 and the multi-basic cleavage site of the hemagglutinin in the virulence of H5N1 avian influenza virus in chickens and ducks. PLoS ONE. 2012;7:e30960. doi: 10.1371/journal.pone.0030960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp GB, Kawaoka Y, Jones DJ, Bean WJ, Pryor SP, Hinshaw V, Webster RG. Coinfection of wild ducks by influenza A viruses: distribution patterns and biological significance. J Virol. 1997;71:6128–6135. doi: 10.1128/jvi.71.8.6128-6135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp GB, Kawaoka Y, Wright SM, Turner B, Hinshaw V, Webster RG. Wild ducks are the reservoir for only a limited number of influenza A subtypes. Epidemiol Infect. 1993;110:161–176. doi: 10.1017/s0950268800050780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge KF, Zhou NN, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti KG, Norwood M, Senne D, Sims L, Takada A, Webster RG. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- Smallman-Raynor M, Cliff AD. Avian influenza A (H5N1) age distribution in humans. Emerg Infect Dis. 2007;13:510–512. doi: 10.3201/eid1303.060849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XH, Xiao H, Huang Y, Fu G, Jiang B, Kitamura Y, Liu W, Liu D, Gao GF. Serological surveillance of influenza A virus infection in swine populations in Fujian province, China: no evidence of naturally occurring H5N1 infection in pigs. Zoonoses Public Health. 2010;57:291–298. doi: 10.1111/j.1863-2378.2009.01270.x. [DOI] [PubMed] [Google Scholar]
- Songserm T, Amonsin A, Jam-on R, Sae-Heng N, Pariyothorn N, Payungporn S, Theamboonlers A, Chutinimitkul S, Thanawongnuwech R, Poovorawan Y. Fatal avian influenza A H5N1 in a dog. Emerg Infect Dis. 2006;12:1744–1747. doi: 10.3201/eid1211.060542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez DL, Perdue ML, Cox N, Rowe T, Bender C, Huang J, Swayne DE. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Okada H, Itoh T, Tada T, Mase M, Nakamura K, Kubo M, Tsukamoto K. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. J Virol. 2009;83:7475–7486. doi: 10.1128/JVI.01434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süss J, Schäfer J, Sinnecker H, Webster RG. Influenza virus subtypes in aquatic birds of eastern Germany. Arch Virol. 1994;135:101–114. doi: 10.1007/BF01309768. [DOI] [PubMed] [Google Scholar]
- Tada T, Suzuki K, Sakurai Y, Kubo M, Okada H, Itoh T, Tsukamoto K. NP body domain and PB2 contribute to increased virulence of H5N1 highly pathogenic avian influenza viruses in chickens. J Virol. 2011;85:1834–1846. doi: 10.1128/JVI.01648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano R, Nidom CA, Kiso M, Muramoto Y, Yamada S, Shinya K, Sakai-Tagawa Y, Kawaoka Y. A comparison of the pathogenicity of avian and swine H5N1 influenza viruses in Indonesia. Arch Virol. 2009;154:677–681. doi: 10.1007/s00705-009-0353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarendeau F, Crepin T, Guilligay D, Ruigrok RWH, Cusack S, Hart DJ. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 2008;4:e1000136. doi: 10.1371/journal.ppat.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawongnuwech R, Amonsin A, Tantilertcharoen R, Damrongwatanapokin S, Theamboonlers A, Payungporn S, Nanthapornphiphat K, Ratanamungklanon S, Tunak E, Songserm T, Vivatthanavanich V, Lekdumrongsak T, Kesdangsakonwut S, Tunhikorn S, Poovorawan Y. Probable tiger-to-tiger transmission of avian influenza H5N1. Emerg Infect Dis. 2005;11:699–701. doi: 10.3201/eid1105.050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry E, Zicola A, Addie D, Egberink H, Hartmann K, Lutz H, Poulet H, Horzinek MC. Highly pathogenic avian influenza H5N1 virus in cats and other carnivores. Vet Microbiol. 2007;122:25–31. doi: 10.1016/j.vetmic.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Suarez DL, Perkins LEL, Senne DA, Lee JG, Lee YJ, Mo IP, Sung HW, Swayne DE. Characterization of a highly pathogenic H5N1 avian influenza A virus isolated from duck meat. J Virol. 2002;76:6344–6355. doi: 10.1128/JVI.76.12.6344-6355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungchusak K, Auewarakul P, Dowell SF, Kitphati R, Auwanit W, Puthavathana P, Uiprasertkul M, Boonnak K, Pittayawonganon C, Cox NJ, Zaki SR, Thawatsupha P, Chittaganpitch M, Khontong R, Simmerman JM, Chunsutthiwat S. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005;352:333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- Vahlenkamp TW, Teifke JP, Harder TC, Beer M, Mettenleiter TC. Systemic influenza virus H5N1 infection in cats after gastrointestinal exposure. Influenza Other Respi Viruses. 2010;4:379–386. doi: 10.1111/j.1750-2659.2010.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Borm S, Thomas I, Hanquet G, Lambrecht B, Boschmans M, Dupont G, Decaestecker M, Snacken R, van den Berg T. Highly pathogenic H5N1 influenza virus in smuggled Thai eagles, Belgium. Emerg Infect Dis. 2005;11:702–705. doi: 10.3201/eid1105.050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106:3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilenko JL, Sarmento L, Pantin-Jackwood MJ. A single substitution in amino acid 184 of the NP protein alters the replication and pathogenicity of H5N1 avian influenza viruses in chickens. Arch Virol. 2009;154:969–979. doi: 10.1007/s00705-009-0399-4. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO | Cumulative number of confirmed human cases of avian influenza A (H5N1) reported to WHO, n.d. WHO | Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO. WHO.
- Xu X, Subbarao K, Cox N. Genetic Characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999 doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- Yingst SL, Saad MD, Felt SA. Qinghai-like H5N1 from domestic cats, northern Iraq. Emerg Infect Dis. 2006;12:1295–1297. doi: 10.3201/eid1208.060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou NN, Shortridge KF, Claas EC, Krauss SL, Webster RG. Rapid evolution of H5N1 influenza viruses in chickens in Hong Kong. J Virol. 1999;73:3366–3374. doi: 10.1128/jvi.73.4.3366-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yang H, Chen W, Cao W, Zhong G, Jiao P, Deng G, Yu K, Yang C, Bu Z, Kawaoka Y, Chen H. A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J Virol. 2008;82:220–228. doi: 10.1128/JVI.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]