Abstract
In the post-genomic era, the medical/biological fields are advancing faster than ever. However, before the power of full-genome sequencing can be fully realized, the connection between amino acid sequence and protein structure, known as the protein folding problem, needs to be elucidated. The protein folding problem remains elusive, with significant difficulties still arising when modeling amino acid sequences lacking an identifiable template. Understanding protein folding will allow for unforeseen advances in protein design, often referred as the inverse protein folding problem. Despite challenges in protein folding, de novo protein design has recently demonstrated significant success via computational techniques. We review advances and challenges in protein structure prediction and de novo protein design, and highlight their interplay in successful biotechnological applications.
Keywords: Protein structure prediction, de novo protein design, computational biology, biotechnology
Protein folding and design are two sides of the same coin
Proteins are polymeric chains of amino acids that organisms and cells rely on for signaling, pathogen clearing, mobility, catalysis, recognition, shape, ordering, and stability. The precise ordering of the amino acids in a protein’s sequence determines how the protein folds into a 3-dimensional structure, and thus its biological function. As our knowledge of the connection between sequence, structure, and function has advanced, interest has grown in designing proteins on a sequence level to produce novel folds and function. Brute-force experimental approaches to solving protein structures and designing protein sequences for new functions remain time consuming and expensive, and add little to our understanding of the physical principles required for both problems [1].
Protein structure prediction aims to accurately determine the full 3-dimensional structure of a protein given only its amino acid sequence. Structure prediction is very challenging if only low homology templates exist. De novo protein design is the inverse problem [2, 3]; given a rigid or flexible backbone structure, one aims to determine a sequence that will fold into that structure. Different sequences can fold into the same structure, so there is degeneracy in protein design space. The existence and accuracy of protein structures as templates for protein design can significantly impact potential success. For this reason, the ability to produce viable protein templates through protein structure prediction is important for protein design, and for advancement in biotechnology and drug discovery. In this review, we describe advances and challenges in the fields of protein structure prediction and de novo protein design focusing on the interplay necessary for success.
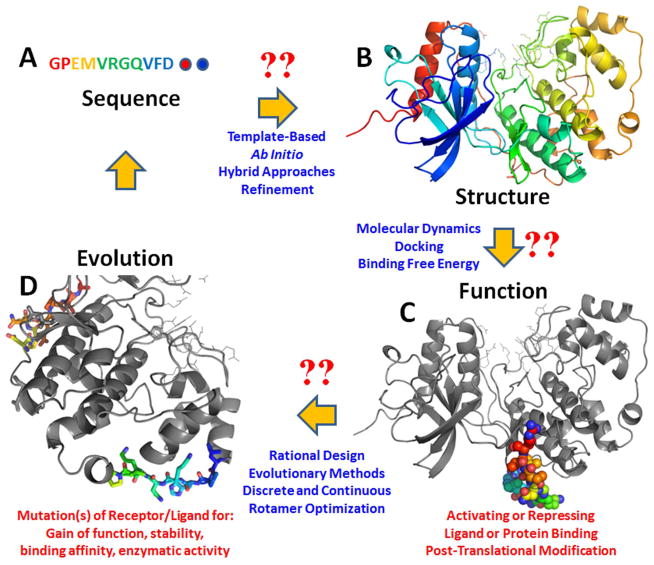
Figure 1 schematically shows the roadmap and key challenges in protein structure prediction and de novo protein design. The last few years have shown impressive applications of computational structure prediction and design to biotechnology, spanning peptide or antibody therapeutics, novel biocatalysts, and self-assembling nanomaterials.
Figure 1.
Roadmap of key challenges in understanding how to predict protein sequence to structure to function and design. Structure prediction begins with a primary amino acid sequence (A) and aims to predict the full 3-dimensional structure (B) of that sequence. (C) Other proteins, peptides, small molecules, or cofactors may form critical interactions with the protein structure critical to its function. Docking with or without binding free energy calculations may be required to find the most probable conformation for a ligand bound to a receptor protein. Understanding how structure leads to function remains a challenge. The protein structure may be subsequently post-translationally modified, and as most methods have focused in predicting the structures of canonical amino acid containing proteins, the literature is lacking in the ability accurately represent post-translationally modified protein structures. The solution or accurate prediction of a protein’s 3-dimensional structure allows it to be used in a design context. (D) Biotechnological applications of protein design shown in the literature include designing/redesigning the receptor protein via site-specific mutations to change its binding affinity toward a ligand, change its fold, increase its stability, and create new or alternative enzymatic activity. The ligand of a peptide can be amenable to similar design strategies to design new sequences to bind more strongly to the receptor and compete with its native binding partner (antagonism) or to bind to and activate through a series of specific interactions with the receptor a particular downstream function (agonism). Upon design of the receptor or ligand peptide with new sequences, the cycle begins again as even a few mutations can cause structural conformation and topology changes. The structure shown in the figure is the mitogen activated kinase ERK2. The ligand bound is the kinase interaction motif of phosphatase MKP3.
State-of-the-art advances and challenges in protein structure prediction and refinement
The consistent determination of structure from sequence is one of nature’s greatest unsolved problems and has recently passed the 50 year milestone [4]. Accurately predicting the three dimensional structure of a protein involves a series of steps performed on a sequence of amino acids: secondary structure prediction (identifying local interactions between amino acid residues), structural alignment to candidate template structures, conformational sampling, and selection (Figure 2A and Box 1). A predicted structure may then undergo refinement, in an attempt to improve the accuracy of that structure [5]. Historically, most refinement methods degrade rather than improve the accuracy of the predicted structure, making protein structure refinement a substantial unsolved problem in its own right [5, 6]. We review recent progress and challenges and refer you to the reviews by Zhang [7] and Floudas [8] for prior advances.
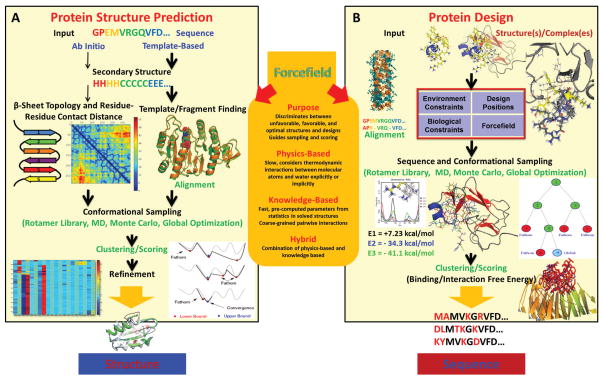
Figure 2.
Detailed view of connections and differences between (A) protein structure prediction and (B) protein design. Dynaneomics image used with permission.
BOX 1. Protein Structure Prediction and De Novo Protein Design are Related Problems.
Protein structure prediction (Figure 2A) begins with a sequence and produces a structure. Two paths are often followed: ab initio and template-based. Ab initio methods attempt to predict the structure from first principles without a template. Some methods utilize secondary structure and contact predictions as constraints. The most expensive step is the conformational sampling in the presence or absence of constraints. Template-based methods begin with a sequence, predict the secondary structure, and attempt to find a template structure and/or fragments from existing structures in the PDB that will fold into the target sequence. These methods rely on the ability to identify suitable templates and then align the target sequence properly to the template sequence. Both methods use advanced sampling techniques such as MD, rotamer optimization, Monte Carlo, and global optimization. After sampling, both methods may cluster or rescore the structures, and may subject them to a refinement stage to increase prediction accuracy. For sequences of ~30% identity or more to a template, one can expect that the predicted structure is a reasonable estimate of the topology. Below 30%, accurate prediction is more challenging.
Protein design (Figure 2B) begins with a structure or complex and produces new sequences. Design positions are chosen to be mutated. Next, the sequence may be aligned to other homologous sequences to produce biological constraints on the sequence space. The solvent accessible surface area (SASA) of each residue being designed can be taken into consideration to further constrain the design space. Sequence design is then performed and can be done using a single state or multiple states. In this step the structure being designed can remain fixed, with only side-chain rotamers changing, or may be completely flexible. The algorithms for sampling come from the same classes of techniques used in protein folding. Designed sequences may then be clustered and evaluated with a more detailed scoring function. Design produces one or many sequences that are predicted to fold into the input structure, often with enhanced biophysical characteristics.
Forcefields are the glue connecting structure prediction and design. They describe the interactions between atoms in a system, guide sequence and structural search, and discriminate between optimal and suboptimal solutions. An improved description of atomistic interactions in forcefields benefits both areas. Improving our ability to predict structures will improve our ability to model complexes of druggable targets and design new sequences.
The Critical Assessment of Techniques for Protein Structure Prediction (CASP) [9] occurs biennially and recently completed its 10th experiment. For the CASP competition, prediction targets are categorized into two groups depending on the availability of structural templates; (1) template-based modeling, in cases for which templates are available and (2) free modeling, in cases for which templates are not available. Table 1 shows the top 5 structure prediction servers in the template-based modeling (TBM) and free modeling (FM) categories (www.predictioncenter.org/casp10/). The average of the top 5 server methods in the free modeling category represents ~1/2 the accuracy of the models produced in template-based modeling. These servers may not apply the same prediction protocol for all targets and instead may perform different pipelines based on the predicted difficulty of the target [10]. The Zhang-Server was assessed to be the best overall server in CASP10 in both TBM and FM. Additionally, Table 1 lists the top 5 protein structure refinement methods assessed in CASP10. The methods listed in Table 1 represent the current state-of-the-art in protein structure prediction and protein structure refinement.
Table 1.
Top performing protein structure prediction servers in Template-Based Modeling and Free Modeling categories and methods in the refinement category independently assessed in CASP10.
| Template-Based Modeling1,3 | Free Modeling2,3 | Refinement4 | ||||||
|---|---|---|---|---|---|---|---|---|
| Rank | Server | Average GDT_TS | URL | Server | Average GDT_TS | URL | Group (Method) | URL |
| 1 | Zhang-Server | 53.90 | http://zhanglab.ccmb.med.umich.edu/I-TASSER/ | Zhang-Server | 26.78 | http://zhanglab.ccmb.med.umich.edu/I-TASSER/ | FEIG (MD) | This method is not available as a server. |
| 2 | Protein Modeling System (PMS) | 48.78 | Server to be released. | BAKER-ROSETTA SERVER | 24.17 | http://robetta.bakerlab.org/ | Seok (GalaxyRefine) | http://galaxy.seoklab.org/refine/ |
| 3 | HHpred-thread | 49.33 | http://toolkit.tuebingen.mpg.de/hhpred | Protein Modeling System (PMS) | 23.66 | Server to be released. | Mufold | http://mufold.org/ |
| 4 | RaptorX-YZ | 50.78 | http://raptorx.uchicago.edu/ | TASSER-VMT | 23.77 | http://cssb.biology.gatech.edu/skolnick/webservice/TASSER-VMT/index.html | KnowMIN (KobaMIN) | http://csb.stanford.edu/kobamin/ |
| 5 | BAKER-ROSETTA SERVER | 47.78 | http://robetta.bakerlab.org/ | Pcons-net (Metaserver) | 24.55 | http://pcons.net | FLOUDAS (Princeton_TIGRESS) | http://atlas.princeton.edu/refinement/ |
TBM Rankings were taken from the slides of the TBM assessor from the CASP10 website (www.predictioncenter.org/casp10/).
FM rankings were taken based on “Best Model” free modeling rankings based on SUM Z-score.
If multiple servers from the same lab are represented in the top 5, the highest ranking server is chosen.
Refinement rankings are taken from the CASP10 website and consider both the top human and server groups (http://predictioncenter.org/casp10/doc/presentations/ranking_CASP10_refinement_DJ.pdf).
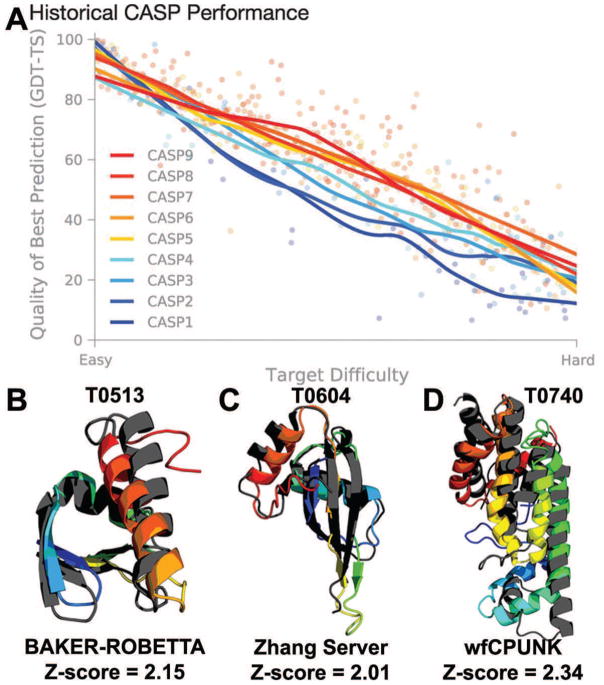
Kryshtafovych et al. constructed a difficulty scale [4] based on the similarity of the sequence and structure of the target to that of the closest template available in the Protein Data Bank (PDB) during CASPs 1–9 [11]. “Easy” targets typically corresponded to those which there are directly identifiable templates through sequence homology. “Hard” targets may have excellent structural templates in the PDB, but their sequences are often so dissimilar to the target protein that it is nearly impossible to identify them. Additionally, “Hard” targets may have no template at all and may represent a new fold. Figure 3 highlights CASP performance for “Easy” and “Hard” targets over the last 18 years, and several top free modeling predictions in the last 3 CASPs. Despite the progress attained for easy targets with identifiable templates, predictors face challenges accurately predicting structures for sequences with difficult to identify templates [12].
Figure 3.
(A) Historical performance of best prediction’s GDT_TS vs. target difficulty over the last 18 years in CASPs 1–9. Target difficulty accounts for both sequence and structural similarity of the target to known template structures available at the time of prediction [12]. Evidently, the quality of the best “Easy” target predictions has been consistently accurate. Conversely, “Hard” targets, most lacking identifiable templates, have not advanced significantly in this time period and still remain the biggest challenge (Adapted from [4] with permission from AAAS, and original data from [12], with permission from John Wiley and Sons). High-ranking blind free modeling predictions submitted to CASP8, 9, and 10 for targets (B) T0513 by BAKER-ROBETTA, (C) T0604 by Zhang Server, and (D) T0740 by wfCPUNK. The native structure is shown in dark grey and the prediction as a rainbow, with the N-terminus in blue and the C-term in red. GDT_TS Z-scores are reported for Model 1 for T0513 and T0604 and for All Models for T0740, with larger values indicating a larger separation from the rest of the predictions. The wfCPUNK prediction resulted from a collaboration between the Floudas, Liwo, and Scheraga labs as part of the collaborative folding experiment WeFold (http://www.wefold.org). The predictions shown in (B) and (C) used template information, whereas the prediction in (D) was strictly ab initio. The targets shown were among the most difficult in the respective CASP experiments.
Template-Based Modeling
TBM has served as a reliable prediction method given an appropriate template structure. This approach utilizes the input sequence and attempts to identify a structure(s) whose sequence(s) can be aligned with the target sequence to infer information about secondary structure and tertiary structure (including topology and residue-residue contacts).
The top performing servers in the TBM category in CASP10 are exhibited in Table 1 and described below. Zhang-Server utilizes a combination of LOMETS[13], I-TASSER [14–16], and QUARK [17]. I-TASSER identifies templates via LOMETS, performs fragment-assembly via replica-exchange Monte Carlo simulations, and refinement using REMO [18] and fragment-guided MD (FG-MD)[19]. Protein Modeling System (PMS) uses conformational space annealing (CSA) with Lorentzian energetic restraints in MODELLER, combining physical and knowledge-based energy terms [20]. HHpred-thread is very fast and accurate, and includes improvements with three statistical scores to compare the target’s sequence profile with template structure and sequence profiles [21]. RaptorX-YZ is an enhancement to RaptorX [22] using machine learning to predict contacts between residues for use as restraints. BAKER-ROSETTASERVER aligns the candidate sequence to multiple templates, assembles fragments using coarse-grained insertion, utilizes Monte Carlo search for both coarse-grained and all-atom sampling of favorable backbone and rotameric states, and energy minimization in both torsion and Cartesian space. Top-scoring models are relaxed according to the Rosetta all-atom forcefield [23]. Commonalities in these top-performing methods are that both PMS and RaptorX utilize MODELLER in model building, and both BAKER-ROSETTASERVER and Zhang-Server utilize Monte Carlo fragment assembly to aid in sampling.
TASSER-VMT was introduced by Zhou and Skolnick [24], and uses the improved SP3 alternative target-template alignment combined with other alignment methods as input to TASSER simulations. They introduced GOAP, a statistical potential with orientation-dependent correction terms for evaluating model quality, recognizing 226 native structures of 278 targets stemming from 11 commonly-used decoy sets [25].
Free Modeling
Free modeling is the prediction of structures for sequences that have no distinguishable template in the PDB. These predictions are considered to be “Hard” and success on this front remains limited (Figure 3A) and represents the “holy grail” of protein folding. In discussing the challenges in free modeling, it is important to point out the difference between “indistinguishable” and “non-existent” templates. The PDB contains over 92,000 solved structures that offer a wide-variety of templates and often several candidate templates for a target sequence. Zhang and Skolnick showed for a set of non-homologous proteins that they can always find similar folds to the native with an average RMSD of 2.5 Å [26].
The ability to predict such difficult targets relies on the ability to select the proper template from structures contained in the PDB and this still remains very challenging evidenced by the low average GDT_TS of even the top predictors in CASP10 (see Table 1) and overall in CASPs 1–9 (see Figure 3A) [12]. Interestingly, none of the best free modeling methods used strictly ab initio methods; all utilize template information. Also, Zhang-Server, using an interplay of I-TASSER (which uses templates) and QUARK [17] (which is denoted as first principlies) outperformed QUARK alone.
Molecular Dynamics Driven Folding
Duan and Kollman folded Villin headpiece starting from an unfolded state using molecular dynamics without the simulation having knowledge of the native contacts [27]. Since that seminal result, a number of studies have reported the ability to simulate the folding of small proteins.
Scheraga and coworkers, using their developed UNRES coarse-grained molecular dynamics package, recently summarized notable first principles predictions made during CASP10 [28]. They were able to predict the correct packing symmetry for a target with a new fold. Recent advances in implementations, extensions, and applications of UNRES are reviewed by Liwo et al. [29]. Shaw and coworkers used equilibrium MD simulations to study the general folding landscape of 12 fast-folding small proteins [30]. In 8 out of the 12 studied, a structure within 2 Å of the native was observed. Shaw and coworkers were able to fold Ubiquitin [31], a 76-residue long protein contained in most eukaryotic organisms having a folding time on the millisecond timescale. These successes have required adjustments in force fields (CHARMM22* [32]), total simulation time on the order of milliseconds, explicit treatment of solvent, and specialized hardware (Anton [33]).
Conformational sampling has been suggested as a major limitation to predicting high-resolution structures [34], while it has been recently claimed that sampling is not the main issue, but instead it is forcefield inaccuracy that needs improvement [35]. At this point, there is limited evidence for the recent claim, and as the conformational search in even the simplest biophysical model is NP-complete [36], we view that both conformational sampling and forcefield development are key limitations.
Contact and β-Sheet Topology Prediction
Jones and coworkers introduced the contact prediction method PSICOV, which utilizes sparse inverse covariance estimation to predict contacts yielding a long-range L/5 contact precision ≥ 0.5 [37]. The approach has the limitation that it requires at least 500 sequences in the multiple sequence alignments for convergence. Marks and Sander introduced EVFold, which predicts contacts based on maximum entropy and co-evolutionary couplings for contact predictions, but a similar challenge is faced in that 1000 sequences are required in the multiple sequence alignments to produce accurate contacts [38].
Success in contact prediction can substantially influence conformational search. Optimization-driven methods based on first-principles were developed for the prediction of inter-helical contacts in α-helical proteins [39] and both α/β and α+β proteins [40]. After input to the global-optimization framework ASTRO-FOLD [41, 42], the contacts reduced the RMSD range of the sampled conformers by one-half [40]. Subramani and Floudas introduced BeST, for the prediction of β-sheet topologies with high precision and recall [43]. Baker and coworkers demonstrated in CASP10 with Rosetta-based methods [23] that given correct contacts for approximately one in twelve residues, this enabled the search for and construction of the correct topology [44], implying that if one can predict contacts with high positive predictive value, one can construct accurate topologies.
Successful Protein Designs with Biotechnological Applications
Protein design is the inverse folding problem [2, 3] (Figure 2B, Box 1). Given a target fold, can we design a sequence to fold into that structure? Several notable examples are highlighted in Table 2. We present an overview of recent computational protein designs with biotechnological applications and describe the interplay with structural modeling necessary for success of the designs as appropriate. We refer the reader to [45, 46] for excellent recent reviews of methodological advances and applications in de novo protein design.
Table 2.
Summary of recent successful computational de novo designed and redesigned systems and their biotechnological applications. Full details are provided in the corresponding references.
| Biotechnological Application | Summary | Structure Modeling | Forcefield(s) | De Novo Design Method | Discriminating Prediction Metric(s) | Ref |
|---|---|---|---|---|---|---|
| Design of disease therapeutics | Computationally redesigned peptide sequence to bind to breast cancer biomarker | Cyclic peptides docked to NMR-derived conformers of target biomarker CRIP1 | Physics | Eris | Change in binding free energy ΔΔG | [50] |
| Bioactive peptide cytotoxic to tumor cells | Design based on primary structure alone | Physics | Resonant Recognition Model (RRM) | Electron-ion interaction potential | [51] | |
| Grafting 4E10 HIV epitope to new protein scaffolds | Matching 4E10 to putative protein scaffolds and docking to mAb 4E10 | Hybrid | RosettaDesign | Rosetta energy | [52] | |
| HIV-1 Entry Inhibitors targeting gp41 | Designed peptides and docked poses using TINKER and Rosetta Ab Initio | Physics, Knowledge-based and Hybrid | Components and subcomponents of the Protein WISDOM protocol | Distance-dependent energy followed by Fold Specificity and Approximate Binding Affinity (K*) | [53] | |
| Non-natural amino acid inhibitors of amyloid fibril formation by capping fibril ends | D-amino acid containing sequence designed with Rosetta | Hybrid | RosettaDesign | Shape complementarity, total binding energy between inhibitor and scaffold, solubility | [56] | |
| Engineered peptides that stabilize amyloid-β oligomers | Molecular Dynamics generated oligomer structure | Physics | Rational | Contact map and secondary structure analysis of MD | [57] | |
| Engineered crosslinking antibody with non-canonical amino acid | Non-canonical amino acid mutations | Hybrid | Rational | Rosetta interface score | [58] | |
| Supercharged thermally resistant antibodies | Homology model of anti-MS2 antibody and supercharged surface mutations | Hybrid | RosettaDesign | Rosetta energy | [59] | |
| Design of antibodies targeting a peptide from hepatitis C, fluorescein, and VEGF | Combinations of Backbones of CDR Structures | Physics | OptCDR and IPRO | Mixed-Integer Linear Optimization formulation to select structures followed by interaction energy between designs | [60] | |
| Inhibitors of hemagglutinin from the 1918 H1N1 pandemic virus that bind to and inhibit multiple other subtypes | Helical protein binders using Rosetta with hot-spot residue identification and shape-complementarity identification | Hybrid | RosettaDesign | Shape complementarity, electrostatic complementarity, RosettaDock, Rosetta energy | [80] | |
| Complement C3aR receptor agonists and antagonists | C3a-derived structures using TINKER | Physics, Knowledge-based | Components and subcomponents of the Protein WISDOM protocol | Distance-dependent energy followed by Fold Specificity | [81] | |
| Complement system inhibitors of the Compstatin-family targeting C3 | Peptide inhibitors of C3 | Physics, Knowledge, and Hybrid | Components and subcomponents of the Protein WISDOM protocol | Distance-dependent energy followed by Fold Specificity and K* | [82, 83] | |
| Design of self-assembling proteins/peptides | Computational design of a protein crystal | Mathematically created idealized homotrimeric coiled-coiled protein | Physics | Site-specific amino acid probabilities calculated using a statistical thermodynamic method | AMBER energy function | [72] |
| Design of a symmetric β-strand mediated homodimer | Computationally identified protein scaffold with symmetric protein-protein docking with Rosetta | Hybrid | Symmetric design with side-chain/backbone minimization using Rosetta | Rosetta-based energy metrics with visual inspection | [73] | |
| Self-assembling 24- and 12-subunit nanomaterials with different symmetries | Quaternary structures and interfaces of designed subunits through Rosetta with symmetric docking | Hybrid | RosettaDesign | Interface shape complementarity and energy | [74] | |
| Design of novel enzymes | Design of enzymes for Retro-Aldol, Kemp elimination, Diels-Alder, and organophosphate hydrolysis reactions | Idealized catalytic sites for transition state stabilization hashed onto template protein structures using RosettaMatch | Hybrid | RosettaDesign | Catalytic geometry, computed transition-state-binding energy, and consistency in side-chain conformations | [64–66, 68] |
| Increased Diels-Alderase activity in de novo designed enzyme | Active-sites reshaped by human players using Foldit | Hybrid | Interactive puzzles that use human intuition and pattern recognition in the online multi-player game Foldit | High-scoring (Low-Energy) sequences | [67] | |
| Redesigned cofactor binding site of CbXR to switch enzymatic cofactor specificity from NADPH to NADH | Homology model of Candida boidinii xylose reductase | Physics | Iterative Protein Redesign and Optimization (IPRO) | Interaction Energy between designed sequences of CbXR and cofactors | [69] | |
| Redesign of GrsA-PheA specificity to non-native amino acid substrates | Penultimate Rotamer Library for rotameric states | Physics | K* | Statistical mechanics-based metric using rotameric ensembles to approximate the binding constant Kd | [70] |
Design of Proteins and Peptides for Therapeutic Applications
Over 200 peptide, protein, or antibody therapeutics have been marketed as of 2010 [47]. Computational approaches have recently been applied to design new proteins and peptides for therapeutic applications. Elucidation of the sequences, structures, and interaction patterns of several disease-related proteins have allowed for the application of computational approaches for peptide therapeutic design.[48]. Craik et al. [49] predict that by 2020 we will see more prevalence of peptides as drugs, while outlining the challenges to meeting that outcome. Here we review timely applications by target.
Cancer
Generally, therapeutic proteins/peptides can (1) interfere with signal transduction cascades, (2) arrest the cell cycle through modulation of cyclin-dependent kinase activity, or (3) directly induce apoptosis by modulation of the proteins controlling apoptosis [48]. CRIP1 is an early biomarker for breast cancer. Hao et al. used phage display to identify peptide sequences that bound to CRIP1. Subsequently they computationally redesigned the scaffold sequence to optimize the binding free energy to increase its affinity for CRIP1, finding experimentally that it improved the IC50 27.5x over the phage-displayed sequence [50]. Cosic and coworkers used the Resonant Recognition Model (RRM) to design a short therapeutic peptide with myxoma virus antitumor/cytotoxic activity [51]. RRM represents a protein sequence as a series of numbers which can be analyzed by Fourier transformation and converted into a discrete spectrum, where a significant correlation to biological activity has been identified [51].
Human Immunodeficiency Virus
Correia et al. developed a computational method using side-chain grafting and Rosetta to transplant a continuous structural epitope, 4E10, into scaffold proteins for conformational stabilization and immune presentation [52]. The method produced epitope-containing designs that bind stronger to monoclonal antibody (mAb) 4E10 than 4E10 alone, and was found to inhibit neutralization by HIV+ sera. Floudas and coworkers designed HIV-1 entry inhibitors starting from the structure of the C14linkmid peptide in complex with the hydrophobic core of gp41 [53]. C14linkmid is a cross-linked peptide derived from the C-terminal heptad repeat gp41. A global optimization based sequence selection was performed with a distance-dependent forcefield originally developed for protein folding [54] to select candidate sequences from the vast combinatorial space. These sequences were re-ranked using fold specificity calculations, which sample conformations near the template structure with substitutions dictated by the the newly designed sequences. It aims to determine how favorably a new sequence folds into the fold of the design template. A subset of top-ranked sequences identified in the fold-specificity stage was evaluated using approximate binding affinity calculations, which approximate the binding equilibrium constant. The best design had an IC50 between 29–253 μM for different HIV-1 donors and mutants. This de novo design approach was made into an interactive web interface, Protein WISDOM [55].
Alzheimer’s
Eisenberg and colleagues performed computationally-guided design to predict and experimentally validate peptide inhibitors of fibril formation by the tau protein associated with Alzheimer’s, as well as an amyloid promoting the sexual transmission of HIV [56]. The designs bind to the end of the steric-zipper and inhibit elongation. Focusing on the tau protein inhibitor methodology, for a rotameric, fixed-backbone sequence optimization, they inverted the chirality of the design target to enable use of the Rosetta suite of tools. They designed L-amino acid sequences that favorably interact with a fixed-atom D- version of the scaffold. Subsequently, the scaffold was reverted to its native L- form, and D-amino acid containing peptides were used as inhibitors experimentally. The designed D- peptides were then verified for shape complementarity, noting that D-Leu2 of the peptide was designed to clash with the target VQIVYK on the opposite sheet, and upon alanine substitution, inhibitory activity ceases. Introducing a tight-binding interface and clashes destroying the ability of a cascade of amyloid-forming sequences to propagate was effective for inhibition. Pande and coworkers, guided by observations made in simulations of Aβ42, designed a non-canonical and D-amino acid containing peptide that organizes Aβ42 into stable oligomers [57].
Antibody Therapeutics
Gray and coworkers utilized Rosetta to introduce a non-canonical amino acid (NCAA) as an oxidizable crosslinker into an antibody complementarity determining region (CDR), with the best design experimentally cross-linking 52% of the available antigen [58]. Ellington and coworkers developed a “supercharging” protocol to substitute multiple surface residues with charged amino acids into proteins, using it to design an antibody with enhanced thermal inactivation resistance and a 30-fold affinity improvement [59]. Pantazes and Maranas introduced OptCDR for the design of antibodies to bind a targeted antigen epitope [60]. They applied it to design antibodies targeting a peptide from the capsid of hepatitis C, fluorescein, and VEGF, and validated the approach with computational metrics and binding energies. They recently introduced the Modular Antibody Parts (MAPs) database [61]. MAPs works in the spirit of template-based modeling where the templates are prototype structures of the random variable (V), diversity (D), and joining (J) regions in the database, resulting in gene combinations with the fewest amino acid changes from the target. Using this database, they were able to predict antibody tertiary structures with an average all-atom RMSD of 1.9Å on a testing set of 260 antibodies [61]. Upon successful prediction of a target structure, such antibodies can be computationally affinity matured using the Iterative Protein Redesign and Optimization (IPRO) framework [62]. IPRO is an iterative framework which optimizes side-chain substitutions in user-determined design positions using a mixed-integer optimization model where subsequently the backbone of the protein being designed is allowed to adjust through local minimizations to the new side-chains.
Design or Redesign of Enzymes and Biocatalysts
Baker reviewed the challenges and utility in succeeding in this endeavor [63]. Jiang et al. developed a computational method for constructing an active site for multistep reactions, designing 32 enzymes, spanning different protein folds and having detectable retro-aldolase activity for 4-hydroxy-4-(6-methoxy-2-naphthyl)-2-butanone, which is not found in biological systems [64]. The method designs active sites for these reactions with superimposed transition states of the reactions involved. Notably, designs identified using explicit water molecules were more successful, achieving enhancements of up to four orders of magnitude compared to the uncatalyzed reaction [64]. They also designed eight enzymes with different catalytic motifs for catalysis of Kemp elimination reactions [65]. Siegel et al. computationally designed stereoselective enzyme catalysts for the Diels-Alder reaction, prior to which none existed [66]. The most active design was confirmed to match the X-ray structure, noting that the success in design presumably was related to the success in modeling the designed catalytic site. Human game-players, through the visual interface of the online multiplayer game, Foldit, were able to “hands-on” remodel and redesign structures and side-chains of a 24-residue helix-turn-helix motif [67]. Based on this crowdsourced design, an 18-fold increase in enzymatic activity over their previously designed enzyme was achieved. The players were guided in the design by scores, which were inversely proportional to the Rosetta energy, and visual intuition. Khare et al. computationally redesigned mononuclear zinc metalloenzymes to catalyze non-native organophosphate hydrolysis activity with the experimental structures largely matching the designed ones [68]. Common themes from the catalyst design work were the correct modeling of the reaction transition states, which require quantum chemical calculations.
Maranas and coworkers computationally redesigned Candida boidinii xylose reductase (CbXR) to experimentally switch its cofactor specificity from NADPH to NADH with the IPRO procedure [69]. There is no experimentally solved structure for CbXR, so they used a homology model to perform the design blind of the true native structure. A 104-fold specificity change was observed to NADH, due primarily to changes in hydrogen bond and local charge interactions. Seven of ten predictions had significant xylose reductase activity utilizing NADH; the remaining two variants had duel cofactor specificity [69].
Donald and coworkers redesigned the specificity of nonribosomal peptide synthetase enzyme gramicidin S synthetase A (GrsA-PheA) from Phe to Leu, Arg, Glu, Lys, or Asp [70]. The computational redesign used physics-based energy evaluations of rotamerically sampled sequence space through the statistical mechanics based K* algorithm to approximate the binding constant Kd for the different analogues [71]. This study suggested that structure-based computational design can identify different mutants than those that have evolved, and that the designs could be used for charged amino acid adenylation [70].
Self-Assembling Proteins/Peptides
Controlling ordered (i.e., crystals) or disordered (i.e., hydrogels) self-assembly of proteins is a critical test of our understanding of both structure and interactions, having applications in biologically inspired materials. Lanci et al. computationally designed a protein crystal starting from an idealized homotrimeric parallel coiled-coil template and redesigned the interfaces [72]. They utilized strictly physics-based energy functions to discriminate favorable interfaces. Stranges et al. took two monomeric proteins’ solvent-exposed β-strands and redesigned them to form an intermolecular β-sheet symmetric homodimer with near atomic-level accuracy [73]. This design demonstrated the creation of unique stabilizing interactions at an interface. King et al. designed symmetric self-assembling complexes to atomic level accuracy [74]. They performed symmetric docking of subunits followed by redesign at the interfaces to design cage-like nanomaterials with tetrahedral or octahedral point group symmetry. The designed structures were confirmed experimentally by crystallography and electron microscopy to high agreement. The control over such self-assembling can be used to design advanced functional materials and molecular machines [74].
Other Applications
Hecht and coworkers designed de novo artificial sequences using a binary code strategy that encoded function and enabled cell growth after knocking out several naturally occurring genes required for cell viability [75]. The binary code strategy postulates that a simple code of alternating polar and nonpolar residues patterned in different ways can yield alpha helices or beta strand structures. They used this strategy to design a series of helical bundles which rescued E. coli cells with essential genes conditionally knocked out, and showed how a simplistic design strategy can produce proteins of novel function sufficient to sustain life. Piana et al. computationally designed the fastest folding β-protein [76]. They noted that the prior fastest β-protein, FiP35, was about an order of magnitude slower than its helical counterpart. The reduced folding time of the predicted design was experimentally confirmed to be approximately 3 times faster than the previous record-holder.
Concluding remarks and future perspectives
One can be successful in accurately predicting protein structures from sequence alone if templates can be identified and properly aligned. However, there is no method yet that can consistently predict structures “template-free”, possibly because conformational sampling and forcefields to guide sampling/selection are still imperfect [34, 35]. Even if accurate conformations are sampled, no method exists to accurately score those models more favorably from other decoys. Interestingly, it has been suggested that all the puzzle pieces needed to construct any structure are available, despite the fact that no method is currently able to properly assemble them in a blind predictive capacity [26, 77]. In our opinion, improvement in forcefields, the ability to accurately predict residue-residue contacts, β-sheet topologies, alignments to non-homologous templates, and effective conformational sampling methods are the key elements to solving the protein folding problem.
Transmembrane proteins are a class of targets that remain challenging for protein folding and design, despite being of significant interest to the pharmaceutical industry. These proteins are extremely difficult to solve experimentally due to their insoluble nature, and therefore few template structures exist for membrane proteins, although they account for the majority of current drug targets [78]. Further advances in the modeling of the membrane protein environment are needed to allow for improved structural models and evaluation of designed ligands.
In the de novo design paradigm, one has 20#DesignPositions sequences to evaluate. Doing this exhaustively computationally is largely impractical and even more so experimentally for even a few design positions. Proteins as potential therapeutics are hindered by proteolytic cleavage, poor solubility, and poor permeability. For these reasons, most have extracellular targets and often must be injected in order to be clinically successful. Using post-translational modifications (PTMs) and non-canonical amino acids (NCAAs) can help with these challenges, because modified peptides are less likely to be recognized by proteases, and these peptide modifications can be selected to fine-tune bioavailability. Design of modified peptide sequences adds complexity, since by considering the over 400 known PTMs for design, the combinatorial problem increases significantly to > 420#DesignPositions combinations [79]. The methods to model PTMs and NCAAs are still at an early stage of development, and represent a challenge in protein structure prediction and de novo protein design. Looking forward, we have just touched the surface of the allowable chemical space of proteins and their potential biotechnological applications.
Outstanding Questions.
How can we predict structures of sequences that are not homologous to any known protein?
How can we accurately predict the β-sheet topology?
How can we accurately predict medium and long-range tertiary contacts?
How can we consistently and substantially refine predicted protein structures to be closer to the native?
How can we predict structures of membrane proteins?
How can we predict the effects of the many PTMs and NCAAs on the structures of proteins?
How can we design soluble, passively permeable, metabolically stable peptides and proteins as therapeutics?
How do we incorporate PTMs and NCAAs into design, and address the massive increase in combinatorial complexity?
Highlights.
Interplay between accurate protein structure prediction and successful de novo protein design
Reviews current state-of-the-art structure protein prediction methods and challenges
Reviews features of successful de novo protein designs
Biotechnology applications in therapeutics, biocatalysts, and nanomaterials are summarized
Acknowledgments
CAF acknowledges support from the National Institutes of Health grant number R01GM052032 and the National Science Foundation. GAK is grateful for support by a National Science Foundation Graduate Research Fellowship under grant number DGE-1148900. We thank members of the Computer-Aided Systems Lab for helpful discussions. We apologize to the many researchers whose work could not be cited due to space limitations.
Glossary Box
- Global Distance Test Total Score (GDT_TS)
This is a metric that approximately represents the % of residues located in the correct position after structural alignment. This is a more robust metric than RMSD.
- Root-Mean Squared Deviation (RMSD)
This is a metric which measures the average distance between two structurally aligned sets of atoms. It is often used a metric for the quality of a prediction, and often computed with the α-carbon atoms. A predicted structure with RMSD to the native is ≤ 3Å is considered to be good enough to perform subsequent computational studies.
- Local Meta-Threading Server (LOMETS)
Generates structure predictions using high scoring alignments of a target sequence to a template using information from 10 threading programs.
- Iterative Threading Assembly Refinement (I-TASSER)
Structure prediction method using multiple threading alignments to templates and fragment assembly.
- QUARK
A protein structure prediction program that assembles fragments without any global template information.
- MODELLER
Protein structure homology modeling program that generates structures satisfying spatial constraints.
- REMO
Program that constructs a full protein model using only α-carbon traces.
- SP3
Fold recognition method that combines structural information through sequence profiles of structure fragments, secondary structure predictions, and dynamic programming to generate an alignment of a target sequence to a template.
- Generalized Orientation-Dependent All-Atom Statistical Potential (GOAP)
A distance-dependent statistical potential that scores models to aid in selecting near-native conformations of a target protein. It utilizes information about the relative plane orientation of interacting pairs of atoms.
- Z-score
Computed as , it is a metric denoting the separation of a value from counterparts. It is useful for assessing the significance of top structure predictions compared to the entire population of predictions from other methods.
- Rotamer
Statistically abundant side-chain conformation.
- Molecular Dynamics (MD)
An algorithm for solving the equations of motion iteratively over time and used to sample conformational space in a physically meaningful way.
- Monte Carlo (MC)
An algorithm reliant on randomly sampling the sequence or structural space according to a probability distribution.
- Cartesian minimization
Refers to a process operating on the variables as 3-dimensional vectors of x,y,z coordinates in order to reduce a conformer’s potential energy.
- Torsion minimization
Refers to a process operating on a reduced set of variables representing torsion angles which control the distance between the first and fourth atom in a series of four atoms in order to reduce a conformer’s potential energy.
- Non-deterministic polynomial time complete (NP-complete)
A difficult class of decision problems that have not been proven to be solvable with an algorithm within polynomial time ~ O(nk).
- Hot-spot
Key interactions at the interface of a protein-protein complex. Many hot-spots include salt-bridges where oppositely charged side-chains attract, hydrogen bonds, and/or ideal van der Waals interactions subject to shape complementarity.
- IC50 or EC50
IC50 is a metric for the half-maximal inhibitory concentration in a competitive binding assay. EC50 is metric for the concentration of compound at half the maximal value on a dose-response curve. Both curves are usually sigmoidal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pantazes RJ, et al. Recent advances in computational protein design. Curr Opin Struct Biol. 2011;21:467–472. doi: 10.1016/j.sbi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Drexler KE. Molecular engineering: An approach to the development of general capabilities for molecular manipulation. Proc Natl Acad Sci U S A. 1981;78:5275–5278. doi: 10.1073/pnas.78.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pabo C. Molecular technology. Designing proteins and peptides. Nature. 1983;301:200. doi: 10.1038/301200a0. [DOI] [PubMed] [Google Scholar]
- 4.Dill KA, MacCallum JL. The Protein-Folding Problem, 50 Years On. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 5.MacCallum JL, et al. Assessment of protein structure refinement in CASP9. Proteins: Struct, Funct, Bioinf. 2011;79:74–90. doi: 10.1002/prot.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacCallum JL, et al. Assessment of the protein-structure refinement category in CASP8. Proteins: Struct, Funct, Bioinf. 2009;77:66–80. doi: 10.1002/prot.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Floudas CA. Computational methods in protein structure prediction. Biotechnol Bioeng. 2007;97:207–213. doi: 10.1002/bit.21411. [DOI] [PubMed] [Google Scholar]
- 9.Moult J, et al. A large-scale experiment to assess protein structure prediction methods. Proteins: Struct, Funct, Bioinf. 1995;23:ii–iv. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y. Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins: Struct, Funct, Bioinf. 2013 doi: 10.1002/prot.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryshtafovych A, et al. CASP9 results compared to those of previous casp experiments. Proteins: Struct, Funct, Bioinf. 2011;79:196–207. doi: 10.1002/prot.23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy A, et al. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy A, et al. A protocol for computer-based protein structure and function prediction. Journal of Visualized Experiments. 2011:e3259. doi: 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins: Struct, Funct, Bioinf. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins: Struct, Funct, Bioinf. 2009;76:665–676. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, et al. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joo K, et al. Protein structure modeling for CASP10 by multiple layers of global optimization. Proteins: Struct, Funct, Bioinf. 2013 doi: 10.1002/prot.24397. [DOI] [PubMed] [Google Scholar]
- 21.Hildebrand A, et al. Fast and accurate automatic structure prediction with HHpred. Proteins: Struct, Funct, Bioinf. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 22.Peng J, Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins: Struct, Funct, Bioinf. 2011;79(Suppl 10):161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leaver-Fay A, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H, Skolnick J. Template-based protein structure modeling using TASSERVMT. Proteins: Struct, Funct, Bioinf. 2012;80:352–361. doi: 10.1002/prot.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, Skolnick J. GOAP: A Generalized Orientation-Dependent, All-Atom Statistical Potential for Protein Structure Prediction. Biophys J. 2011;101:2043–2052. doi: 10.1016/j.bpj.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 28.He Y, et al. Lessons from application of the UNRES force field to predictions of structures of CASP10 targets. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1313316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liwo A, et al. Coarse-grained force field: general folding theory. Physical chemistry chemical physics: PCCP. 2011;13:16890–16901. doi: 10.1039/c1cp20752k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindorff-Larsen K, et al. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 31.Piana S, et al. Atomic-level description of ubiquitin folding. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piana S, et al. How robust are protein folding simulations with respect to force field parameterization? Biophys J. 2011;100:L47–49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw DE, et al. Millisecond-scale molecular dynamics simulations on Anton. In. High Performance Computing Networking, Storage and Analysis, Proceedings of the Conference on; IEEE; 2009. pp. 1–11. [Google Scholar]
- 34.Bradley P, et al. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 35.Raval A, et al. Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins: Struct, Funct, Bioinf. 2012;80:2071–2079. doi: 10.1002/prot.24098. [DOI] [PubMed] [Google Scholar]
- 36.Berger B, Leighton T. Protein folding in the hydrophobic-hydrophilic (HP) model is NP-complete. J Comput Biol. 1998;5:27–40. doi: 10.1089/cmb.1998.5.27. [DOI] [PubMed] [Google Scholar]
- 37.Jones DT, et al. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 38.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajgaria R, et al. Towards accurate residue–residue hydrophobic contact prediction for α helical proteins via integer linear optimization. Proteins: Struct, Funct, Bioinf. 2009;74:929–947. doi: 10.1002/prot.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajgaria R, et al. Contact prediction for beta and alpha-beta proteins using integer linear optimization and its impact on the first principles 3D structure prediction method ASTRO-FOLD. Proteins: Struct, Funct, Bioinf. 2010;78:1825–1846. doi: 10.1002/prot.22696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramani A, et al. ASTRO-FOLD 2.0: An enhanced framework for protein structure prediction. AIChE J. 2012;58:1619–1637. doi: 10.1002/aic.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klepeis JL, Floudas CA. ASTRO-FOLD: a combinatorial and global optimization framework for Ab initio prediction of three-dimensional structures of proteins from the amino acid sequence. Biophys J. 2003;85:2119–2146. doi: 10.1016/s0006-3495(03)74640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramani A, Floudas CA. β-sheet Topology Prediction with High Precision and Recall for β and Mixed α/β Proteins. PLoS One. 2012;7:e32461. doi: 10.1371/journal.pone.0032461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DE, et al. One contact for every twelve residues allows robust and accurate topology-level protein structure modeling. Proteins: Struct, Funct, Bioinf. 2013 doi: 10.1002/prot.24374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samish I, et al. Theoretical and computational protein design. Annu Rev Phys Chem. 2011;62:129–149. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- 46.Fung HK, et al. Computational De Novo Peptide and Protein Design: Rigid Templates versus Flexible Templates. Ind Eng Chem Res. 2008;47:993–1001. [Google Scholar]
- 47.Vlieghe P, et al. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Pirogova E, Istivan T. Bioinformatics of Human Proteomics. Springer; 2013. Toward Development of Novel Peptide-Based Cancer Therapeutics: Computational Design and Experimental Evaluation; pp. 103–126. [Google Scholar]
- 49.Craik DJ, et al. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 50.Hao J, et al. Identification and Rational Redesign of Peptide Ligands to CRIP1, A Novel Biomarker for Cancers. PLoS Comput Biol. 2008;4:e1000138. doi: 10.1371/journal.pcbi.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Istivan TS, et al. Biological effects of a de novo designed myxoma virus peptide analogue: evaluation of cytotoxicity on tumor cells. PLoS One. 2011;6:e24809. doi: 10.1371/journal.pone.0024809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Correia BE, et al. Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Bellows ML, et al. Discovery of Entry Inhibitors for HIV-1 via a New De Novo Protein Design Framework. Biophys J. 2010;99:3445–3453. doi: 10.1016/j.bpj.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajgaria R, et al. Distance dependent centroid to centroid force fields using high resolution decoys. Proteins: Struct, Funct, Bioinf. 2008;70:950–970. doi: 10.1002/prot.21561. [DOI] [PubMed] [Google Scholar]
- 55.Smadbeck J, et al. Protein WISDOM: A Workbench for In silico De novo Design of BioMolecules. Journal of Visualized Experiments. 2013:e50476. doi: 10.3791/50476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sievers SA, et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature. 2011;475:96–100. doi: 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajadas J, et al. Rationally Designed Turn Promoting Mutation in the Amyloid-β Peptide Sequence Stabilizes Oligomers in Solution. PLoS One. 2011;6:e21776. doi: 10.1371/journal.pone.0021776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J, et al. Structure-based non-canonical amino acid design to covalently crosslink an antibody–antigen complex. J Struct Biol. 2013 doi: 10.1016/j.jsb.2013.1005.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miklos Aleksandr E, et al. Structure-Based Design of Supercharged, Highly Thermoresistant Antibodies. Chem Biol. 2012;19:449–455. doi: 10.1016/j.chembiol.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pantazes RJ, Maranas CD. OptCDR: a general computational method for the design of antibody complementarity determining regions for targeted epitope binding. Protein Eng Des Sel. 2010;23:849–858. doi: 10.1093/protein/gzq061. [DOI] [PubMed] [Google Scholar]
- 61.Pantazes RJ, Maranas CD. MAPs: a database of modular antibody parts for predicting tertiary structures and designing affinity matured antibodies. BMC Bioinformatics. 2013;14:168. doi: 10.1186/1471-2105-14-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saraf MC, et al. IPRO: an iterative computational protein library redesign and optimization procedure. Biophys J. 2006;90:4167–4180. doi: 10.1529/biophysj.105.079277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 2010;19:1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang L, et al. De Novo Computational Design of Retro-Aldol Enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rothlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 66.Siegel JB, et al. Computational Design of an Enzyme Catalyst for a Stereoselective Bimolecular Diels-Alder Reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eiben CB, et al. Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat Biotechnol. 2012;30:190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khare SD, et al. Computational redesign of a mononuclear zinc metalloenzyme for organophosphate hydrolysis. Nat Chem Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khoury GA, et al. Computational design of Candida boidinii xylose reductase for altered cofactor specificity. Protein Sci. 2009;18:2125–2138. doi: 10.1002/pro.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C-Y, et al. Computational structure-based redesign of enzyme activity. Proceedings of the National Academy of Sciences. 2009 doi: 10.1073/pnas.0900266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lilien RH, et al. A Novel Ensemble-Based Scoring and Search Algorithm for Protein Redesign and Its Application to Modify the Substrate Specificity of the Gramicidin Synthetase A Phenylalanine Adenylation Enzyme. J Comput Biol. 2005;12:740–761. doi: 10.1089/cmb.2005.12.740. [DOI] [PubMed] [Google Scholar]
- 72.Lanci CJ, et al. Computational design of a protein crystal. Proceedings of the National Academy of Sciences. 2012;109:7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stranges PB, et al. Computational design of a symmetric homodimer using β-strand assembly. Proceedings of the National Academy of Sciences. 2011;108:20562–20567. doi: 10.1073/pnas.1115124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.King NP, et al. Computational Design of Self-Assembling Protein Nanomaterials with Atomic Level Accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fisher MA, et al. De Novo Designed Proteins from a Library of Artificial Sequences Function in Escherichia Coli and Enable Cell Growth. PLoS One. 2011;6:e15364. doi: 10.1371/journal.pone.0015364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Piana S, et al. Computational Design and Experimental Testing of the Fastest-Folding β-Sheet Protein. J Mol Biol. 2011;405:43–48. doi: 10.1016/j.jmb.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 77.Skolnick J, et al. Further Evidence for the Likely Completeness of the Library of Solved Single Domain Protein Structures. The Journal of Physical Chemistry B. 2012;116:6654–6664. doi: 10.1021/jp211052j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Overington JP, et al. How many drug targets are there? Nat Rev Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 79.Khoury GA, et al. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Scientific Reports. 2011:1. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fleishman SJ, et al. Computational Design of Proteins Targeting the Conserved Stem Region of Influenza Hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bellows-Peterson ML, et al. De Novo Peptide Design with C3a Receptor Agonist and Antagonist Activities: Theoretical Predictions and Experimental Validation. J Med Chem. 2012;55:4159–4168. doi: 10.1021/jm201609k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellows ML, et al. New Compstatin Variants through Two De Novo Protein Design Frameworks. Biophys J. 2010;98:2337–2346. doi: 10.1016/j.bpj.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorham RD, Jr, et al. Novel compstatin family peptides inhibit complement activation by drusen-like deposits in human retinal pigmented epithelial cell cultures. Exp Eye Res. 2013 doi: 10.1016/j.exer.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]