Abstract
Protein-protein interaction (PPI) is one of the most important functional components of a living cell. Recently, researchers have been interested in investigating the correlation between PPI and microRNA, which has been found to be a regulator at the post-transcriptional level. Studies on miRNA-regulated PPI networks will not only facilitate an understanding of the fine tuning role that miRNAs play in PPI networks, but will also provide potential candidates for tumor diagnosis. This review describes basic studies on the miRNA-regulated PPI network in the way of bioinformatics which includes constructing a miRNA-target protein network, describing the features of miRNA-regulated PPI networks and overviewing previous findings based on analysing miRNA-regulated PPI network features.
Keywords: miRNA, Regulation, Protein-protein interaction
Review
Brief introduction to MicroRNA and protein-protein interaction networks
MicroRNAs (miRNAs) are a subset of small (~22 nucleotide – structural units of RNA and DNA - in length) non-coding RNA molecules, which comprise 1% of genes in animal genomes. The miRNAs are mainly found in intergenic regions (IG region), although some are also located in intronic regions [1,2]. miRNAs can repress gene expression by interacting with three prime untranslated regions (3′ UTR) which are sequences that are not translated into proteins in the 3′end of the target mRNAs. Since Rosalind Lee and Rhonda Feinbaum discovered the first two precursors (pre-miRNAs) - lin-4 and let-7, products of C. elegans genes, miRNAs have been keenly studied for several years [1-6]. miRNAs play an important role in controlling nematode developmental timing and repressing mRNA translation [3,4] at the post-transcriptional level in a gene regulatory network (the gene regulatory network can be divided into the transcription level and post-transcription level [7], during which transcription factors (TF) and miRNAs contribute the most, respectively). Research has also indicated that miRNAs have a close relationship with tumorigenesis [8-10]. Recent research showed evidence that miRNAs can affect cancer initiation and progression [11,12].
MiRNAs have a complete complementarity region (2-8nt) which is called the “seed sequence” in their 5′ end. Most miRNAs are highly conserved among species [13,14]. In RNA regulation, the 5′ region and coding region play important roles in binding to mRNA targets. Thus, a many-to-many relationship structure can be established in miRNAs and their target mRNAs, that is: a miRNA can regulate more than one target mRNA and each target mRNA also can be regulated by more than one miRNA[15]. miRNAs have the ability to post-transcriptional degrade their mRNA target or inhibit translation. Both mechanisms fine-tune mRNA expression by distinguishing sequence motifs in the 3′UTR of mRNA, for example, miR-182 binds with its direct targets, fibroblast growth factor 9 (FGF9) and neurotrimin (NTM) in response to injury stimuli [16]. The length of 3′ UTR indicates the density of miRNA binding sites. It has been shown that the mRNA with a longer 3′ UTR always participates in more sophisticated functions, whereas mRNAs with a shorter length 3′ UTR are generally involved in basic biological processes [17]. Some special mRNAs are regulated by miRNAs in their five prime untranslated regions (5′ UTR). Ørom et al. found that miR-10a binds 5′UTR of ribosomal protein mRNAs to enhance their translation [18]. Recent studies show that miRNAs can also bind with their targets in coding regions [19].
The interaction between miRNA and mRNA provides a new way to determine gene functions. In studies on miRNA-mRNA interaction, how to accurately find the target mRNA genes for miRNAs is the most important question. Various approaches have emerged since the discovery of miRNAs. Targeting mRNA genes in plants can easily be done because most miRNAs have perfect and stable complementarities [20]. miRNAs in animals, however, are allowed to have mismatches and gaps in the functional duplex, which generates uncertainty in the target site prediction in miRNAs [21]. Efficient target prediction approaches are discussed later in this paper.
In recent years, researchers have studied miRNA-regulated networks, including miRNA co-regulated networks, miRNA-mRNA networks, miRNA-transcription factor networks and miRNA-protein interaction networks. Currently, research on miRNA-regulated protein-protein interaction networks is in its initial stage. Barriers to this research not only exist because of the mystery of miRNAs, but also due to the complexity of protein-protein interactions.
Protein-Protein Interaction (PPI) is one of the most important tasks required for a living cell to carry out its biological functions such as DNA replication, transcription, translation, signal transduction [22]. PPI can be simulated as an undirected static network structure. The properties of the PPI network can be denoted by a series of elements, edges, nodes, cluster coefficients and so on [23]. The topological features are discussed later in this paper.
The research on PPI networks has developed rapidly since the yeast two-hybrid system was first described in Field and Song’s publication [24]. Based on Y2H, Schwikowski, Uetz and Field released a PPI network map in yeast [25] in 2000, then a human PPI network map was reported in 2005 [26]. Before this, PPI network research generally centred on other species such as yeast- Saccharomyces cerevisiae, and worms- C. elegans. Currently, proteome-wide studies of PPI networks in different species mostly concentrate on PPI network detection and prediction [24,27-30], signal transduction pathways [31-33], protein function prediction based on PPI networks and protein complex prediction in PPI networks [34-37].
Currently, the research on miRNA-regulated PPI networks can be divided into two main areas: a) basic studies on the correlation between miRNAs and general PPI networks. This subject mainly uses bioinformatics approaches and statistical means to explore the relationship between miRNA or cluster miRNAs and PPI network topological properties linked by miRNA target genes, which aims to find rules in miRNA-regulated gene expression beside seed matching. Although basic studies on miRNA-regulated PPI networks always suffer from partial coverage and false positives and negatives, it is undeniable that the studies are important. The basic studies of miRNA regulation in PPI networks attempt to discover new correlations and tendencies between miRNAs and their target proteins in a relatively fast way, which provides part of the theoretical support to laboratory experiments; and b) identification of the impact of miRNA regulation on PPI networks in diseases. As an essential component of PPI networks and prime candidates of miRNA targeting modulators in animal cells [38], signalling transduction pathways have been extensively explored in recent years. The correlation between miRNAs and signalling pathways was mostly proved by biochemical analysis. For example, Rogler et al. [31] found that through down-regulating the target gene Smads (including Smad3, 4 and 5), which are the key genes in transforming growth factor-beta signalling of miR-23b cluster miRNAs (including miR-23b, miR-27b, miR-24-1, miR-10a, miR-26a, and miR-30a), miR-23b miRNAs can be promoted in growth and consequently repress bile duct gene expression in fetal hepatocytes. Jie et al.[32] pointed out that miR-146a expression induced by epidermal growth factor receptor (EGFR) signalling can repress human gliomagenesis by down-regulating its target gene, NOTCH1. Recent research [33] shows that miR-128 can play a similar role by targeting oncogenic receptor tyrosine kinases signalling. In this review, we describe the current computational developments in basic studies on the correlation between miRNAs and general PPI networks, which may be helpful to gain new insights into current miRNA-regulated PPI network studies.
Understanding the role of miRNA in regulating target genes and proteins
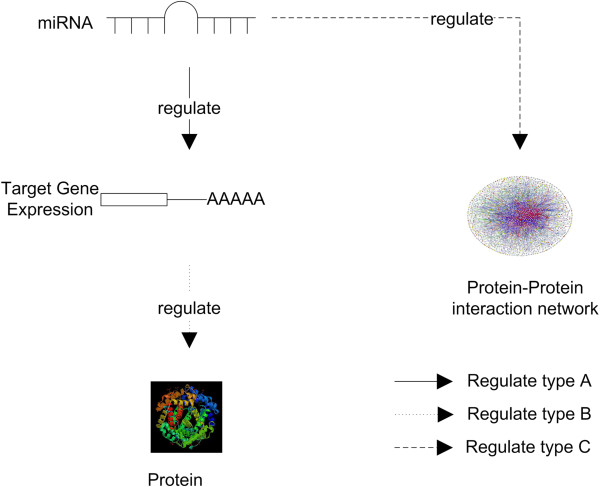
MiRNAs are involved in fine-tuning gene expression and biological processes at the post-transcription level by regulating their targets, such as signalling proteins, enzymes, transcription factors (TFs) [39]. For the mechanism of miRNA regulation, it has been accepted that in mature miRNAs directly associated with AGO1 (a member of the Argonaute protein family, the main component of RNA-induced silencing complex) in the RNA-induced silencing complex (RISC), the process of target selection occurs in RISC by miRNA strand unwinding, which leads to the repression of mRNA translation, stability and localization [38,40,41]. However, how miRNAs behave in RISC or how miRNAs play a regulation role is still unknown. As shown in Figure 1, we summarized the possible types how miRNAs regulation in PPI network from previous publications. In terms of regulating proteins, miRNAs regulate protein-coding targets to affect protein synthesis. miRNAs are reported as a regulator to fine-tune their target genes. The action of under or over expression of miRNAs can directly repress gene translation [42], and according to [43], miRNAs regulate more than 60% of genes which are related to protein synthesis, although this kind of down-regulation impacts most of their targets by no more than 50% [38,43]. In terms of regulating the PPI network, miRNAs may serve as a regulator by maintaining the stablity of PPI networks [44]. It has been reported that protein-protein interactions express a dynamic state rather than maintaining a static state to keep stability in the external environment [44,45]. Protein abundance, which can be tuned by miRNAs indirectly, is one of the most effective factors in the dynamic robustness of the PPI network [42]. In this case, it can be concluded that miRNAs act as an indirect regulator in PPI network stability. Furthermore, the most important sub-graph of the PPI network is the signalling pathway, hence investigation into the relationship between miRNAs and PPI networks could facilitate a deeper understanding of miRNA-regulated signalling pathways.
Figure 1.
Types of miRNA regulation. Type A: MiRNA regulating gene expression. miRNAs can lead to mRNA cleavage and degradation or mRNA translational repression. Type B: MiRNA regulating target protein. The action of under or over expression of miRNAs can directly repress gene translation. Type C: MiRNA regulating on PPI networks. miRNA shows as an indirect regulator to affect dynamic PPI network stability.
Resources to construct a reliable miRNA-protein network
The construction of a miRNA-protein network (Figure 2a) using highly reliable resources is important for the commencement of a miRNA-regulated PPI network study. As shown in Figure 2, a miRNA–target protein network is constructed by miRNA-target interactions and PPI network data. During the process, the selection of miRNA target predicting approaches and filtering approaches is important to obtain highly reliable data. According to Figure 2a, we divide the resources into the miRNA database (detecting miRNAs based on High Throughput Sequencing (HTS) and miRNA expression profiles included), miRNA target predicting approaches, miRNA-targets interaction database, miRNA-targets interaction filtering tools, PPI database (integrated PPI databases included) and filtering tools for the PPI database, as shown in Table 1. To complete the resources, we also mention platform resources in Table 1. In this section, we introduce several of the tools.
Figure 2.
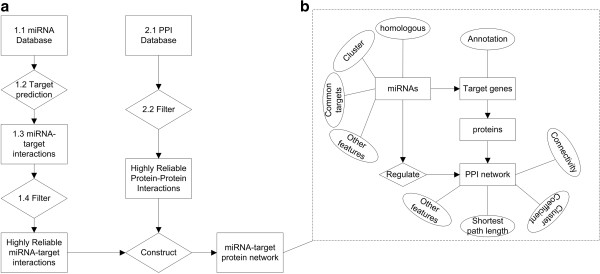
Constructing a miRNA-target protein network. a) Process to construct a miRNA-target protein network: the process labels correspond to Table 1 - column 2. b) Relationships within miRNA-target protein network and its features.
Table 1.
Resources to construct a reliable miRNA-target protein network
| Name | Process | Main feature | URL | Ref. |
|---|---|---|---|---|
| BioGrid |
2.1 |
Protein-protein interaction database |
http://thebiogrid.org/ |
[46] |
| Cytoscape |
Platform |
Social or molecular networks analysis and visualization. |
http://www.cytoscape.org/ |
[47] |
| Database of Interacting Proteins (DIP) |
2.1 |
Protein-protein interaction |
http://dip.doe-mbi.ucla.edu/dip/ |
[48] |
| Ingenuity system |
Platform |
Signalling and metabolic pathways analysis; molecular network analysis etc. |
http://www.ingenuity.com/ |
|
| Human Protein Reference Database (HPRD) |
2.1 |
Protein-protein interaction |
http://www.hprd.org/ |
[49] |
| String |
2.1 |
Protein-protein interaction |
http://string-db.org/ |
[50] |
| The MIPS mammalian protein-protein interaction database |
2.1 |
protein-protein interaction |
http://mips.helmholtz-muenchen.de/proj/ppi/ |
[51] |
| Protein Interaction Network Analysis (PINA) |
2.2 |
PPI network construction, filtering, analysis, visualization and management |
http://cbg.garvan.unsw.edu.au/pina/ |
[52] |
| HitPredict |
2.1 |
Integrated PPI database |
http://hintdb.hgc.jp/htp/ |
[53] |
| iRefIndex |
2.1 |
Integrated PPI database |
http://www.irefindex.org/wiki/index.php?title=iRefIndex |
[54] |
| SynechoNET |
1.1 |
Integrated PPI database |
http://bioportal.kobic.re.kr/SynechoNET/ |
[55] |
| PMRD |
1.1 |
Plant miRNA database |
http://bioinformatics.cau.edu.cn/PMRD/ |
[56] |
| Gene ontology |
Platform |
Gene annotation, develop controlled vocabulary of genes |
http://www.geneontology.org/ |
[57] |
| MiRTarBase |
Platform |
miRNA-target interactions |
http://mirtarbase.mbc.nctu.edu.tw/ |
[58] |
| PicTar |
1.2 |
miRNA target prediction |
http://pictar.mdc-berlin.de/ |
[59] |
| RNAhybrid |
1.2 |
miRNA target prediction |
http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/ |
[60] |
| TargetScan |
1.2 |
miRNA target prediction |
http://www.targetscan.org/ |
[61] |
| GeneSet-2miRNA |
1.2 |
miRNA target predicting with mRNA expression profile |
http://mips.helmholtz-muenchen.de/proj/gene2mir/ |
[62] |
| MMIA |
1.2 |
miRNA target predicting with mRNA expression profile |
http://129.79.244.122/~MMIA/ |
[63] |
| miRanda |
1.2 |
miRNA target predicting & miRNA expression profiles |
http://www.microrna.org/ |
[64] |
| MiRTif |
1.4 |
miRNA target interaction filter |
http://mirtif.bii.a-star.edu.sg/ |
[65] |
| miRBase |
1.1 |
miRNA sequences and annotations |
http://www.mirbase.org/ |
[66] |
| The human microRNA disease database (HMDD) |
1.1 |
miRNA sequences and annotations |
http://202.38.126.151/hmdd/mirna/md/ |
[49] |
| miRExpress |
1.1 |
Extract miRNA expression profiles based on HTS results |
http://mirexpress.mbc.nctu.edu.tw/ |
[67] |
| TarBase |
1.2 |
Experimental supported miRNA target |
http://diana.cslab.ece.ntua.gr/tarbase/ |
[68] |
| miRDeep |
1.1 |
Detect novel miRNA based on HTS |
https://www.mdc-berlin.de/690237/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/miRDeep/index.html |
[69] |
| miRTools |
1.1 |
Detect novel miRNA based on HTS |
http://centre.bioinformatics.zj.cn/mirtools/ |
[70] |
| starBase |
1.3 |
Decoding microRNA-target and protein-RNA interaction |
http://starbase.sysu.edu.cn/ |
[71] |
| IPA | 1.4 | Comprehensive software on biological analysis. Support miRNA target filtering | http://www.ingenuity.com/products/training.html |
HTS technology provides high performance and low-cost in detecting miRNA sequencing and therefore replaces traditional Sanger sequencing. The most famous HTS technologies are 454 sequencing [79] from Roche, Solexa sequencing [80] from Illumina, SOLiD sequencing [81] from ABI and SMRT [82] from Pacific Biosciences. Currently, there are several platforms based on HTS, established for scientific use, such as High-Throughput Genome Sequencing from the Gene Bank and the Princeton University High Throughput Sequencing Database. Specific to miRNAs, deep sequencing tools have been developed to analyse HTS results and then to detect novel miRNAs or extract miRNA expression profiles, for example, miRExpress [67], miRTools [70] and miRDeep [69].
Current experimental approaches to miRNA target identification mainly focus on the use of large scale mRNA expression profiling. The common way to uncover miRNA targets is to directly test miRNA expression levels on different mRNA profiling [83] or to use different phenotypes to test expression levels [84] based on microarray [85]. Various computational approaches [11,86-92] provide fast and low-cost support to experiments. Sequence complementarity, evolutionary conservation and free energy among miRNA-mRNA duplex are the most common features to identify miRNA targets. The most frequently used applications are TargetScan [61], PicTar [59], miRanda [64], RNAHybrid [60] etc. The combination of computational approaches with mRNA expression profiles have proved efficient in recent years, and it has been shown that they can effectively minimize the false positives of miRNA target prediction.
For miRNA target filtering, previous studies have shown that considering conservation in strains combined with performing seed matches [93,94] or evaluating the accessibility of binding sites [95] can facilitate miRNA target predictions. Target filtering approaches are then developed to post-process the results of prediction applications. miRTif [65], for example, filters the predicted targets by SVM scores to evaluate the accessibility of binding sites and the properties of miRNA-mRNA interactions, such as the value of seed complementarity and binding energy. Recent studies show that combining both conservation and accessibility [96] can achieve better results in filtering miRNA targets.
In PPI data filtering, computational approaches are widely used to improve the PPI data obtained from laboratory experiments. Some approaches check whether the interactions support genomic features such as sequence, structure and annotation information to check the possibility of real interactions [97]. Alternatively, computational approaches integrate several databases to obtain more reliable interaction data, for example, HitPredict [53] integrated the databases of IntAct, BIOGRID and HPRD, collecting 168,458 highly reliable interactions across nine species. iRefIndex [54] and SynechoNET [55] are integrated databases as well. For single databases, checking whether interacting proteins have homolog across one or more species has proved an effective way to obtain high quality interactions [98].
After the filtering procedure, we can obtain highly reliable miRNA-target and protein-protein interaction networks. For certain miRNAs, miRNA-regulated PPI sub-networks can be easily constructed by combining their target sites and PPI networks according to the requirement. The sub-network may include miRNAs, target proteins, proteins associated with target proteins and the associations between proteins.
Features of miRNA-regulated PPI networks
When a miRNA-targeted protein network is available, the task is to analyse the features of the miRNA-regulated PPI network (Figure 2b). A miRNA-regulated PPI network is defined to identify the featured sub-networks of the miRNA-target protein network. Here, we firstly describe the PPI network features.
The features of PPI networks are defined with a focus on topological characteristics, such as connectivity, cluster coefficients, shortest path length and so on. PPI network features are commonly used to predict unannotated protein functions combined with Gene Ontology [99].
Connectivity or degree denotes the interacted number of a node. Proteins with high degree are called hub proteins [100]. Betweenness centrality is the proportion of the shortest path length passing through a vertex protein and all the shortest path lengths from one certain protein to another certain protein. The nodes which have higher betweenness centrality are called network bottlenecks. Close Centrality is then defined to measure the sum of the shortest paths between the target protein and other proteins [23,73]. (Local) Cluster coefficient measures the cluster tendency of a node in a network, which equals the ratio of all the real interaction numbers in a cluster and the number of possible connections. For a node i, Cluster Coefficient Ci = 2ni/k (k-1), where k is the number of edges in node i, and n is the edge number for all k neighbours connecting to each other.
In miRNA-regulated PPI networks, miRNAs are classified by several properties: a) the miRNA family or homologous miRNAs, which denote the miRNA group whose conserved seed regions are common [61]; b) clustered miRNAs, which are miRNAs whose pairwise chromosomal distances are no more than 3000nt [101]; and c) miRNAs with common targets. miRNAs can directly or indirectly down-regulate 100–200 genes on average [102], and each gene can be targeted by multi-miRNAs. Therefore, it is possible for miRNAs to regulate common target genes. There are still other classification criteria such as gene expression level, target mRNA and protein stability and the impact on target mRNA degradation, which are also essential in further analysis.
The purpose of the basic study on miRNA-regulated PPI networks is to find new rules on miRNA regulation or protein interaction, based on constructed sub-networks between miRNAs and protein interaction networks. As described above, miRNAs and protein-protein interaction networks both have different features, and by combining the analysis of these features, it is possible to obtain useful information or rules on both miRNA regulation or protein interaction, such as the relationship between miRNAs and protein complexes, how miRNAs coordinate to regulate targets and so on.
Current findings in the study of miRNA-regulated PPI networks
As listed in Table 2, we have divided the previous findings into three areas: correlation between protein connectivity and miRNA regulation complexity; miRNA-regulated specific proteins, the coordination role of miRNAs in regulating PPI networks; and identifying miRNA-regulated PPI networks in special diseases.
Table 2.
Current findings in the study of miRNA-regulated PPI networks
| Research area | Description |
|---|---|
| Correlation between protein connectivity and miRNA regulation complexity |
A. There is positive correlation between miRNA target site types and its regulated protein connectivity. B. MiRNA target propensity may be due to high protein connectivity. C. MiRNA regulation propensity changes due to different hub proteins [72]. |
| miRNA targeted proteins have short distance and higher modularity than randomly selected proteins [73]. | |
| MiRNA-regulated specific proteins in PPI networks |
A. MiRNAs that target a lower number genes have the propensity to regulate commonly expressed proteins rather than tissue-specific proteins. B. Commonly expressed proteins and tissue-specific proteins are always regulated together by a miRNA, and the numbers of protein expressed are close in both proteins [44]. |
| The coordination role of MicroRNAs: miRNA clusters regulate PPI networks |
miRNAs in the same clusters have the tendency to coordinate to regulate protein functions in protein-protein interaction networks [74]. |
| The coordination role of MicroRNAs: miRNAs coordinate to regulate protein complex |
A. MiRNAs coordinate to regulate protein complexes in posttranscriptional level. B. Correlations between the proteins exist in the same complex regulated by miRNAs [75]. |
| The coordination role of MicroRNAs: miRNA crossingtalking with transcription factors |
Crosstalk motifs between miRNAs and transcription factors motif demonstrate higher network properties in miRNA-regulated PPI networks [7] |
| Identifying miRNA-regulated PPI networks in special diseases | A. In gastric cancer [76]: six miRNA-regulated protein networks are identified in gastric cancer based on the human PPI network; it is suggested that miR-148a may resist tumor extension. B. In human ovarian cancer [77]: six miRNAs (hsa-mir-20a, hsa-mir-24-2, hsa-mir-34a, hsa-mir-21, hsamir-17 and hsa-mir-hsa-mir-155) and six TFs (BRCA1, SP1, ESR1, SMAD3, PO2F1 and TFE2) play key roles in ovarian cancer progression. C. In aging-related diseases [78]: 35 genes related to diseases associated with aging were identified. |
Correlation between protein connectivity and miRNA regulation complexity
Liang and Li [72] found that multiple miRNA-regulated proteins have more interacting neighbours in a PPI network. They observed that the number of miRNA target site types increases with the increasing connectivity of a protein. Based on observations, they developed the prediction formula for the average number (y) of target site types in the 3′-UTR. If x is the range of mRNA-expressed tissues (0 < x < 73), y = 0.0225x + 1.774. Liang and Li subsequently revealed that miRNA propensity [103] to regulate certain biological processes may be due to the higher connectivity of proteins in the PPI network. Finally, they pointed out that regulation by miRNA plays a more important role in low clustering coefficient hub proteins (connectivity > 8) which are intended to coordinate several functions than those hub proteins with high clustering coefficients.
Hsu, Juan and Huang [73], who extended Liang and Li’s study, proposed that miRNA-regulated proteins are always the hub or bottleneck in the PPI network. To clarify this issue, they firstly defined four PPI features [23]: degree, cluster coefficient, betweenness centrality and closeness centrality. Two sub-networks were then defined referring to the relationships between miRNAs and the PPI network. In the experiment, the target genes were detected by TargetScan, and then the target proteins with the target genes regulated by certain miRNAs were collected as a sub-network L0, and all the interactions which contain target proteins were collected as sub-network L1. Z score and p value were then computed in sub-network L0 and L1. By comparing randomly selected protein sub-graphs, the paper suggested that miRNA target proteins have shorter distance and higher modularity than randomly selected proteins.
MiRNA-regulated specific proteins in PPI networks
Zhu et al.[44] explored the relationships between miRNA and tissue-specific proteins in the human PPI network. They selected 10 main human tissues and defined proteins which were expressed in less than 3 tissues as tissue-specific proteins, and proteins which were expressed in all 10 tissues were commonly expressed proteins. Based on the findings of [104], which determined that tissue-specific proteins have the propensity to interact with commonly expressed proteins, Zhu et al. further found that miRNAs which target a lower number of genes have the propensity to regulate commonly expressed proteins rather than tissue-specific proteins in the human PPI network. From observations of miRNA regulation in all 10 tissues, they pointed out that miRNA regulation maintained consistency, that is, commonly expressed proteins and tissue-specific proteins were always regulated together by a miRNA, and the numbers of protein expressed are close in both proteins.
The coordination role of MicroRNAs in regulating PPI networks
Firstly, clustered miRNAs may coordinate to regulate the PPI network. Yuan et al. [74] revealed that miRNAs in the same clusters coordinate to regulate protein functions in protein-protein interaction networks. 55 clusters were identified at the miRNA family level according to miRBase. The miRNAs in the same clusters were defined as sc-miRNAs. The coordination strength of sc-miRNAs is measured by the number of clusters which regulate the two interacting proteins and the coordination range is represented by the number of interactions which are regulated by the miRNAs in the same clusters. The measurement of coordination indicates that interacting proteins have a tendency to be regulated by sc-miRNAs. Furthermore, the authors also point out that the tendency is influenced by protein interaction features in the protein-protein interaction network, for example, distance (shortest path between proteins) and connectivity (protein neighbour numbers).
Secondly, miRNAs coordinate to regulate protein complex. Sass et al.[75] used protein complexes which are verified based on the PPI network to investigate the relationships between miRNAs and protein complexes at a system level. The significance of the associations between miRNAs and their target protein complexes were ranked based on the P-value resulting from Fisher’s exact test. According to the test, the complexes which were regulated by single miRNAs or miRNAs in the same clusters were ranked higher than the ones which were regulated by multi-miRNAs in different clusters. The paper also proved that there are correlations between proteins in the same complex which are regulated by miRNAs.
Thirdly, miRNAs may coordinate with other regulators such as transcription factors (TFs). Based on previous findings, Cui et al.[39,105] found a correlation between transcription factors and miRNAs, and Lin et al. [7] extended this investigation to the human PPI network. Via the analysis of large gene/proteins of the regulators (TFs or miRNAs), the roles that regulators serve in the PPI network can be identified. Four types of motifs are defined in the paper for comparison purposes. These are single-regulation motifs, co-regulation motifs, crosstalk motifs and independent motifs. All these motifs contain both regulators and their target genes/proteins. As illustrated in Figure 3, for synergistic regulators r0 and r1, which have two common targets, and their targets t0 and t1, single regulation motif = {r0, t0} or {r1, t1} co-regulation motif = {r0, t0} ∩ {r1, t1}; crosstalk motif = {r0, t0} ∪ {r1, t1} − {r0, t0} ∩ {r1, t1}; if t0 and t1 do not have intersections, independent motif = {r0, t0} and {r1, t1}. PPI enrichment analysis and property analysis of the PPI network such as degree, closeness, density and so on are conducted to compare the significance of the four motifs in the PPI network. The results showed crosstalk motifs between miRNAs and TFs motifs, which demonstrated higher network properties such as higher degree, closeness, density etc., play a more important role in regulating the PPI network.
Figure 3.
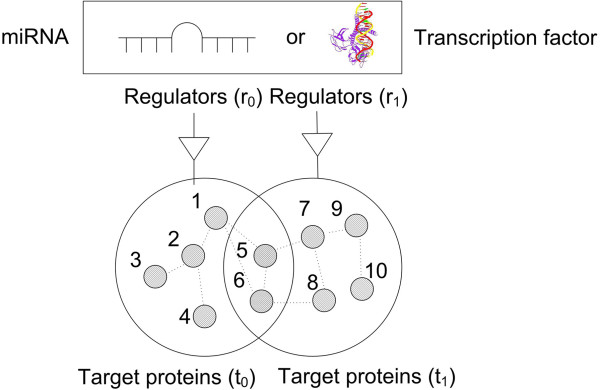
Cooperating regulation motifs of miRNA and transcription factors: four regulator motifs: single regulation motif = {r 0 , 1,2,3,4,5,6} or {r 1 , 5,6,7,8,9,10}; co-regulation motif = {r 0 , r 1 , 5,6} crosstalk motif = {r 0 , r 1 ,1,2,3,4,7,8,9,10}; If {5} or {6} does not exist, independent motif = {r 0 , 1,2,3,4,5,6} and {r 1 , 5,6,7,8,9,10}.
Identifying miRNA-regulated PPI networks in special diseases
Since the importance of miRNA-regulating PPI networks has been acknowledged, researchers have started to identify special miRNA-regulated networks in PPI networks related to disease. For example, Tseng [76] proposed an integrative method to identify miRNA-regulated protein networks specialized in gastric cancer. They combined miRNA targets and mRNA expression profiles and identified 16 miRNA-regulated protein networks in gastric cancer based on the human PPI network. Specially, they further investigated the network and functions of miR-148a which may resist tumor extension. Schmeier et al. [77] analysed miRNA-regulated PPI networks involving TFs in human ovarian cancer. They firstly generated miRNA-regulated networks based on 162 miRNAs and TFs related to ovarian cancer and human PPI data extracted from five databases, and then developed a node ranking algorithm in a weighted directed network to identify network hubs. Through the analysis of miRNAs and TFs linking to high ranking hubs, the authors determined some “key players” including miRNAs and TFs in ovarian cancer. Tacutu et al. [78,106] identified 35 genes related to aging related diseases (ARDs) from the miRNA-regulated PPI networks which they constructed from miRNA-regulated genes and the Common Gene Signature (in [78,107-109]; CGS denotes a PPI network based on the overlap between the PPI Human Longevity Network-http://www.netage-project.org[106] and human aging-related disease PPI networks).
Conclusion
Through analysing miRNA-regulated PPI networks based on miRNA features and PPI network features, it can be seen that: a) proteins in PPI networks are different in miRNA regulation. miRNAs have the propensity to regulate higher connected proteins. Additionally, the target proteins have shorter and higher modularity than other random proteins. Especially for hub proteins, miRNAs seem to play more important roles in higher cluster coefficient proteins than lower ones. For proteins in special tissues, miRNAs that target a lower number of genes have the propensity to regulate commonly expressed proteins rather than tissue-specific proteins. Additionally, protein complexes tend to be regulated by miRNAs; b) miRNAs may cooperate to regulate target proteins with others. The collaborator can be other miRNAs in a cluster or it can also be TFs. Interacting proteins have the tendency to be regulated by miRNAs in the same cluster. On the other hand, the crosstalk between miRNAs and TFs plays a more important role in regulating proteins than other types, such as single-regulation, co-regulation and independent regulation. Finally, through analysing miRNA-regulated PPI networks in cancer, several key miRNAs associated with cancers can be identified. These findings suggest that by combining miRNAs and PPI network features, it is possible to obtain useful information or rules on both miRNA regulation and protein interaction.
Research on miRNA-regulated PPI networks is continuing and still has a long way to go. In one aspect, there are still other ways to further investigate miRNA regulation roles in the PPI network. For example, it is reported [99] that there is low connectivity among hub proteins in PPI networks, so is this a possible explanation for miRNA target propensity? We also notice that current studies on PPI networks are mostly focused on static PPI networks, however, PPIs may be expressed as a dynamic state to maintain stability in the external environment, hence, it would be interesting to study the relationship between miRNA-regulated dynamic PPI networks or even at a later stage, on data hub proteins. On the other hand, studies on miRNA-regulated networks can also be extended into another three aspects (if applicable): a) the improvement of current miRNA target prediction approaches by integrating protein-protein interaction data. Liang and Li [72] found a convincing relationship between miRNA target sites and PPI networks and then mentioned that integrating PPI networks may result in the improvement of miRNA target prediction; b) the improvement of the detection of signalling pathways by integrating miRNA target data. miRNA regulating pathways related to cancer have been the subject of a great deal of recent research. Conversely, it is also possible to use this regulatory mechanism to infer signal pathways, and hence provide theoretical support to current miRNA-regulated signalling transduction pathway research; c) the improvement of protein function prediction approaches by integrating protein-protein interaction data and miRNA target data. Current protein function prediction algorithms mostly rely on designed mathematical algorithms and single resources (e.g. only using the PPI network database), which results in lower applicability. miRNAs play a fine-tuning role in gene expression, the integrating miRNA-regulated data can result in the current protein function prediction to be more biologically meaningful, and in turn, result in the improvement of accuracy.
Key points
•We divide current miRNA-regulated PPI networks into: 1) basic studies of miRNA-regulated PPI networks; and 2) identification of miRNA-regulated networks in diseases which covers most parts of current miRNA-regulated PPI network studies.
•We provide relatively complete resources to construct a miRNA-target protein network.
•We introduce the features of miRNA-regulated PPI networks and based on this, we provide a brief explanation on the methodologies of several studies on miRNA-regulated PPI networks.
•We review previous findings and propose several potential computational research directions which may be further explored in the future.
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
WZ conceived and designed the study, YPP surprised the work. WZ and YPP wrote reviewed and edited the paper. All authors read and approved the final manuscript.
Authors’ information
Wei Zhu is a Ph.D candidate in the Department of Computer Science and Computer Engineering at La Trobe University. The focus of his thesis is knowledge discovery in protein-protein interactions.
Yi-Ping Phoebe Chen is the Professor and Chair & Director of Research in the Department of Computer Science and Computer Engineering, La Trobe University, Melbourne, Australia. She is currently working on knowledge discovery technologies and is especially interested in their application to genomics and biomedical science. Her research focus is to find best solutions for mining, integrating and analysing complex data structures and functions for scientific and biomedical applications.
Contributor Information
Wei Zhu, Email: w6zhu@students.latrobe.edu.au.
Yi-Ping Phoebe Chen, Email: phoebe.chen@latrobe.edu.au.
References
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome research. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MJ. New human and mouse microRNA genes found by homology search. FEBS Journal. 2005;272(1):59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. The EMBO journal. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Chen YP. Exploring cross-species-related miRNAs based on sequence and secondary structure. IEEE transactions on bio-medical engineering. 2010;57(7):1547–1553. doi: 10.1109/TBME.2010.2043734. [DOI] [PubMed] [Google Scholar]
- An J, Choi KP, Wells CA, Chen YP. Identifying co-regulating microRNA groups. Journal of bioinformatics and computational biology. 2010;8(1):99–115. doi: 10.1142/S0219720010004574. [DOI] [PubMed] [Google Scholar]
- Lin CC, Chen YJ, Chen CY, Oyang YJ, Juan HF, Huang HC. Crosstalk between transcription factors and microRNAs in human protein interaction network. BMC systems biology. 2012;6:18. doi: 10.1186/1752-0509-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. Journal of pharmacological sciences. 2006;101(4):267–270. doi: 10.1254/jphs.CPJ06013X. [DOI] [PubMed] [Google Scholar]
- Tzur G, Israel A, Levy A, Benjamin H, Meiri E, Shufaro Y, Meir K, Khvalevsky E, Spector Y, Rojansky N. et al. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PloS one. 2009;4(10):e7511. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziere P, Enright AJ. Prediction of microRNA targets. Drug discovery today. 2007;12(11–12):452–458. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer research. 2008;68(14):5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Guo L, Lu ZH. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5(6):e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics, proteomics & bioinformatics. 2009;7(4):147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40(20):10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10(431) doi: 10.1186/1471-2164-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Molecular Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Coller HA. The code within the code MicroRNAs target coding regions. Cell Cycle. 2010;9(8):1533–1541. doi: 10.4161/cc.9.8.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS biology. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761 Suppl):C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Zhang SH, Jin GX, Zhang XS, Chen LN. Discovering functions and revealing mechanisms at molecular level from biological networks. Proteomics. 2007;7(16):2856–2869. doi: 10.1002/pmic.200700095. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nature biotechnology. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Fields S. High-throughput two-hybrid analysis: the promise and the peril. Febs J. 2005;272(21):5391–5399. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- Zhu W, Hou J, Chen YP. Exploiting multi-layered information to iteratively predict protein functions. Mathematical biosciences. 2012;236(2):108–116. doi: 10.1016/j.mbs.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-β/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50(2):570–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- Mei J, Bachoo R, Zhang CL. MicroRNA-146a inhibits glioma development by targeting Notch1. Mol Cell Biol. 2011;31(17):3584–3592. doi: 10.1128/MCB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, Liu C, Zong H, Rowitch DH, Barres BA. et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31(15):1884–1895. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic acids research. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AD, Przulj N, Jurisica I. Protein complex prediction via cost-based clustering. Bioinformatics. 2004;20(17):3013–3020. doi: 10.1093/bioinformatics/bth351. [DOI] [PubMed] [Google Scholar]
- Cho YR, Hwang W, Ramanathan M, Zhang A. Semantic integration to identify overlapping functional modules in protein interaction networks. BMC bioinformatics. 2007;8:265. doi: 10.1186/1471-2105-8-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Wong L, Chua HN. Complex discovery from weighted PPI networks. Bioinformatics. 2009;25(15):1891–1897. doi: 10.1093/bioinformatics/btp311. [DOI] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Molecular systems biology. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Yang L, Du Z. MicroRNA regulation and tissue-specific protein interaction network. PloS one. 2011;6(9):e25394. doi: 10.1371/journal.pone.0025394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP. et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430(6995):88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30(1):303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3(10):e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen L, Kolehmainen M, Wong G. Prediction of microRNA targets in caenorhabditis elegans using a self-organizing map. Bioinformatics. pp. 1247–1254. [DOI] [PubMed]
- Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stumpflen V, Mewes HW. et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21(6):832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6(1):75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- Patil A, Nakai K, Nakamura H. HitPredict: a database of quality assessed protein-protein interactions in nine species. Nucleic Acids Res. 2011;39(Database issue):D744–749. doi: 10.1093/nar/gkq897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Kang S, Kim BC, Oh J, Cho S, Bhak J, Choi JS. SynechoNET: integrated protein-protein interaction database of a model cyanobacterium Synechocystis sp. PCC 6803. BMC Bioinformatics. 2008;9(1):S20. doi: 10.1186/1471-2105-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yu J, Li D, Liu F, Zhou X, Wang T, Ling Y, Su Z. PMRD: plant microRNA database. Nucleic Acids Res. 2010;38(Database issue):D806–813. doi: 10.1093/nar/gkp818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology: the gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM. et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic acids research. 2011;39(Database issue):D163–169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M. et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW. GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Research. 2009;37:W323–W328. doi: 10.1093/nar/gkp313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Li M, Choi KM, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Research. 2009;37:W356–W362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang YP, Li KB. MiRTif: a support vector machine-based microRNA target interaction filter. BMC Bioinformatics. 2008;9(12):S4. doi: 10.1186/1471-2105-9-S12-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010;39(Database issue):D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40(Database issue):D222–229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nature Biotechnology. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, Wu J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38(Web Server issue):W392–397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202–209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li WH. MicroRNA regulation of human protein protein interaction network. RNA. 2007;13(9):1402–1408. doi: 10.1261/rna.634607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CW, Juan HF, Huang HC. Characterization of microRNA-regulated protein-protein interaction network. Proteomics. 2008;8(10):1975–1979. doi: 10.1002/pmic.200701004. [DOI] [PubMed] [Google Scholar]
- Yuan X, Liu C, Yang P, He S, Liao Q, Kang S, Zhao Y. Clustered microRNAs’ coordination in regulating protein-protein interaction network. BMC systems biology. 2009;3:65. doi: 10.1186/1752-0509-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass S, Dietmann S, Burk UC, Brabletz S, Lutter D, Kowarsch A, Mayer KF, Brabletz T, Ruepp A, Theis FJ. et al. MicroRNAs coordinately regulate protein complexes. BMC systems biology. 2011;5:136. doi: 10.1186/1752-0509-5-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CW, Lin CC, Chen CN, Huang HC, Juan HF. Integrative network analysis reveals active microRNAs and their functions in gastric cancer. BMC systems biology. 2011;5:99. doi: 10.1186/1752-0509-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeier S, Schaefer U, Essack M, Bajic VB. Network analysis of microRNAs and their regulation in human ovarian cancer. BMC systems biology. 2011;5:183. doi: 10.1186/1752-0509-5-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Budovsky A, Wolfson M, Fraifeld VE. MicroRNA-regulated protein-protein interaction networks: how could they help in searching for pro-longevity targets? Rejuvenation research. 2010;13(2–3):373–377. doi: 10.1089/rej.2009.0980. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G. et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5(7):613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera BM, Lockstone HE, Taylor JM, Wills QF, Kaisaki PJ, Barrett A, Camps C, Fernandez C, Ragoussis J, Gauguier D. et al. MicroRNA-125a is over-expressed in insulin target tissues in a spontaneous rat model of Type 2 Diabetes. Bmc Medical Genomics. 2009;2 doi: 10.1186/1755-8794-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA. et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Zhou X. Computational methods for the identification of microRNA targets. Open Access Bioinformatics. 2010;2:29–39. doi: 10.2147/OAB.S6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Computational identification of microRNAs and their targets. Computational Biology and Chemistry. 2006;30(6):395–407. doi: 10.1016/j.compbiolchem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Min H, Yoon S. Got target?: computational methods for microRNA target prediction and their extension. Experimental and Molecular Medicine. 2010;42(4):233–244. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri K, Chatterjee R. MicroRNA detection and target prediction: integration of computational and experimental approaches. DNA and Cell Biology. 2007;26(5):321–337. doi: 10.1089/dna.2006.0549. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. Computational analysis of miRNA targets in plants: current status and challenges. Brief Bioinform. 2010;12(2):115–121. doi: 10.1093/bib/bbq065. [DOI] [PubMed] [Google Scholar]
- Wang X. Computational prediction of microRNA targets. Methods Mol Biol. 2010;667:283–295. doi: 10.1007/978-1-60761-811-9_19. [DOI] [PubMed] [Google Scholar]
- Hammell M. Computational methods to identify miRNA targets. Seminars in cell & developmental biology. 2010;21(7):738–744. doi: 10.1016/j.semcdb.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews Drug discovery. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA. et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci USA. 2008;105(14):5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Marin RM, Vanicek J. Optimal use of conservation and accessibility filters in MicroRNA target prediction. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0032208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Nakamura H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC bioinformatics. 2005;6:100. doi: 10.1186/1471-2105-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Nakamura H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics. 2005;6:6. doi: 10.1186/1471-2105-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296(5569):910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Gursoy A, Keskin O, Nussinov R. Topological properties of protein interaction networks from a structural perspective. Biochemical Society transactions. 2008;36(Pt 6):1398–1403. doi: 10.1042/BST0361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Molecular systems biology. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Yu Z, Pan Y, Purisima EO, Wang E. MicroRNAs preferentially target the genes with high transcriptional regulation complexity. Biochemical and biophysical research communications. 2007;352(3):733–738. doi: 10.1016/j.bbrc.2006.11.080. [DOI] [PubMed] [Google Scholar]
- Tacutu R, Budovsky A, Fraifeld VE. The NetAge database: a compendium of networks for longevity, age-related diseases and associated processes. Biogerontology. 2010;11(4):513–522. doi: 10.1007/s10522-010-9265-8. [DOI] [PubMed] [Google Scholar]
- Chen YPP, Chen F. Using bioinformatics techniques for gene identification in drug discovery and development. Current Drug Metabolism. 2008;9(6):567–573. doi: 10.2174/138920008784892056. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen YPP. Mining frequent patterns for AMP-activated protein kinase regulation on skeletal muscle. BMC Bioinformatics. 2006;7:394. doi: 10.1186/1471-2105-7-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Wang M, Chen YPP, Wang H, Han P, Sun H. Prediction of pre-miRNA with multiple stem-loops using pruning algorithm. Computers in Biology and Medicine. 2013;43(5):409–416. doi: 10.1016/j.compbiomed.2013.02.003. [DOI] [PubMed] [Google Scholar]