Abstract
Objectives
Methicillin-resistant Staphylococcus aureus (MRSA) is an important pathogen. Its resistance to multiple antibiotics and its prevalence in healthcare establishments make it a serious threat to human health that requires novel interventions. Manuka honey is a broad-spectrum antimicrobial agent that is gaining acceptance in the topical treatment of wounds. Because its mode of action is only partially understood, proteomic and genomic analysis was used to investigate the effects of manuka honey on MRSA at a molecular level.
Methods
Two-dimensional gel electrophoresis with dual-channel imaging was combined with matrix-assisted laser desorption ionization–time of flight mass spectrometry to determine the identities of differentially expressed proteins. The expression of the corresponding genes was investigated by quantitative PCR. Microarray analysis provided an overview of alterations in gene expression across the MRSA genome.
Results
Genes with increased expression following exposure to manuka honey were associated with glycolysis, transport and biosynthesis of amino acids, proteins and purines. Those with decreased expression were involved in the tricarboxylic acid cycle, cell division, quorum sensing and virulence. The greatest reductions were seen in genes conferring virulence (sec3, fnb, hlgA, lip and hla) and coincided with a down-regulation of global regulators, such as agr, sae and sarV. A model to illustrate these multiple effects was constructed and implicated glucose, which is one of the major sugars contained in honey.
Conclusions
A decreased expression of virulence genes in MRSA will impact on its pathogenicity and needs to be investigated in vivo.
Keywords: quorum sensing, biofilms, wounds, 2D-DIGE, microarrays
Introduction
The advent of antibiotics generated confidence that effective means of treating infections were available, but the emergence of antibiotic resistance has altered our perceptions. In 1961, when methicillin resistance in Staphylococcus aureus was discovered, its future global impact was unimaginable. Now the widespread prevalence of strains with multiple antibiotic resistance determinants has made methicillin-resistant S. aureus (MRSA) a serious threat to human health. The limited development of antimicrobial agents in recent times has compounded the situation1 and increased the necessity to search for alternative remedies to complement or replace antibiotics. Natural compounds isolated from plants have historically been used as templates for successful antimicrobial therapies.2 Before the discovery of antibiotics, honey was used for thousands of years in the topical treatment of wounds. During the past decade, it has been formulated into a range of modern wound dressings and has been reintroduced into conventional medicine.
Honey is a broad-spectrum antimicrobial agent with bactericidal activity against a number of wound pathogens.3–7 Its high osmolarity, acidity, generation of hydrogen peroxide on dilution and insect-derived antimicrobial peptides contribute to antibacterial activity, yet not all honeys are equivalent.8 Manuka honey is a distinctive honey that is produced in New Zealand and is used as a medical-grade honey in the manufacture of wound dressings licensed for clinical use in Australasia, Europe and North America. Whereas the antibacterial effect of many honeys on dilution is confined to the generation of hydrogen peroxide, manuka honey possesses additional antibacterial factors. Methylgloxal9,10 and leptosin11 were recently identified, and further bioactive factors may yet be discovered.
The bactericidal effects of manuka honey on wound pathogens have been investigated in vitro, especially in staphylococcal species. The role of manuka honey in preventing cell division in S. aureus was deduced from electron micrographs of honey-treated cells, where elevated numbers of cells with entire septa were found to accumulate. Autolysins (also known as murein hydrolases) were implicated in this effect.12 Similarly an inability to complete the cell cycle was later observed in MRSA treated with manuka honey; despite an increased expression of the autolysin (atl) gene, murein hydrolase activity was at undetectable levels. Failure to cleave peptidoglycan was thought to contribute to the persistence of the septa and to the failure to divide and complete the cell cycle.13 In addition, a decreased expression of universal stress protein A indicated that MRSA was unable to accommodate the stresses caused by exposure to manuka honey.14
Transcriptome analysis of Escherichia coli exposed to manuka honey revealed multiple cellular effects.5 More recently, a proteomic study found that 12 proteins were differentially expressed in S. aureus following treatment with manuka honey,15 but those effects were not investigated by transcriptome analysis. Both of these studies showed that manuka honey had a distinct mode of action that involved multiple cellular processes.
In chronic wounds, an association between the persistence of the wounds and the presence of biofilms16 has increased the urgency to find effective antimicrobial agents that inhibit not only planktonic bacterial cells, but also those contained in antibiotic-tolerant biofilm communities. The aim of this study was therefore to investigate changes in protein and gene expression in MRSA caused by treatment with manuka honey, with a view to elucidating the mechanisms that influence pathogenicity.
Materials and methods
Bacterial strains and growth conditions
The test bacterium used in this study was EMRSA-15 NCTC 13142. This was grown at 37°C with shaking at 120 rpm in tryptone soya broth (TSB; Oxoid Cambridge, UK), with or without 10% (w/v) sterile medical-grade manuka honey (Manukacare 18+, Comvita, UK).
Two-dimensional (2D) electrophoresis
For the preparation of cell extracts, cells were grown in 50 mL of TSB with or without 10% (w/v) manuka honey. At 4 h, the culture was harvested by centrifugation at 10 000 g for 5 min. The supernatant was discarded and the cells were resuspended in 10 mL sterile water. The cells were then disrupted at 4°C using 0.1 mm glass beads in a bead beater (BioSpec, Bartlesville, USA) using three homogenization cycles of 60 s each. The liquid phase was gently decanted from the beads and the beads were discarded. Insoluble or aggregated proteins in the retained supernatant were sedimented by a 4 min centrifugation at 13 000 g. The supernatant was then transferred into clean tubes and stored at −80°C.
The 2D gel electrophoresis was performed using an immobilized pH gradient technique adapted from published methods.17,18 Briefly, the 24 cm pH 3–10 strips were rehydrated in the Ettan IPGphor3 IEF system (GE Healthcare, Little Chalfont, UK). Prior to electrophoresis, 160 μg of protein from each of the control and honey-treated cells was incubated with fluorochromes Cy5 and Cy3 respectively according to manufacturer's instructions (CyDye DIGE Fluors; GE Healthcare, Little Chalfont, UK). Protein extracts were then combined and soluble proteins were loaded onto the rehydrated IPG strip before being isoelectrically focused for 60 000 Vh. The IPG strip was next equilibrated in buffer containing 1% DTT for 15 min (reduction) and then in buffer containing 2.5% iodoacetamide for 15 min (alkylation).
2D gels were prepared (270 × 210, 1 mm 10% SDS polyacrylamide gels) and proteins were separated after embedding the IPG into gel using 1% agarose (Sigma, Dorset, UK). 2D electrophoresis was performed using an Ettan IPGPhor system (GE Healthcare, Little Chalfont, UK) and the proteins were stained with Coomassie blue.
The difference gel electrophoresis (DIGE)-labelled gels were scanned using a Typhoon Trio variable-mode imager (GE Healthcare, Little Chalfont, UK) with 580BP 30Cy3, TAMRA, Alexa Fluor 546 and 670BP 30Cy5 emission filters, and then saved as .ds files. These files were then analysed using Decyder 2D version 6.5 software (GE Healthcare, Little Chalfont, UK). Gels were checked for spot resolution, and exclusion filters were set at slope 3.2, Area 120, Volume 25 000 and Peak height 100–6500. Spot-difference analysis allowed the identification of spots with a 2-fold or more up- or down-regulation compared with the control.
Sample preparation of spots for mass spectrometry (MS) analysis
Gel plugs (1.5 mm diameter) of spots of interest were manually excised and placed in a 96-well plate. Peptides were then recovered following trypsin digestion using a modified version of that described by Shevchenko et al.19 Sequencing grade modified trypsin (Promega, UK) was used at 6.25 ng/μL in 25 mM NH4HCO3 and incubated at 37°C for 3 h. Finally, the dried peptides were resuspended in 5 μL of 50% (v/v) acetonitrile in 0.1% (v/v) trifluoroacetic acid (TFA) for MS analysis, and an aliquot corresponding to 10% of the material (0.5 μL) was spotted onto a 384-well MS plate. The samples were allowed to dry and were overlaid with 0.5 μL of α-cyano-4-hydroxycinnamic acid [Sigma, Dorset, UK; prepared by mixing 5 mg of matrix with 1 mL of 50% (v/v) acetonitrile in 0.1% (v/v) TFA].
MS analysis
MS was performed using matrix-assisted laser desorption ionization–time of flight (MALDI TOF) MS (4800 MALDI TOF-TOF Analyzer; Applied Biosystems, Foster City, CA, USA) with a 200 Hz solid state laser operating at a wavelength of 355 nm.20,21 MALDI mass spectra and subsequent MS/MS spectra of the eight most abundant MALDI peaks were obtained following routine calibration. Common trypsin autolysis peaks and matrix ion signals and precursors within 300 resolutions of each other were excluded from the selection, and the peaks were analysed with the strongest peak first. For positive-ion reflector mode spectra, 800 laser shots were averaged (mass range 700–4000 Da, focus mass 2000). In MS/MS positive-ion mode, 4000 spectra were averaged with 1 kV collision energy (the collision gas being air at a pressure of 1.6 × 10−6 Torr) and default calibration.
Combined PMF and MS/MS queries were performed using the MASCOT Database search engine v2.1 (Matrix Science Ltd, London, UK)22 embedded in Global Proteome Server (GPS) Explorer software v3.6 (Applied Biosystems) on the Swiss Prot database (download date 16 December 2009). Searches were restricted to bacterial taxonomy with trypsin specificity (with one missed cleavage allowed), the tolerances set for peptide identification searches at 50 ppm for MS and 0.3 Da for MS/MS. Cysteine modification by iodoacetamide was employed as a fixed modification, with methionine oxidation as a variable modification. Search results were evaluated by manual inspection, and conclusive identification confirmed whether there were high-quality tandem MS (good y-ion) data for two or more peptides (E value P < 0.05 for each peptide; overall P < 0.0025) or one peptide (only if the E value had a value of P < 0.0001).
Extraction of RNA for real-time PCR and microarray analysis
Cells were grown with and without manuka honey as described above. RNA was isolated using a Promega SV Total RNA isolation kit, and cDNA was prepared using an Applied Biosystems High-Capacity cDNA Reverse Transcription Kit, both according to the manufacturer's instructions. The RNA was treated with an extra DNAse treatment using Ambion DNA-free according to the manufacturer's instructions to avoid a carry-over of genomic DNA (Invitrogen, Paisley, UK). Real-time PCR was performed on all samples, using Fast SYBR Green (Applied Biosystems, Foster City, CA, USA), with the procedures suggested by the manufacturer on a CFX96 real-time PCR system (Bio-Rad). Primers for quantitative PCR (qPCR) (Table 1) were designed using NCBI Primer-BLAST to be 20–24 bases long, with a GC content of more than 50% and a melting temperature of around 60°C. All reactions were carried out in triplicate, and the expression of genes was analysed with reference to the expression of the housekeeping gene acetyl coenzyme A (yqiL).
Table 1.
Primers used in this study
| Target gene | Direction | Primer sequence (5′– 3′) |
|---|---|---|
| yqiL | forward | GACGTGCCAGCCTATGATTT |
| yqiL | reverse | ATTCGTGCTGGATTTTGTCC |
| pykA | forward | TGCAGCAAGTTTCGTACGTC |
| pykA | reverse | GGGATTTCAACACCCATGTC |
| clpC | forward | GTTGGTGCTCCTCCAGGATA |
| clpC | reverse | ACTTGAACCACCGAATCCAG |
| argF | forward | CCAAGCAGAATTCGAAGGA |
| argF | reverse | GGATGCGCACCTAAATCAAT |
| adh | forward | GTTGCCGTTGGTTTACCTGT |
| adh | reverse | TTCAGCAGCAAATTCAAACG |
| menB | forward | CTGGGGAAGGTGATTTAGCA |
| menB | reverse | ACCGCCACCTACAGCATAAC |
| pur7 | forward | GAAGCGCATTTTCTCAACAA |
| pur7 | reverse | CCCTTACCTGCCATTGTGTC |
| pdp | forward | GCAATGCGCTTGAGTTACAA |
| pdp | reverse | TATTGAGCTTGTGGCAAACG |
| fabG | forward | CCGGGACAAGCAAACTATGT |
| fabG | reverse | CCAAAACGTGCTAACGGAAT |
| glmM | forward | AGGTGTCGCAAACCAAGAAC |
| glmM | reverse | TCGCGACCTACAAGTACACG |
| argF | forward | GCCCATTCGAAGAAAACGTA |
| argF | reverse | ACCTAATGCTGGCGCTAATG |
| scdA | forward | CGAAAGCAGCGGATATTTTT |
| scdA | reverse | GCGAACCTGGTGTATTCGTT |
For microarray analysis, RNA was isolated as above, hybridized, stained and scanned on Affymetrix arrays according to the manufacturer's instructions for prokaryotic target preparation. All experiments were carried out with three biological replicates and the mean values are presented here. The fold changes have been corrected and normalized to account for background noise. Genes showing more than a 2-fold differential regulation at a significance of P = 0.001 using a Bayesian t-test were examined.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO)13 and are accessible through GEO Series accession number GSE31592 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31592).
Results
The mode of action of manuka honey on MRSA was investigated initially by proteomic analysis, identifying proteins with differential expression. The expression of genes encoding for those identified proteins was then determined using qPCR and whole-genome effects were explored using microarray analysis.
Proteomic analysis
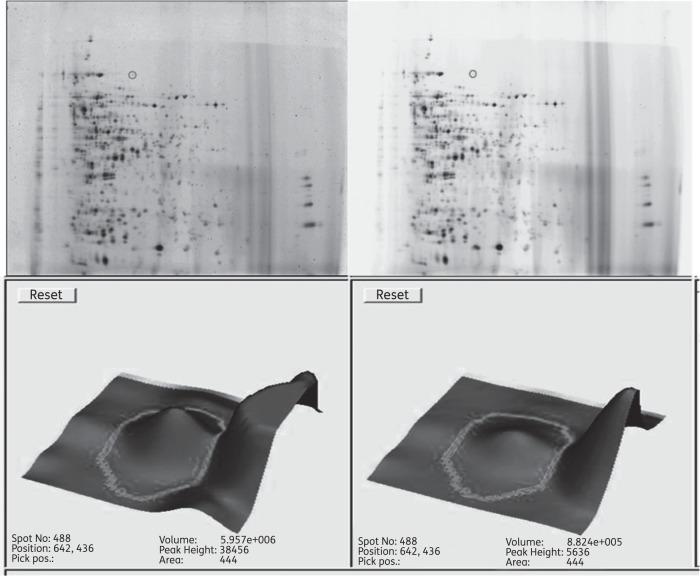
Proteins extracted from MRSA treated with and without a bactericidal concentration (10% w/v) of manuka honey for 4 h were separated by 2D -DIGE and 15 proteins whose levels of expression were altered by a factor of at least two were selected for further characterization by MS. Figure 1 shows an example of a gel with a protein spot that was characterized by MS as an ATP-binding subunit of an ATP-dependent Clp protease (ClpC). Five proteins were measured at increased levels compared with untreated cells (Table 2) and 10 proteins were decreased (Table 3). Three protein spots were characterized as being pyruvate kinase (Table 2), two were characterized as phosphoglucosamine mutase (Table 3) and one was tentatively identified as transaldolase. Therefore 15 samples represented 12 different proteins, of which 11 were successfully characterized. They included proteins involved in carbohydrate metabolism, cell wall biosynthesis and the stress response. Two were pertinent to two of our previous studies: the cell wall-related protein (ScdA) where cell division was affected13 and the Clp protease (ClpC) where inability to cope with stress was observed.14
Figure 1.
2D-DIGE gel showing spots of interest with proteins isolated from untreated MRSA (Cy5; left) and proteins from honey-treated MRSA (Cy3; right). The expression of protein spot 488 (grey circle) was analysed using Decyder 2D version 6.5 software (GE Healthcare, Little Chalfont, UK) and was down-regulated 2-fold. It was picked and identified as Clp protease using the Applied Biosystems 4800 MALDI TOF/TOF Analyzer (Foster City, CA, USA).
Table 2.
Proteins (determined by MS) in MRSA that increased 2-fold or more following exposure to manuka honey
| Accession number | Protein name/function | Best MS sequence | Mascot score | Second best MS sequence | Mascot score | Third best MS sequence | Mascot score |
|---|---|---|---|---|---|---|---|
| C2GBD3_STAAU | possible transaldolase | EITEAVTEGVPTYVSVFAGR | 1.20E-10 | EIPDASISFEVFADDLETMEK | 4.70E-11 | LNVEVFADGADIEEMK | 5.20E-07 |
| KYPK_STAAR 551 | pyruvate kinase | STDALLNNAVATAVETGR | 1.9E-013 | ENVDFIAASFVR | 1.5E-011 | IHLVGDEIANGQGIGR | 1.8E-006 |
| KYPK_STAAR | pyruvate kinase | KSTDALLNNAVATAVETGR | 1.7E-018 | STDALLNNAVATAVETGR | 7.3E-015 | ENVDFIAASFVR | 5.8E-011 |
| KYPK_STAAR | pyruvate kinase | KSTDALLNNAVATAVETGR | 1.5E-015 | STDALLNNAVATAVETGR | 5.7E-013 | IHLVGDEIANGQGIGR | 7.6E-009 |
| MENB_STAAR | naphthoate synthase | VGSFDAGYGSGYLAR | 3.30E-10 | GHGGYVGEDQIPR | 9.60E-08 |
Table 3.
Proteins (determined by MS) in MRSA that were found to be decreased following exposure to manuka honey
| Accession number | Protein name/function | Best MS sequence | Mascot score | Second best MS sequence | Mascot score | Third best MS sequence | Mascot score |
|---|---|---|---|---|---|---|---|
| ALF2_STAAR | fructose-bisphosphate aldolase | DVLNNDKEVYDPR | 1.20E-07 | ||||
| ADH_STAAR | alcohol dehydrogenase | NADFGDVTGVTLGHEGIGK | 4.30E-15 | LVLDGIEVVGSLVGTR | 7.70E-14 | KLEEINDIFEEMENGTITGR | 7.20E-12 |
| FABG_STAAR | 3-oxoacyl reductase | EVVSQFGSLDVLVNNAGITR | 6.40E-15 | GVDSFAIQANVADADEVK | 0.00033 | FGQDTDIANTVAFLASDK | 0.00027 |
| PUR7_STAAR | phosphoribosylamino-imidazole-succinocarboxamide synthase | TETGQILLADEISPDTCR | 5.70E-13 | NNTGSLIETYQIFLNK | 3.60E-07 | ATNANFDKDVYR | 3.00E-07 |
| PDP_STAAR | pyrimidine-nucleoside phosphorylase | VEEGESLLTIHSNR | 1.10E-06 | LPQAQYQIEYK | 8.90E-05 | ||
| OTCC_STAAR | ornithine carbamoyltransferase | ENFGYLEGINLTYVGDGR | 6.40E-18 | AEFEGLIDFAITLK | 1.90E-09 | AAFTVASIDLGAHPEFLGK | 7.70E-09 |
| GLMM_STAAR | phosphoglucosamine mutase | VVETESDFGLAFDGDGDR | 3.70E-12 | ||||
| GLMM_STAAR | phosphoglucosamine mutase | VVETESDFGLAFDGDGDR | 6.10E-07 | ||||
| SCDA_STAAR | cell wall-related protein | LNEVEQTNTPGSLNPK | 5.80E-08 | NVDLNELLQR | 6.20E-07 | VHGPNHPYLVELK | 1.00E-06 |
| CLPC_STAAR 488 | Clp protease | DAAVHAQEFENAANLR | 6.9E-009 | FAGFGGSSDGQDYETIR | 0.00011 |
qPCR analysis
Excluding transaldolase, the effect of manuka honey on the expression of the genes that coded for the 11 identified proteins was investigated quantitatively with PCR. It was found that changes in gene expression (Table 4) did not necessarily concur with the altered levels of protein expression observed using proteomic analysis (Tables 2 and 3). The expression of five corresponding proteins and genes did show the same trend (Table 4). Two genes (fbaA and pdp) had unchanged levels of expression and four genes showed increased expression (Table 4) in comparison with their respective proteins (Table 3). Hence six genes showed higher levels of expression by qPCR after honey treatment, although their proteins appeared diminished by 2D-DIGE. One explanation might be that changes in protein structure were caused directly by exposure to manuka honey or indirectly by altered post-translational modification.
Table 4.
Changes in gene expression in MRSA following treatment with bactericidal concentrations as determined by qPCR
| Gene | Gene product | Function | Fold change |
|---|---|---|---|
| pykA | pyruvate kinase | glycolysis | +6 |
| fbaA | fructose-bisphosphate aldolase | glycolysis | no change |
| adh | alcohol dehydrogenase | fermentation | −2 |
| menB | naphthoate synthetase | anaerobic electron transport | +5 |
| fabG | 3-oxoacyl reductase | fatty acid biosynthesis | +4 |
| purC | phosphoribosylaminoimidazole-succinocarboxamide synthase | purine biosynthesis | −2 |
| pdp | pyrimidine-nucleoside phosphorylase | pyrimidine biosynthesis | no change |
| argF | ornithine carbamoyltransferase | virulence | −2 |
| glmM | phosphoglucosamine mutase | cell wall | +4 |
| scdA | cell wall related protein | cell wall | +2 |
| clpC | Clp protease | stress | +16 |
Fold changes are shown in relation to untreated MRSA cells, and genes in bold show the same trend as proteins identified by 2D-DIGE.
Microarray analysis
Microarray analysis was undertaken to resolve the anomalies found with proteomic analysis and qPCR and to investigate wider changes in gene expression. Raw data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and are accessible through GEO series accession number GSE31592. Genes were sorted into The Institute for Genomic Research (TIGR) categories using the information available in the J. Craig Ventnor Institute Comprehensive Microbial Resource. Only changes relevant to our study are discussed in detail here.
Not all of the genes investigated by qPCR were among the 463 most highly differentially expressed genes identified by microarray analysis. This could be due in part to the fact the microarray chip was constructed from four strains of S. aureus—N315 (National Institute of Technology and Evaluation, Japan), Mu50 (National Institute of Technology and Evaluation, Japan), NCTC 8325 (University of Oklahoma, lab strain), and COL (TIGR)—which differed from the epidemic strain of MRSA that is prominent in the UK and that was used in this study. A specific chip for our test organism was not available. At the level of significance chosen here (P = 0.001), analysis indicated that 290 genes had increased levels of transcription and 173 genes showed decreased levels of transcription following exposure of MRSA to manuka honey compared with untreated MRSA. Of the 11 proteins/genes that had been investigated by proteomics and qPCR, down-regulation of alcohol dehydrogenase (adh) and up-regulation of pyruvate kinase (pykA) was confirmed.
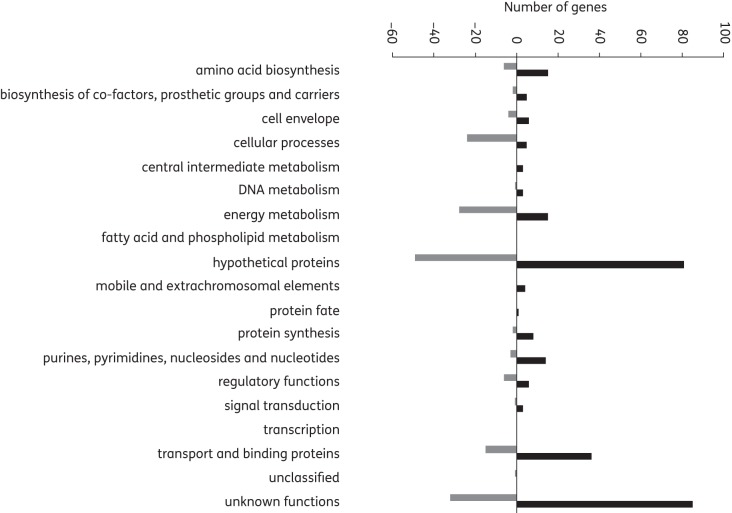
Globally, genes involved in the biosynthesis of amino acids, proteins, co-factors, prosthetic groups and carriers, purines, pyrimidines, nucleosides and nucleotides and cell envelope, as well as transport and binding proteins, were among the 290 genes found to have increased levels of expression in MRSA, while genes with decreased levels of expression mainly included those involved in energy metabolism and cellular processes (Figure 2). This suggests that growth and division was compromised in MRSA by manuka honey.
Figure 2.
Changes in gene transcription (microarray data) classified by main functions. Genes with altered levels of transcription after exposure of MRSA to 10% (w/v) manuka honey for 4 h at 37°C (compared with untreated cells) were divided into categories based on main functions according to the J. Craig Ventor Institute Comprehensive Microbial Resource.
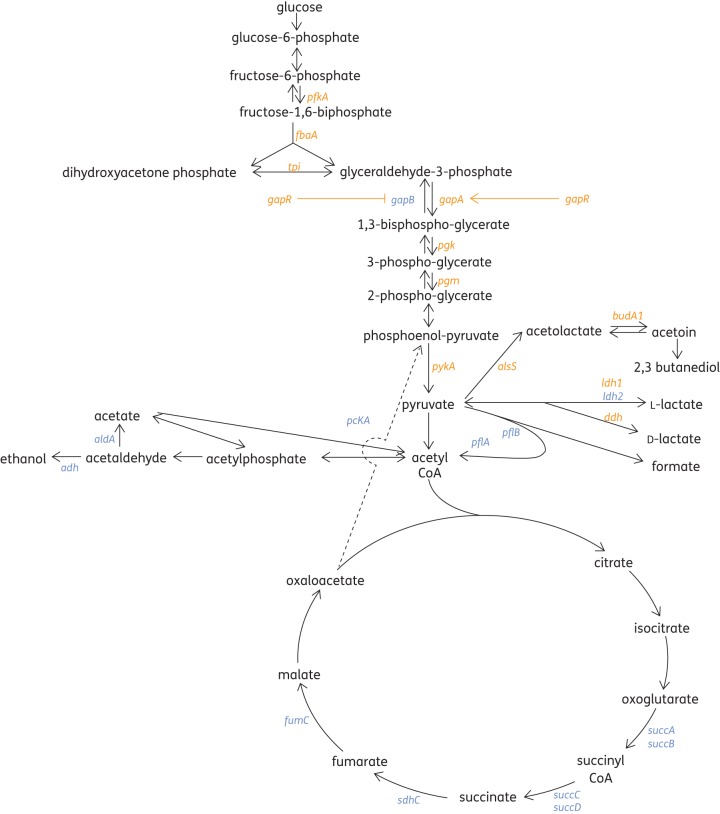
By mapping the affected genes to central pathways, a number of genes concerned with carbohydrate metabolism were found to be altered in MRSA by exposure to manuka honey (Figure 3). Essentially, glycolysis was promoted by increased transcription of pfkA, tpi, gapA, pgk, pgm and pykA (≥2-fold) and gluconeogenesis was restricted by a decreased transcription of gapB (33-fold). This effect was likely to be due to an increased expression of gapR (6.8-fold), which has been shown to regulate gapA and gapB reciprocally in response to glucose concentration, with glucose inducing gapA and repressing gapB.23 Since glucose accounts for approximately 33% of honey by weight, this effect was not unexpected. We have previously shown, however, that the antibacterial effect of manuka honey is not attributable solely to its sugar content.3
Figure 3.
Effect of manuka honey on the expression of genes concerned with carbohydrate metabolism in MRSA treated with and without 10% (w/v) manuka honey for 4 h at 37°C, as determined by transcriptome analysis. Up-regulated genes are shown in orange and down-regulated genes are shown in blue.
All of the tricarboxylic acid (TCA) cycle genes reported by microarray assay were found to be repressed, but those involved in fermentation showed varying patterns of expression (Figure 3). Essentially, manuka honey seemed to promote glycolysis and fermentation at the expense of oxidative metabolism. This would lead to the accumulation of acidic end-products and reduce the supply of ATP available to drive active transport and biosynthetic pathways.
The most notable changes in gene expression provided by the microarray data indicated that manuka honey had a marked effect on the expression of important MRSA virulence determinants, such as haemolysins, leucocidin, lipase and fibronectin-binding protein (Table 5). The largest change was seen in the gene coding for enterotoxin type C3 (sec3), which was down-regulated by a factor of 109. Another important observation was that three genes within the accessory gene regulator (agrB, agrC and agrD) exhibited decreased levels of expression, as did the genes within the two component histidine kinase regulators (saeS and saeR). Hence genes within two global regulatory operons (agrABCD and saeSR) were found to be repressed in MRSA exposed to manuka honey. Both regulate not only virulence, but also biofilm formation in staphylococci.24
Table 5.
Genes of potential clinical significance in MRSA identified by microarray analysis following treatment with and without 10% (w/v) manuka honey
| Function | Gene | Gene product | Fold change |
|---|---|---|---|
| Virulence | sec3 | enterotoxin type C3 | −109 |
| fnb | fibronectin binding protein | −54.5 | |
| hlgA | gamma haemolysin component A | −54 | |
| lip | lipase | −44.4 | |
| hla | alpha-haemolysin | −27.5 | |
| SA1813 | leucocidin protein | −23 | |
| sspA | serine protease | −3.8 | |
| hlgB | gamma haemolysin component B | −2.7 | |
| hlgC | gamma haemolysin component C | −2.3 | |
| Virulence regulators | sarV | HTH type regulator | +2 |
| agrB | accessory gene regulatory operon: membrane-associated protein | −5 | |
| agrC | accessory gene regulatory operon: membrane-associated autoinducer peptide sensor | −2.6 | |
| agrD | accessory gene regulatory operon: pro-autoinducer peptide | −2.4 | |
| saeS | histidine kinase sensor protein | −11 | |
| saeR | response regulator | −13 | |
| Cell envelope and cell division | ftsL | cell division protein | −2.6 |
| scpA | segregation and condensation protein A | −2.1 | |
| mecR1 | methicillin-resistance regulator protein | −3 | |
| lrgB | antiholin | +22 | |
| cidB | holin | −1.9 | |
| Stress | sod | superoxide dismutase | +1.6 |
| acpD | FMN-dependent NADH azoreductase | +4 | |
| mscL | large-conductance mechanosensitive channel | +2.5 | |
| asp23 | alkaline shock protein 23 | −1.4 | |
| SACOL1759 | putative universal stress protein | −4.4 |
The elevated expression of sarV (Table 5), another regulator gene,25 suggested that manuka honey might increase levels of autolysins and proteases in MRSA. Three genes that code for proteins involved in cell wall functions and division (ftsL, cidB and scpA) were found in transcriptome data to have decreased levels of expression, while lrgB, which functions as an antiholin in reducing the extracellular activity of murein hydrolase,26 was increased 22-fold. Some markers of the stress response were also changed (Table 5).
Discussion
Investigations into the impact of manuka honey on transcription in E. coli5 and on protein expression in S. aureus15 have demonstrated multiple effects on bacterial function. The differential expression of the proteins and genes identified in this study largely differed from those previously reported15 and may perhaps reflect the different strains, exposure times and honey concentrations employed in each study. Here, a bactericidal concentration of manuka honey was used [10% (w/v)], which was approximately twice the MIC and at least eight times lower than that normally used in the topical treatment of wounds. Data generated indicate multiple effects that impact on the ability of MRSA to grow and divide, as well as on its pathogenicity.
Virulence factors
The role of virulence determinants of S. aureus in human disease is well established.24 Virulence genotyping of strains of S. aureus isolated from diabetic foot ulcers showed that sec was one of the genes significantly more frequently associated with strains isolated from infected ulcers compared with non-infected ones.27 Reducing the transcription of sec in wounds by applying manuka honey can, therefore, be expected to confer a clinical benefit, but this effect will have to be investigated in vivo.
Reduced expression of the enterotoxin gene in MRSA in the presence of honey has considerable relevance for the food industry. Honey has been used as a food preservative for many years on the premise that it prevents microbial growth, but it may have an additional benefit in reducing staphylococcal pathogenicity, with the risk of staphylococcal food poisoning possibly being decreased by incorporating honey into perishable foodstuffs. Further investigation is needed to determine whether the repression of the enterotoxin gene is limited to manuka honey, or whether a wider variety of honeys produced for human consumption have a similar effect.
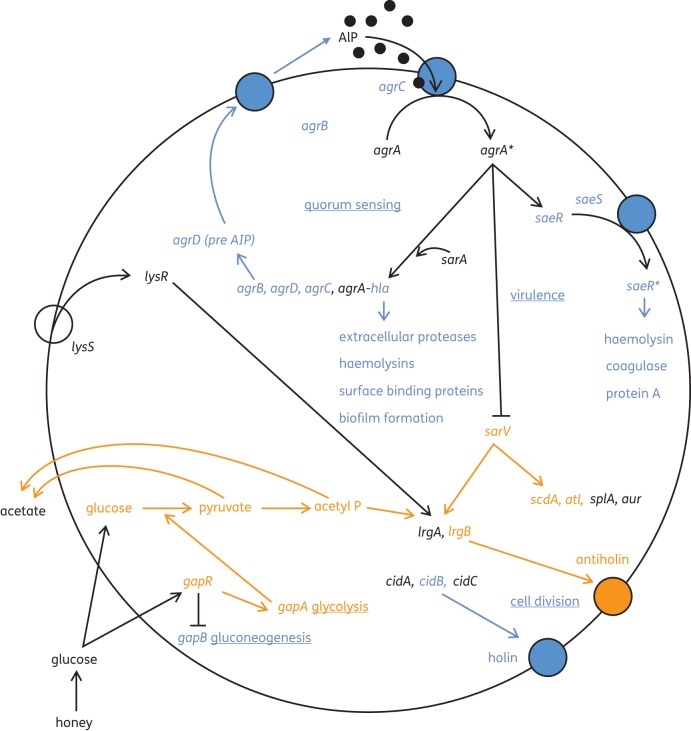
Many cell surface and extracellular proteins that contribute to virulence have been identified in S. aureus (Table 6); the expression of these genes is usually controlled by global virulence regulators including the accessory gene regulator (agrABCD) and a staphylococcal accessory regulator (sarA).28,29 In this study, a decreased transcription of three genes within the agr operon (Table 5) was found in treated MRSA. The agr locus (Figure 4) is a quorum-sensing gene cluster containing five genes (agrB, agrD, agrC, agrA and hla),30 four of which facilitate the production and detection of an autoinducing peptide (AIP) to regulate the expression of genes coding for virulence factors.31,32 Although certain honeys have been shown to inhibit quorum sensing in Gram-negative bacteria,33–35 this is the first indication that manuka honey inhibits quorum sensing in Gram-positive bacteria. Thus, the repression of some of the regulatory genes within the agr cluster is likely to account for the decreased expression of virulence factors under their control.
Table 6.
S. aureus virulence factors controlled by the global regulators agr, sarA, sarE and sae
| Virulence factors (gene) | Virulence factors (gene) |
|---|---|
| Toxins | Surface proteins |
| alpha haemolysin (hla) | bone sialoprotein-binding protein |
| beta haemolysin (hlb) | clumping factor A (clfA) |
| delta haemolysin (hld) | clumping factor B (clfB) |
| gamma haemolysin (hlgA-C) | collagen-binding protein (can) |
| enterotoxin A (sea) | extracellular fibrinogen |
| enterotoxin B (seb) | binding protein (efb/fib) |
| enterotoxin C (sec3) | fibronectin-binding protein A (fnbA) |
| enterotoxin E-I (entE-I) | fibronectin-binding protein B (fnbB) |
| exfoliative toxin A (etaA) | lactoferrin-binding protein |
| exfoliative toxin B (etaB) | laminin-binding protein |
| leucocidin P-V (lukS/F) | lectin-like protein |
| toxic shock syndrome toxin-1 (tst) | MHC-II analogous protein (map) |
| Enzymes | plasminogen-binding protein protein A (spa) |
| alkaline/phosphatase | Sdr A-D (sdrA, B, C, D) |
| beta lactamase (blaZ) | thrombospondin-binding protein |
| coagulase (coa) | vitronectin-binding protein |
| cysteine protease (sspB) | |
| fatty acid modifying enzyme (FAME) | |
| glycerol ester hydrolase (geh) | |
| hyaluronate lyase (hysA) | |
| lipase/butyrylesterase (lip) | |
| metalloprotease/aureolysin (aur) | |
| thermonuclease (nuc) | |
| PI-phospholipase C (plc) | |
| staphopain/proteasell (scp) | |
| staphylokinase (sak) | |
| V8 serine protease (sspA) |
Those factors in bold were found to be down-regulated by treatment with honey in this study.
Figure 4.
A model to show how manuka honey affected genes of clinical importance in MRSA. Up-regulated genes are shown in orange, down-regulated genes are shown in blue and genes with unknown levels of transcription are shown in black. Affected cellular processes are underlined.
Biofilm genes
Manuka honey has been demonstrated to prevent the formation of biofilms and to disrupt established staphylococcal biofilms in vitro,36,37 although the underlying mechanism has been unknown. Alpha-haemolysin (hla), a protein that elicits host cell lysis by disrupting host cell membranes, was found to be down-regulated 27.5-fold by manuka honey (Table 5). Mutants of S. aureus defective in hla have been shown to be unable to form biofilms due to a requirement for cell–cell interaction mediated by this toxin.38 Although agr has been implicated in the regulation of hla, another regulator (sae) has also been suggested.39 This global regulator of virulence in S. aureus is the two-component saeSR system, which is thought to be activated by agr and is essential for the expression of staphylococcal adhesins.40 Here, decreased expression of the histidine kinase sensor (saeS) and regulator (saeR) genes in manuka honey-treated cells (Table 5) might explain this observation since mutation of saeRS has been shown to increase protease activity and restrict biofilm formation.41 The adhesion of bacteria to host cells, wound beds or indwelling medical devices is an important prerequisite to both infection and the initiation of biofilm formation, and interventions that prevent or disrupt biofilms in wounds may improve wound healing outcomes. The recent demonstration of an association between wound chronicity and the presence of biofilm16,42 reinforces the clinical importance of anti-biofilm effects.
Reduced transcription of the gene coding for fibronectin-binding protein by MRSA after treatment with manuka honey (Table 5) is likely to impact on the ability of the bacterial cells to adhere to host fibronectin within a wound and will further reduce the opportunity to initiate infection and biofilm formation. Manuka honey has already been shown to attenuate the efficacy of binding in Streptococcus pyogenes by reducing the expression of two fibronectin-binding proteins, Sof and Sfbl.6 In the case of S. aureus, the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase was also demonstrated to function as a cell wall transferrin-binding protein and fibronectin-binding protein.43 Microarray data showing decreased expression of fnb and gapB genes (Table 5) give a strong indication that manuka honey impairs the efficacy of ligand binding required for adherence that confers enhanced virulence and biofilm formation in MRSA.
Cell wall functions and cell division
The agr operon has also been linked to the control of β-lactam resistance via the regulation of autolysis;44 hence the reversion of oxacillin resistance to oxacillin susceptibility owing to a reduced expression of mecR1 caused by manuka honey45 may have resulted from a down-regulation of agr. Autolysis is important in bacterial growth and cell division and is controlled by complex mechanisms. The increased expression of one pertinent transcriptional regulator (sarV) was seen (Table 5), which positively regulates the transcription of scdA, lrgB, atl, splA and aur.25 Increased levels of ScdA (Table 3) and scdA were found (Table 4) and the expression of lrgB was increased 22-fold (Table 5). An increased transcription of atl was reported in MRSA in response to treatment with manuka honey, but the gene product (murein hydrolase) exhibited undetectable activity in both cell-free and extracellular extracts.45 An inability to degrade peptidoglycan at the cell equator due to a diminished activity of murein hydrolase helps to explain why MRSA failed to execute cell division and complete its cell cycle.
Cell wall-related protein (ScdA) is a di-iron protein involved in the repair of proteins that result from conditions of oxidative stress and is also required for cell division.46 Changes in the expression of scdA affect peptidoglycan cross-linking, and cells with depleted levels of ScdA have been reported to form large aggregated clumps of cells with aberrant septum placement. The diminished rate of autolysis was attributed to structural changes in peptidoglycan rather than altered murein hydrolase activity.46 Transcription of sarV in S. aureus has been shown to be repressed by sarA and mgrA25 and sarA is normally repressed by agrA. Altered levels of agr A were not discovered in this study, yet three of the genes of the agr locus exhibited decreased levels of expression (Table 5). If agrA had been repressed too, sarV would have been derepressed and transcription of the regulator gene would have been allowed. Increased levels of expression of sarV, scdA and lrgB (Table 5) support this hypothesis.
Bacterial growth and division is intimately linked to the controlled synthesis and cleavage of peptidoglycan in the cell wall by autolysins (or murein hydrolases). The cidABC operon works in conjunction with the lrgAB operon to regulate murein hydrolase, as well as antibiotic tolerance.26,47 The export of murein hydrolases is regulated by the cidABC operon, which promotes murein hydrolase activity and increases sensitivity to penicillin, while the lrgAB operon decreases murein hydrolases activity and penicillin sensitivity.26,47 An increased transcription of lrgB (22-fold) and a decreased transcription of cidB (1.9) (Table 5) were observed in this study. Since the products of the cidABC operon act as holins to facilitate the export of murein hydrolase, and the products of the lrgAB operon act as antiholins to restrict export, the deduction that manuka honey interferes with the cell cycle45 is strengthened. Further evidence of an impact on cell division was provided by the diminished transcription of two other genes whose products are involved in cell division: scpA (for segregation) and ftsL (in septum formation) (Table 5).
The transcription of cidABC and lrgAB has been shown to be influenced by the metabolism of glucose when fermentation end-products such as acetic acid and lactate accumulate and acidic stress increases.48 In this study, increased glycolysis together with a reduced expression of pflB and a reduced oxidation of acetyl CoA by the TCA cycle might be expected to induce such conditions.
Phosphoglucosamine mutase (GlmM) demonstrated decreased protein expression (Table 3) and increased gene expression (Table 4). GlmM converts glucosamine 6-phosphate into glucosamine 1-phosphate, which is an essential precursor of peptidoglycan, lipopolysaccharide and teichoic acids.49 Hence perturbations in this enzyme will affect the cell wall composition, reducing both cell stability and resistance to antibiotics, and inhibition has been linked to dramatic morphological changes.50
Stress
Exposure of MRSA to manuka honey has already been shown to reduce levels of universal stress protein A.14 Here, further evidence of stress was found, with an up-regulation of sod, acpD and mscL and a down-regulation of asp23 and a putative stress protein (Table 5). ATP-dependent ClpC showed reduced levels by proteomics, but the equivalent gene (clpC) was found to be up-regulated by qPCR. This might have been brought about by altered post-transcriptional events that may have affected its migration pattern during 2D-DIGE, perhaps causing the dissociation of the two parts of the enzyme. This protease complex comprises an ATPase specificity factor and a proteolytic domain51 and has been demonstrated to play a role in bacterial adaptation to multiple stresses by the degradation of accumulated misfolded proteins. In S. aureus, clpC has been shown to regulate the TCA cycle, which is required for metabolism and growth in recovery from the stationary phase and cell death.52 Within S. aureus, it is also thought to be essential for virulence, long-term survival and intracellular replication; mutants lacking clp showed attenuated virulence in mouse models.53
Summary
In this study, we compared patterns of protein expression in MRSA cells treated with and without manuka honey, examined the expression of pertinent genes by qPCR and investigated the whole-genome response by microarray analysis. In order to understand the complex effects of manuka honey on MRSA, changes in gene expression and cellular processes were mapped (Figure 4).
Essentially, the glucose in honey promoted glycolysis via gapA to generate pyruvate, acetate and acetyl phosphate. An accumulation of acetate is capable of activating the LytSR protein kinase system to induce the transcription of lrgAB, and acetyl phosphate has been postulated to activate lytR alone to promote lrgAB transcription.54 Only an up-regulation of lrgB was detected here, but since both lrgA and lrgB are regulated by LytSR,55 it is reasonable to assume that antiholin was induced and would have limited the export of murein hydrolases. The down-regulation of cidB indicated that the cidABC operon was not transcribed and that holin was in limited supply for the export of murein hydrolase. A repression of quorum sensing was deduced by the reduced expression of three of the genes in the agr operon and this would have resulted in diminished virulence in MRSA and a failure to initiate biofilm formation. Knock-on effects were a repression of saeRS, with a repression of further virulence genes and a derepression of sarV, which promoted the induction of lrgB, scdA and atl. The products of these three genes can be assumed to also limit cell wall functions and cell division (Figure 4), and this supports previous deductions that manuka honey interrupts cell division in MRSA.13
The differential expression of proteins and genes observed in this study provided a valuable insight into the mechanisms by which growth and pathogenicity in MRSA were inhibited and confirms the multifactorial effects of manuka honey on bacterial cells. Although the precise mode of inhibition of quorum sensing was not found, this study provides many leads for further investigation. Collectively, the observations made here support the hypothesis that honey reduces the fitness of MRSA to initiate infections or biofilms in vitro; whether this will be elicited in wounds colonized by MRSA in the human host must be explored. We have addressed the major responses observed, but it is important to remember that one cannot expect to see a complete correlation between gene expression and the corresponding activity level of the gene product. In addition, genes showing the highest fold change might not necessarily coincide with the most important bacterial physiological response. There remains a further opportunity to explain how manuka honey affects other genes in MRSA.
Funding
This work was supported by The Waterloo Foundation.
Transparency declarations
R. C. has received grants from the BSAC, the Society for General Microbiology, the European Wound Management Association, the University of Waikato (in collaboration with the National Honey Board), the Waterloo Foundation, the UWIC Foundation and the Sir Halley Stewart Trust. Sponsorship to attend scientific meetings has been received from Capilano, derma Sciences Inc. and Molnlycke. Consultancy has been undertaken for Advancis Medical, Arnold & Porter, Aspen Medical, Brightwake Ltd, BSN medical, Comvita UK, Derma Sciences Inc., FlenPharma, KCI, Medlock Medical, Medihoney and the North American Center for Continuing Medical Education. Remuneration for presentations has been received from the Tissue Viability Society, the American Professional Wound Care Association, Derma Sciences Inc., Comvita UK, the North American Center for Continuing Medical Education, the World Union of Wound Healing Societies and numerous beekeeping organizations. R. J. and N. B.: none to declare.
Acknowledgements
2D-DIGE, MS and microarray experiments were performed at the Central Biotechnology Service at Cardiff University (http://medicine.cf.ac.uk/cbs). We would like to thank Peter Giles for his help in analysing the microarray data.
References
- 1.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–93. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahady GB. Medicinal plants for the prevention and treatment of bacterial infections. Curr Pharm Des. 2005;11:2405–27. doi: 10.2174/1381612054367481. [DOI] [PubMed] [Google Scholar]
- 3.Cooper RA, Molan PC, Harding KG. The sensitivity to honey of Gram-positive cocci of clinical significance isolated from wounds. J Appl Microbiol. 2002;93:857–63. doi: 10.1046/j.1365-2672.2002.01761.x. [DOI] [PubMed] [Google Scholar]
- 4.Cooper RA, Halas E, Molan PC. The efficacy of manuka honey in inhibiting strains of Pseudomonas aeruginosa from infected burns. J Burns Care Rehab. 2002;23:366–70. doi: 10.1097/00004630-200211000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Blair SE, Cockcetin NN, Harry EJ, et al. The unusual antibacterial activity of medical-grade Leptospermum honey: antibacterial spectrum, resistance and transcriptome analysis. Eur J Clin Microbiol Infect Dis. 2009;10:1199–208. doi: 10.1007/s10096-009-0763-z. [DOI] [PubMed] [Google Scholar]
- 6.Maddocks SE, Salinas Lopez M, Rowlands R, et al. Manuka honey inhibits the development of biofilms of Streptococcus pyogenes and causes reduced expression of two fibronectin binding proteins. Microbiol. 2012;158:781–90. doi: 10.1099/mic.0.053959-0. [DOI] [PubMed] [Google Scholar]
- 7.Majtan J, Majtanova L, Bohora J, et al. Honeydew honey as a potent antibacterial agent in eradication of multi-drug resistant Stenotro-phomonas maltophila isolates from cancer patients. Phytother Res. 2011;25:584–7. doi: 10.1002/ptr.3304. [DOI] [PubMed] [Google Scholar]
- 8.Kwakman PHS, te Velde AA, de Boer L, et al. Two major medicinal honeys have different mechanisms of bactericidal activity. PLoS One. 2011;6:e17709. doi: 10.1371/journal.pone.0017709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavric E, Wittmann S, Barth G, et al. Identification and quantification of methylglyoxal as the dominant antibacterial constituent of manuka (Leptospermum scoparium) honeys from New Zealand. Mol Nutr Food Res. 2008;52:483–9. doi: 10.1002/mnfr.200700282. [DOI] [PubMed] [Google Scholar]
- 10.Adams CJ, Boult CH, Deadman BJ, et al. Isolation by HPLC and characterisation of the bioactive fraction of New Zealand manuka (Leptospermum scoparium) honey. Carbohydr Res. 2008;343:651–9. doi: 10.1016/j.carres.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Kato Y, Umeda N, Maeda A, et al. Identification of a novel glycoside, leptosin, as a chemical marker of manuka honey. J Agric Food Chem. 2012;60:3418–23. doi: 10.1021/jf300068w. [DOI] [PubMed] [Google Scholar]
- 12.Henriques A, Jenkins RE, Burton NF, et al. The intracellular effects of manuka honey on Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2010;29:45–50. doi: 10.1007/s10096-009-0817-2. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins RE, Burton N, Cooper RA. Manuka honey inhibits cell division in methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2011;66:2536–42. doi: 10.1093/jac/dkr340. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins RE, Burton NF, Cooper RA. The effect of manuka honey on the expression of universal stress protein A in methicillin-resistant Staphylo-coccus aureus. Int J Antimicrob Agents. 2011;37:373–6. doi: 10.1016/j.ijantimicag.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 15.Packer JM, Irish J, Herbert BR, et al. Specific non-peroxide antibacterial effect of manuka honey on the Staphylococcus aureus proteome. Int J Antimicrob Agents. 2012;40:43–50. doi: 10.1016/j.ijantimicag.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 16.James G, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Rep Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 17.Bernhardt J, Büttner K, Scharf C, et al. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis. 1999;20:2225–40. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2225::AID-ELPS2225>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Kohler C, von Eiff C, Peters G, et al. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J Bacteriol. 2003;185:6928–37. doi: 10.1128/JB.185.23.6928-6937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shevchenko A, Wilm M, Vorm O, et al. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 20.Medzidhradszky KF, Campbell JM, Baldwin MA, et al. The characteristics of peptide collision-induced dissociation using a high performance MALDI-TOF/TOF tandem mass spectrometer. Anal Chem. 2000;72:552–8. doi: 10.1021/ac990809y. [DOI] [PubMed] [Google Scholar]
- 21.Bienvenut WV, Deon C, Pasguarello C, et al. Matrix-assisted laser desorption/ionization-tandem mass spectrometry with high resolution and sensitivity for identification and characterization of proteins. Proteomics. 2002;2:868–76. doi: 10.1002/1615-9861(200207)2:7<868::AID-PROT868>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Purves J, Cockayne A, Moody PC, et al. Comparison of the regulation, metabolic functions, and roles in virulence of the glyceraldehyde-3-phosphate dehydrogenase homologues gapA and gapB in Staphylococcus aureus. Infect Immun. 2010;78:5223–32. doi: 10.1128/IAI.00762-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarraud S, Mougel C, Thioulouse J, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–41. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manna AC, Ingavale SS, Maloney M, et al. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol 20045; 186:5267–80. doi: 10.1128/JB.186.16.5267-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groicher KH, Firek BA, Fujimoto DB, et al. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sotto A, Lina G, Richard J-L, et al. Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers. Diabetes Care. 2008;31:2318–24. doi: 10.2337/dc08-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Booth MC, Cheung AL, Hatter KL, et al. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65:1150–6. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan F, Foster SJ. The role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–41. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng HL, Novick RP, Kreiswirth B, et al. Cloning, characterisation, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1998;170:4365–72. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novick RP, Ross HF, Projan S, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO. 1993;12:3967–75. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haslinger-Loffler B, Khal BC, Grundmeier M, et al. Multiple virulence factors are required for Staphylococcus aureus-induced apoptosis in endothelial cells. Cell Microbiol. 2005;7:1087–97. doi: 10.1111/j.1462-5822.2005.00533.x. [DOI] [PubMed] [Google Scholar]
- 33.Truchado P, López-Gálvez F, Gil MI, et al. Quorum sensing inhibitory and antimicrobial activities of honeys and the relationship with individual phenolics. Food Chem. 2009;115:1337–44. [Google Scholar]
- 34.Truchado P, Gil-Izquierdo A, Tomás-Barbéran F, et al. Inhibition by chestnut honey of N-acetyl-l-homoserine lactones and biofilm formation in Erwinia carotovora, Yersinia enterocolitica, and Aeromonas hydrophila. J Agric Food Chem. 2009;57:11186–93. doi: 10.1021/jf9029139. [DOI] [PubMed] [Google Scholar]
- 35.Wang R, Starkey M, Hazan R, et al. Honey's ability to counter bacterial infections arises from both bactericidal compounds and QS inhibition. Front Microbiol. 2012;3:144. doi: 10.3389/fmicb.2012.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alandejani T, Marsan JG, Ferris W, et al. Effectiveness of honey on Staphylococccus aureus and Pseudomonas aeruginosa biofilms. Otolaryngol Head Neck Surg. 2009;139:107–11. doi: 10.1016/j.otohns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Merckoll P, Jonassen TØ, Vad ME, et al. Bacteria, biofilm and honey: a study of the effects of honey on ‘planktonic’ and biofilm-embedded chronic wound bacteria. Scand J Infect Dis. 2009;41:341–7. doi: 10.1080/00365540902849383. [DOI] [PubMed] [Google Scholar]
- 38.Caiazza NC, O'Toole GA. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J Bacteriol. 2003;185:3214–7. doi: 10.1128/JB.185.10.3214-3217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goekre C, Fluckiger U, Steinhuber A, et al. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infections resolved by quantitative transcript analysis. Mol Microbiol. 2001;40:1439–47. doi: 10.1046/j.1365-2958.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 40.Harraghy N, Kormanec J, Wolz C, et al. sae is essential for expression of the staphylococcal adhesions Eap and Emp. Microbiology. 2005;151:1789–800. doi: 10.1099/mic.0.27902-0. [DOI] [PubMed] [Google Scholar]
- 41.Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun. 2008;76:1756–65. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bjarnsholt T, Kirketerp-Møller K, Jensen PØ, et al. Why chronic wounds will not heal: a novel hypothesis. Wound Rep Regen. 2008;16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 43.Modun B, Williams P. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999;67:1086–92. doi: 10.1128/iai.67.3.1086-1092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piriz DS, Kayser FH, Berger-Bachi B. Impact of sar and agr on methicillin resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1996;141:255–60. doi: 10.1111/j.1574-6968.1996.tb08394.x. [DOI] [PubMed] [Google Scholar]
- 45.Jenkins RE, Cooper RA. Synergy between oxacillin and manuka honey sensitizes methicillin-resistant Staphylococcus aureus to oxacillin. J Antimicrob Chemother. 2012;67:1405–7. doi: 10.1093/jac/dks071. [DOI] [PubMed] [Google Scholar]
- 46.Brunskill EW, de Jonge BLM, Bayles KW. The Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiology. 1997;143:2877–82. doi: 10.1099/00221287-143-9-2877. [DOI] [PubMed] [Google Scholar]
- 47.Rice KC, Firek BA, Nelson JB, et al. The Staphylococcal aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol. 2003;185:2635–43. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice KC, Nelson JB, Patton TG, et al. Acetic acid induces expression of the Staphylococcal aureus cidABC and lrgAB murein hydrolase regulator operons. J Bacteriol. 2005;187:813–21. doi: 10.1128/JB.187.3.813-821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cordwell SJ, Larsen MR, Cole RT, et al. Comparative proteomics of Staphylococcus aureus and the response of methicillin-resistant and methicillin-sensitive strains to Triton X-100. Microbiology. 2002;148:2765–81. doi: 10.1099/00221287-148-9-2765. [DOI] [PubMed] [Google Scholar]
- 50.Jolly L, Pompeo F, van Heijenoort J, et al. Autophosphorylation of phosphoglucosamine mutase from Escherichia coli. J Bacteriol. 2000;182:1280–5. doi: 10.1128/jb.182.5.1280-1285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michel A, Agerer F, Hauck CR, et al. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol. 2006;188:5783–96. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatterjee IP, Becker M, Grundmeier M, et al. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J Bacteriol. 2005;187:4488–96. doi: 10.1128/JB.187.13.4488-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frees D, Chastanet A, Qazi S, et al. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–62. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 54.Sadykov MR, Bayles KW. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol. 2012;15:211–5. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunskill EW, Bayles KW. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–2. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]