Abstract
Activation of the complement system is primarily initiated by pathogen- and damage-associated molecular surface patterns on cellular surfaces. However, there is increasing evidence for direct activation of individual complement components by extrinsic proteinases as part of an intricate crosstalk between physiological effector systems. We hypothesized that kallikrein-related peptidases (KLKs), previously known to regulate inflammation via proteinase-activated receptors, can also play a substantial role in innate immune responses via complement. Indeed, KLKs exemplified by KLK14 were efficiently able to cleave C3, the point of convergence of the complement cascade, indicating a potential modulation of C3-mediated functions. By using in vitro fragmentation assays, mass spectrometric analysis, and cell signaling responses, we pinpointed the generation of the C3a fragment of C3 as a product with potential biological activity released by the proteolytic action of KLK14. Using mice with various complement deficiencies, we demonstrated that the intraplantar administration of KLK14 results in C3-associated paw edema. The edema response was dependent on the presence of the receptor for C3a but was not associated with the receptor for the downstream complement effector C5a. Our findings point to C3 as one of the potential substrates of KLKs during inflammation. Given the wide distribution of the KLKs in tissues and biological fluids where complement components may also be expressed, we suggest that via C3 processing, tissue-localized KLKs can play an extrinsic complement-related role during activation of the innate immune response.
Keywords: anaphylatoxin, C3a receptor, complement, inflammation, kallikrein-related peptidases, paw edema, trypsin
Introduction
The complement network, consisting of more than fifty plasma-borne and membrane-bound proteins, is an essential component of the innate immune system, which forms a first line of defense against microbial invaders and exerts important functions in immune surveillance and homeostasis (1). The constituents of the network are organized into three major pathways (classical, lectin, and alternative) characterized by distinct ‘molecular recognition motifs’ that trigger activation of the cascade upon detection of damage- or pathogen-associated surface patterns (1,2). All three pathways converge on the proteolytic release of active fragments of complement component 3 (C3), the C3a anaphylatoxin and C3b, the latter of which is an important opsonin and vital component of the C3 convertase. This convertase can in turn amplify the generation of C3b from the available C3 deposits, as well as contribute to cleavage of C5 and generation of the C5a anaphylatoxin and C5b. The main biological activities of complement activation are: (a) the opsonization of pathogens; (b) the anaphylatoxin-derived recruitment of inflammatory cells; (c) bacterial cell lysis as a result of the formation of the membrane-attack complex downstream of C5; and (d) the regulation of adaptive immunity by stimulation of B- and T-cells (1,2). C3a and C5a, and their desarginated derivatives, have received particular attention as major signaling effectors that exert a broad spectrum of biological effects ranging from chemotaxis to immune cell activation, thereby contributing to innate and adaptive immune functions and inflammation. They act primarily via their G protein-coupled receptors C3aR and C5aR (CD88), respectively; a third anaphylatoxin receptor, C5L2, has been described but its role and specificity are still a matter of debate (3).
Apart from the traditional view of activation within the three branches of the complement cascade, a potential extrinsic pathway of initiation of complement activation, mainly at the level of C3 and/or C5, by non-complement proteinases has also been suggested (1,2). For example, generation of potentially biologically active fragments from C3 and C5 by the prototype serine proteinase trypsin has been reported (4,5). Direct activation and/or degradation of C3 or C5 have additionally been shown for other proteinases, including plasma kallikrein (6,7), components of the coagulation cascade like factor Xa and thrombin (8), the neutrophil-released proteinases elastase and cathepsin-G (9), and mast cell tryptase (10). A role for microbial proteinases released during infection in the regulation of anaphylatoxin activity has also been suggested (11).
Despite the presence of circulating enzymes described above that may be able to modulate complement activity in the periphery, little is known about local complement activation in inflamed tissues and tumors, where proteinases in addition to thrombin and tissue-trypsins would be activated. Kallikrein-related peptidases (KLKs) are a family of tissue serine proteinases, distinct from plasma kallikrein (12,13). KLKs have trypsin or chymotrypsin-like activity and belong biochemically to the same enzyme class as trypsin. The human KLK family consists of 15 enzymes, which are upregulated in tissues, mainly of epithelial origin, and in biological fluids, such as ascites fluid and sera from cancer patients. Cascade enzymatic activity of KLKs has been hypothesized to occur in the central nervous system and seminal plasma. In the skin, several KLKs have been isolated from the outermost layers of the stratum corneum and overall epidermis and are implicated in subcutaneous (patho)physiology (14). Amongst the KLK enzymes, KLKs 5, 6, and 14, are characterized by wide tissue distribution, high biological activity, and significant association with the clinical progression and outcome of several types of cancer, such as breast, ovarian, prostate, and skin carcinoma (12). Specifically KLK14, possibly as part of a KLK catalytic cascade, has been linked to epidermal desquamation (i.e., shedding) due to a degradation of the intercellular (corneo)desmosomal adhesion molecules that link adjacent corneocytes (14). Preliminary analyses have also connected KLK14 expression in inflamed skin tissues to the pathogenesis of rosacea, while other KLKs have been associated with atopic dermatitis, psoriasis vulgaris, and Netherton syndrome (13,14). Despite data establishing a role for KLKs in skin homeostasis (i.e., desquamation, matrix remodelling) (12–14), knowledge of their specific contributions to skin inflammatory pathologies is still largely unknown.
It has been reported that C3aR, responsive to C3a signals, can be found on cells that can be localized in skin tissue (i.e., mast cells, neutrophils, and monocytes/macrophages) (15), where KLKs would also be present (14). Keratinocytes, a major source of KLKs (14), are key components of the innate immune response (16), and abundantly secrete C3 (17,18) and express C3aR (19) in the settings of inflammation. Similar to the KLKs (13,14), an association between complement activation and skin inflammation (e.g., dermatitis, psoriasis) has also been made (20). In addition to inflammation, both the complement system and KLKs have independently been attributed roles in the regulation of oncogenesis (12,21). Specifically for skin carcinogenesis, proteolytic activation of C3 may contribute to tumorigenicity and metastasis of human melanoma cells in nude mice (22).
Given the similar tissue distribution and role of the complement and KLK families, we hypothesized that kallikrein-related peptidases can have a major contribution to inflammatory responses taking place in peripheral tissues, including the skin, by modulating the activity/bioavailability of complement components. Our analyses of the C3-targeting function of KLKs, exemplified in this study by the activity of KLK14, show that this interaction can lead to biologically significant responses in ex vivo and in vivo models of peripheral tissue complement activation.
Materials and Methods
Reagents and protein preparations
Chemicals were purchased from Fisher Scientific (Hampton, NH), unless indicated otherwise. Human complement proteins C3a, C3a-desArg, and C5 were from Complement Technology (Tyler, TX). C3 was purified from human plasma as previously described (23) with addition of an extra gel filtration step (Superdex 200, GE Healthcare; flow rate of 1 ml/min in PBS). Fractions were analyzed by SDS-PAGE under reducing conditions, total protein assay, and enzyme-linked immunosorbent assay (ELISA). More specifically, regarding ELISA, our purified preparations of unknown C3 content were diluted 2.5-fold with PBS, pH 7.5 and incubated in 96-well plates for 2 h at room temperature. Plates were subsequently blocked with 1% BSA in PBS and allowed to sit for 30 min at room temperature. Wells were washed 3 times with PBS-Tween-20 (0.05%) before addition of an HRP-conjugated goat anti-human C3 polyclonal detection antibody (50 µl; 1/1000 in PBS; MP Biomedicals, cat. no 0855237) for 30 min at room temperature. The plate was washed 3 times with PBS-Tween-20 (0.05%) and developing solution (50 µl/well of a 100 mM sodium citrate buffer supplemented with 0.5 mg/ml ABTS [Roche Applied Science, Indianapolis, IN, cat. no 10102946001] and 0.03% H2O2 [Fisher, cat. no 7722-84-1]) was added to each well and incubated for 2 min at room temperature. Absorbance of light was measured at a wavelength of 405 nm.
KLK6 was expressed in a mammalian human embryonic kidney cells (HEK) cell system (24) and KLK5 was expressed in Pichia pastoris (25); enzymes were purified and activated as previously described (24,25). Active KLK14 was expressed in Pichia pastoris and purified as described previously (25). In brief, concentrated yeast supernatant, diluted 1:2 with running buffer (10 mm MES, pH 5.3), was loaded onto a 5-ml HiTrap SP HP-Sepharose cation exchange column (Amersham Biosciences). The column was eluted with 10 mM MES buffer, pH 5.3, with a linear gradient of 0–1 M KCl. Fractions were analyzed by a KLK14-specific ELISA, pooled, and concentrated 10 times in 10 mM MES, pH 5.3. The concentration of the purified catalytically active recombinant protein was determined by ELISA and total protein assay. The purity and integrity of the protein (>90%) was verified by SDS-PAGE.
Estimation of trypsin-like serine proteinase activity
The enzymatic activity of the trypsin-like kallikrein-related serine peptidases was measured using the fluorogenic t-butoxycarbonyl-tripeptide-7-amino-4-methylcoumarin (AMC) synthetic substrates QAR-AMC (KLK14) or VPR-AMC (KLKs 5 and 6) (Bachem Bioscience, King of Prussia, PA) (25). In brief, each kallikrein was incubated at 37°C in a microtiter plate, with the optimal activity assay buffer and varying concentrations (within the linear range) of fluorescent substrates in a final volume of 100 µl. The initial rate of AMC release was measured on a Wallac Victor fluorometer (PerkinElmer, Waltham, MA) set at 355 nm for excitation and 460 nm for emission. The fluorescence values of enzyme-free reactions were subtracted from each value. All experiments were performed in triplicate. Enzyme activity was quantified by monitoring the rates of tripeptide-AMC hydrolysis (slopes of the fluorescence curves expressed as fluorescence units (FU)/min). A standard curve with known concentrations of AMC was used to calculate the rate of product formation. Kinetic analysis was performed by nonlinear regression analysis using the Enzyme Kinetics Module 1.1 (Sigma Plot, SSPS, Chicago, IL) and activity of each enzyme was estimated by means of nmoles of AMC-substrate cleaved per min of reaction per mg of enzyme used. The activity of a given concentration of KLK was compared with that of a standard concentration of pure trypsin (5 nM or 2.5 units/ml; Sigma-Aldrich; cat. no T-7418), measured using the same AMC substrate employed for assessing each KLK activity, and subsequently expressed by means of ‘trypsin-like units of activity’. This approach allowed for the direct comparison of activity levels between the different proteinases.
Fragmentation analysis
In vitro fragmentation experiments were performed by incubating native C3 or C5 in PBS, pH 7.5 in the absence or presence of KLKs or trypsin at 37°C in a concentration- and time-dependent manner. The enzyme to substrate ratio was expressed in w/w units; in the case of KLK14 the 1% w/w enzyme to C3 ratio corresponds to a 1:16 molar ratio. Given the comparable molecular weights of KLKs and trypsin, the molar ratios of enzyme to substrate were estimated to be similar allowing the direct comparison of the KLKs with trypsin in facilitating C3 cleavage. SDS-PAGE separation under reducing and denaturing conditions was followed by Coomassie staining to evaluate the fragmentation of complement components by the serine proteinases.
The identity of the low molecular weight C3-derived proteolytic products was further validated by using MALDI-TOF mass spectrometry. In brief, purified C3 (20 µg) was incubated with KLK14 or trypsin at indicated concentrations and times at 37°C, in PBS, pH 7.5. Samples were passed through C18 ZipTip (Millipore, Billerica, MA) and analyzed by MALDI-TOF. Spectra were acquired on a MALDI-Micro MX (Waters, Milford, MA) in linear mode as recently described (8).
C3 (7.4 µg) or C3a (0.5 µg) were also subjected to KLK14 incubation (equal molar ratio at the level of 1:30 of enzyme to C3/C3a, corresponding to a w/w ratio of 0.5% for C3 and 10% for C3a) directly at 37°C or at room temperature, in PBS, pH 7.5, followed by liquid chromatography-coupled mass spectrometry (LC-MSE) analysis. Analytes were separated on a 1.7 µm ultra performance liquid chromatography (UPLC) BEH130 C18 column (Waters, 2.1 µm × 150 mm, cat. no 186003556). The analytical column temperature was maintained at 40°C. Peptides were separated for 8 min using a flow rate of 0.15 ml/min and a linear gradient of 10–60% of solvent B (0.1% formic acid in acetonitrile [ACN], v/v) in combination with solvent A (0.1% formic acid in water, v/v). LC-MSE analysis was performed on a SYNAPT HDMS (Waters) equipped with an electrospray ionization (ESI) source controlled by MassLynx 4.1 software (Waters). The capillary voltage was 3.2 kV, the cone voltage was 30 V, and the source temperature was 120°C. [Glu1]-fibrinopeptide B (Sigma) was used for lock-mass correction with a sampling rate of 30 s. Mass spectra were acquired in positive mode over an m/z range of 100–2,000 Da at a scan rate of 1 s. Fragment sequences were verified by collision-induced dissociation (CID) fragmentation.
The C3a degradation by KLK14 was also followed by studying the proteolytic release of downstream fragments by HPLC (Ettan LC; GE Healthcare). In particular, 4 µg of C3a was incubated at room temperature with 0.4 µg of KLK14 (10% enzyme to C3a ratio in PBS; 1:30 molar ratio) and the reaction was followed for up to 60 min. Reactions were terminated by using 0.02% TFA and samples were run on an analytical column (Vydac MS C18 300 A, 5 µm × 50 mm, ID 1 mm; Grace, Deerfield, IL, cat. no 218MS5105) using a gradient of 15 to 65% solvent B (0.1 % trifluoroacetic acid in ACN, v/v) for 60 min and a flow rate of 100 µl/min (0.1% TFA in water was used as solvent A). Peak fractions (absorbance at 215 nm) were collected and identification of the KLK14-mediated C3a fragments was performed by MALDI-TOF and/or ESI analysis as detailed above.
Detection of C3a by ELISA
Estimation of C3a released by the KLK14-mediated C3 cleavage relative to the one released by trypsin was achieved by incubating KLK14 with purified human C3 (1% w/w for the indicated time points at 37°C, in PBS, pH 7.5) and by detecting C3a using a rabbit polyclonal antibody (UP1896), which was raised against recombinant human C3a and produced in-house according to standard procedures, and a HRP-conjugated goat anti-rabbit IgG (Bio-Rad, Hercules, CA, cat. no 172-1019) in a direct ELISA approach.
C3aR-mediated cell activation assays
Rat basophilic leukemia (RBL-2H3) cells stably expressing the C3a receptor (RBL-C3aR) were obtained as previously described (26); cell dynamics in response to C3a were monitored with the SRU BIND® platform (SRU Biosystems, Woburn, MA), which utilizes photonic crystal technology to sense activation-induced changes in cell morphology and adhesion (27). In brief, the wells of a 384-well BIND CA2 biosensor plate (CA2 is a matrix surface that promotes cell attachment; SRU Biosystems) were hydrated with 50 µl of deionized water for 30 min at room temperature. Water was then substituted with 25 µl of complete medium [Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS)] for 30 min at room temperature. The baseline signal was recorded using the BIND PROFILER instrument (SRU Biosystems). RBL-C3aR cells (25,000 cells per well) were seeded on the plate in DMEM (Invitrogen, Grand Island, NY), supplemented with 10% heat-inactivated FCS, and maintained overnight at 37°C (5% CO2). The plate was allowed to equilibrate at room temperature for 30 min and the cell attachment signal was obtained. Cells were stimulated with C3, C3a, or the KLK14-mediated C3 fragments. Incubation of KLK14 with C3 (0.5% w/w; 1:30 molar ratio) occurred in the absence of serum to avoid the KLK inhibitory effects of serum-contained serpins (28). Cells were subsequently treated with serial dilutions of the original incubation mixture in the FCS-supplemented medium and the changes in cell dynamics (recorded as peak wavelength values [PWV] in pm) were followed for up to 30 min post-ligand incubation. Recorded values were analyzed using the EMS software (SRU Biosystems) and the EC50 of cell responses were calculated.
Paw edema inflammation model
Complement-deficient mice lacking C3 (C3−/−), the C3a receptor (C3aR−/−), or the C5a receptor (C5aR−/−) have been previously described (29–31). Deficient mice had been backcrossed for at least nine generations onto a C57BL/6J background, and their wild-type littermates or C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) were used as controls where indicated. Complement-deficient and control animals (6–8 weeks old; 20–25 g) were obtained and housed at the University of Pennsylvania animal facility under carefully regulated conditions with a 12 h light–dark cycle and provided free access to food and water. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania and were performed in accordance with guidelines established by the Animal Welfare Act (AWA).
Prior to the intraplantar administration of KLK14, a basal measurement of the paw thickness of each mouse was recorded using an electronic micrometer caliper (Mitutoyo, Aurora, IL, cat. no 500-196-20). The compounds used for treatment were diluted in sterile Dulbecco's Phosphate Buffered Saline (DPBS) and administered via intraplantar injection into the hind paw at a final volume of 10 µl per mouse paw (1.2 µg or 22 trypsin-like Units of KLK14 per paw). Control mice were treated with KLK14 inactivated with soybean trypsin inhibitor (STI; Sigma; incubation for 30 min at 37°C using 1:5 molar ratio of enzyme to inhibitor) (32) or buffer alone (n≥4–5 mice per group; three independent experiments). STI was used to indicate that the edema response is mediated due to the proteolytic action of KLK14. As an index of edema formation, paw thickness monitored with the micrometer caliper (Mitutoyo) was followed for up to 4 h post-injections. Paw thickness values at baseline were subtracted from the values obtained at each time point. All values obtained from the paw thickness measurements were analyzed by two-way ANOVA (Prism, GraphPad Software, La Jolla, CA), and the χ2 test. A p value of 0.05 or less was considered to indicate a significant difference between groups.
Results
Kallikrein-related peptidases cleave complement component 3 (C3)
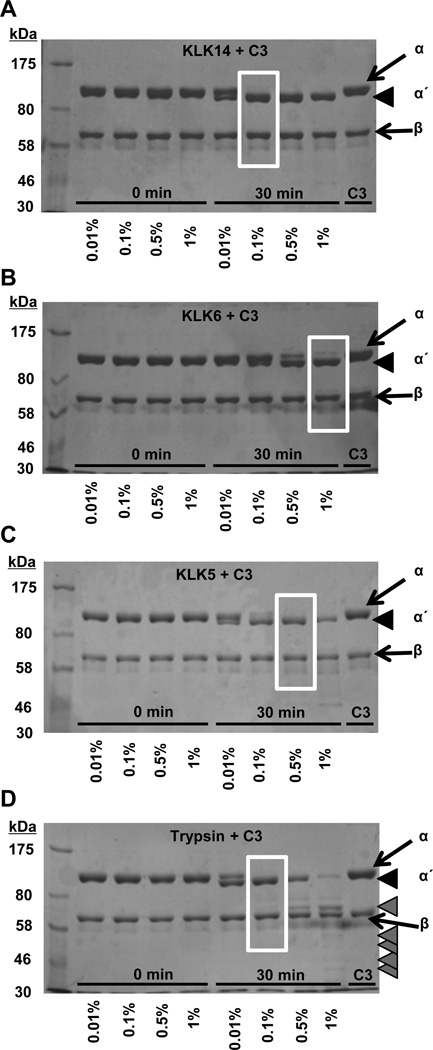
KLK5, KLK6, and KLK14 caused a limited proteolysis of C3 (Fig. 1, Fig. 2; Supplemental Fig. S1). The cleavage of C3 resulted in the generation of a molecular weight species similar to that of C3b that is comprised of the fragmented α-chain (α΄-chain) and intact β-chain fragments. The proteolytic processing pattern for KLKs (Fig. 1A–C) displayed similarities with that of the enzymatically-related serine proteinase trypsin (Fig. 1D) (4,33) that resulted in the disappearance of the C3 α-chain and appearance of an α΄-chain fragment. Coomassie staining of the C3 digestion products detected in reducing gels that separate the α- and β-chains indicated that all three KLK enzymes could cleave C3, but with different efficiencies. For instance, at comparable ratios of enzyme to substrate, both KLK5 and KLK14 cleaved the α-chain (Fig. 1A, 1C; Supplemental Fig. S1), whereas much higher enzyme/substrate ratios were required for KLK6-mediated cleavage (Fig. 1B; Supplemental Fig. S1). In contrast to trypsin, which at higher ratios of enzyme to substrate caused a disappearance of the α΄-chain to yield a constituent migrating just above the β-chain (Fig. 1D, far-right lane), KLK14, even at relatively high enzyme to substrate ratios, was not able to efficiently cleave the α΄-chain further (Fig. 1A).
Figure 1. Proteolysis of complement component C3 by kallikrein-related peptidases and trypsin.
The concentration-dependent KLK14-mediated generation of C3b-like fragments from degradation of C3 is followed (A), in comparison to the proteolytic processing of C3 by KLKs 5, 6, and trypsin (B–D) at the indicated enzyme to C3 ratios (w/w). Samples were run on 7.5% SDS-PAGE gels under reducing and denaturing conditions, and visualized with Coomassie staining. The C3 α- and β-chains are indicated with arrows, while the α΄-chain is indicated with black arrowheads. Lower molecular weight C3 fragments are indicated with grey arrowheads.
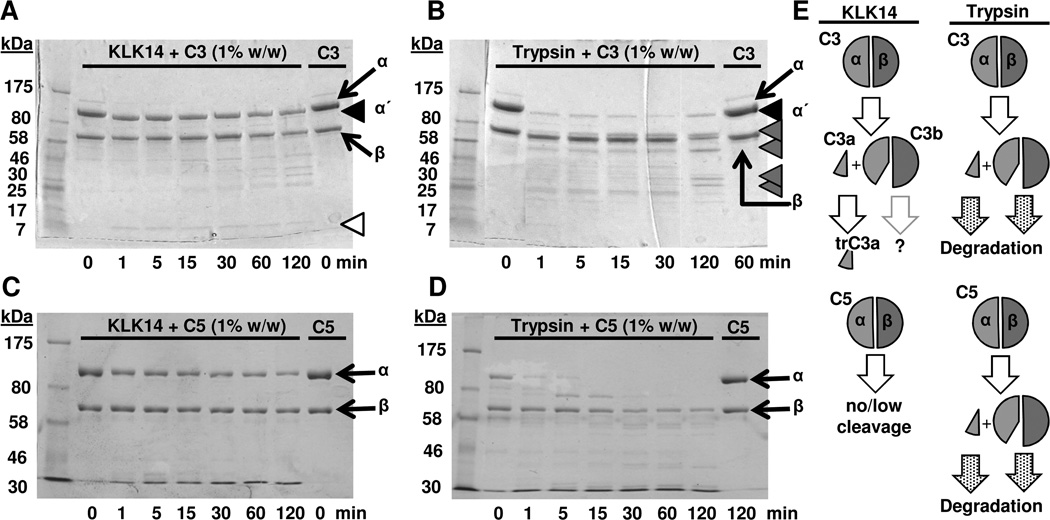
Figure 2. Comparison of limited proteolysis of complement components C3 and C5 by kallikrein-related peptidase 14 and trypsin.
A–B) The time-dependent KLK14-mediated (A) and trypsin-mediated (B) generation of C3b-like fragments from degradation of C3 at indicated enzyme to C3 ratios (w/w) is shown. C–D) Proteolysis of C5 by KLK14 and trypsin is shown for comparison. Samples were run on 4–15% (A–B) or 7.5% (C–D) SDS-PAGE gels under reducing and denaturing conditions, and visualized with Coomassie staining. The C3 α- and β-chains are indicated with arrows, while the α΄-chain is indicated with black arrowheads. Lower molecular weight C3 fragments are indicated with grey arrowheads and the C3a-like fragment in panel A is indicated with a white arrowhead. E) A pictorial representation of the proteolytic processing of C3 and C5 by KLK14 and trypsin is shown. The fragmentation scheme is based on the SDS-PAGE analysis (Fig. 1–2 and Supplemental Fig. S1) and MS data (Fig. 3 and Supplemental Fig. S3), as well as on previous literature (4,5). Cleavage of native C3 or C5 by trypsin results in the release of the C3a and C5a anaphylatoxins, respectively. The remaining parts of the molecule (α΄-chain and β chain) constitute the components of C3b and C5b. In contrast, proteolytic processing by KLK14 leads in generation of C3a, which is further truncated into a smaller fragment the characteristics of which are shown in Fig. 3 and Supplemental Fig. S3.
We elected to focus on KLK14 as a prototype member of the kallikrein-related peptidase family with expression and/or activity documented in variable disease settings. To standardize the activity of KLK14, its proteolytic efficiency was compared to the one exhibited by a standard concentration of pure trypsin (5 nM or 2.5 U/ml), measured using the same QAR-AMC substrate employed for assessing the KLK activity (25). By performing fluorogenic substrate activity analysis for all KLKs and trypsin, we found that our KLK14 preparation had a trypsin-like activity of about 18 U/µg of KLK (25). When compared to the preparation of trypsin we used for our analysis, 1% of KLK14 (0.05 µg; w/w of C3) corresponded to 0.9 U of trypsin-like activity against the same QAR-AMC substrate. By performing similar calculations, the levels of trypsin-like activity exhibited by 0.05 µg of KLKs 5 and 6 were found to be lower than KLK14 (decreased by approximately 5 and 22-fold, respectively, when compared to KLK14; data not shown).
A comparative time course analysis of the C3 cleavage at 1% enzyme concentration revealed a faint band of a low molecular weight species corresponding to C3a that rapidly appeared after 1 min and remained visible throughout the 2-h incubation (Fig. 2A; white arrowhead). In agreement, the α-chain was rapidly converted to the α΄ fragment; the visible weakening of the α΄ band after 1 h with appearance of additional faint bands may indicate a slow yet progressive degradation of the α΄-chain. Cleavage of the β-chain appears possible but would likely occur at a slow rate, as well (Fig. 2A). As observed above, digestion of C3 by trypsin is more complex and leads to the rapid generation of additional fragments at early time-points (Fig. 1D, 2B). While detailed conclusions regarding the comparative enzyme kinetics of the proteolytic enzymes for cleaving C3 cannot be extrapolated from our qualitative degradation analysis, our data clearly show that KLK14 exhibits a proteolytic ability comparable to trypsin in performing the “initial” cleavage of C3 to a C3b-like fragment (Fig. 1); nevertheless, KLK14 does not subsequently cleave C3b further as extensively as trypsin.
KLK14 shows limited cleavage activity towards complement component C5
As both C3 and C5 are degraded by trypsin (4,5,33), the similar levels of C3-fragmentation activity between KLK14 and trypsin raised the possibility for KLK14-mediated cleavage of C5. However, our data indicated that the two enzymes displayed significantly different efficiency for cleaving the downstream component C5 (Fig. 2C–D). More specifically, C5 cleavage by KLK14 was limited compared with the extensive cleavage caused by trypsin. Furthermore, the fragments that were generated by the proteolytic action of KLK14 under the same conditions as used for C3 were not indicative of the release of fragments with molecular weights expected for activated C5 (i.e., C5b of ~170 kDa) (Fig. 2C). This is in contrast to what has been reported for C5 fragmentation by several proteinases of the coagulation system, which lead to the generation of active C5 fragments (8). Although these studies on the KLK-mediated cleavage of C5 may be extended in the future, our data for the cleavage specificity of KLK14 revealed a cleavage pattern distinct from trypsin and indicated that C3 was a preferred substrate when compared with C5 (Fig. 2E). Consequently, our focus for subsequent work remained on C3.
C3 cleavage by KLK14 results in generation of C3a-like species
The KLK14-generated release of a C3b-like molecular weight species (Fig. 1A; Fig. 2A) suggested the possible generation of a corresponding C3a-like anaphylatoxin fragment released from the α-chain of C3 (Fig. 2A, white arrowhead; Fig. 2E). Detection of a C3a-like species within 1 min of C3 incubation in the KLK14-containing reaction mixture using a direct ELISA approach further supported this hypothesis (Supplemental Fig. S2).
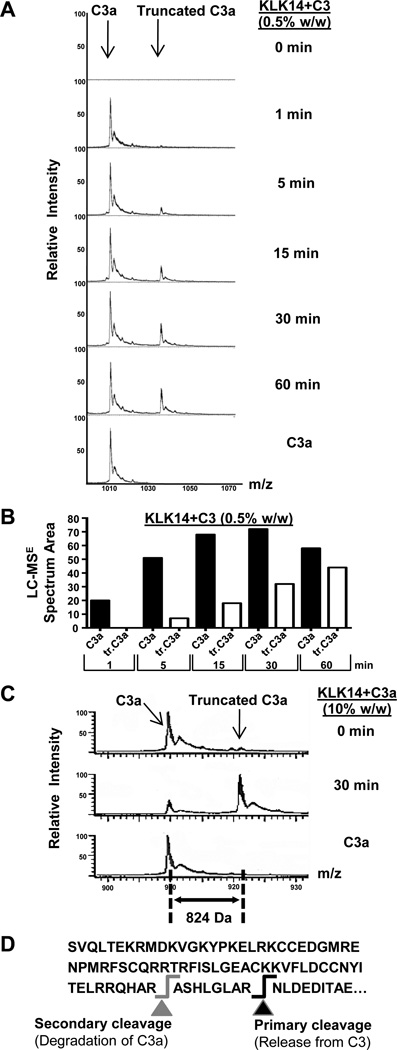
The release of the intact C3a fragment from C3 by KLK14 was verified by chromatographic separation of the proteolysis products followed by MALDI-TOF and/or LC-MSE mass spectrometry (Fig. 3). In particular, our proteomic analyses identified a fragment with a molecular weight of 9,087 Da and charge properties equivalent to those expected for human C3a (Fig. 3) (8). Furthermore, time-resolved analysis revealed that, after release from C3, intact C3a was processed further by KLK14 during incubation into a lower molecular weight species, which was resistant to subsequent proteolysis during the 30–60-min follow-up incubation time (Fig. 3A). Calculation of the peak areas for C3a and its truncated fragment confirmed the fragmentation pattern and indicated a slower rate for the second cleavage (Fig. 3B). Notably, KLK14 was also able to cleave purified human C3a into the same truncation species (Fig. 3C); the time-dependent cleavage of C3a by KLK14 was also followed by HPLC (not shown). The fragment secondarily produced by the proteolytic action of KLK14 had a molecular weight of 8,282 Da. Using LC-MSE analysis, it was shown that this shift in molecular weight corresponds to the removal of the “ASHLGLAR” peptide, with a molecular weight of 824 Da, from the C-terminus of C3a (Fig. 3C, 3D; Supplemental Fig. S3A). The peptide sequence was confirmed by CID fragmentation analysis (Supplemental Fig. S3B). The KLK14-mediated proteolytic fragmentation of C3a may also explain the immunologic detection pattern in the C3a ELISA (Supplemental Fig. S2), which showed a decrease of the initial detection signal exhibited by the KLK14-incubated C3 fragments at and beyond 30 min of reaction; even though a polyclonal anti-C3a antibody has been used, the possibility that the dominant epitope involves the C-terminal region of C3a cannot be excluded. Sustained time-dependent detection of C3a released by KLK14 was enhanced further at lower ratios of enzyme to substrate, down to 0.01% of C3 (w/w/; comparative MALDI-TOF analyses shown for KLK14 in Supplemental Fig. S3A), while in contrast, fragmentation by trypsin similar to that of KLK14 could only be observed when the enzyme to substrate ratio used was lower than 0.01% (w/w/; data not shown). Notably, in our calculations, the comparable molecular weights of KLK14 and trypsin resulted in similar molar concentrations of either trypsin or KLK14 used at the equivalent enzyme to substrate w/w levels.
Figure 3. Mass spectrometric identification of the C3a-like fragments released by KLK14.
A) The release by KLK14 of two distinct molecular weight species, C3a and a lower molecular weight C3a component, was investigated by LC-MSE within the 60-min time-frame of incubation with the enzyme. B) A graphical representation of the LC-MSE spectrum area of the C3 fragments proteolytically released by KLK14 (panel A) is shown. Truncated C3a is indicated as tr.C3a. C) Fragmentation of C3a by KLK14 under similar conditions is also shown for comparison purposes. The molar ratio of enzyme to C3 was similar to that of enzyme to C3a, corresponding to a w/w enzyme to C3a ratio of 0.5% for C3 and 10% for C3a. D) The C-terminal part of C3 is indicated, with the cleavage resulting in the release of “active” C3a fragment represented with black arrow and the KLK14-mediated subsequent cleavage shown with a grey arrow. Panels A and C show representative data from two independent experiments with similar results.
KLK14-mediated C3 fragmentation triggers cell activation
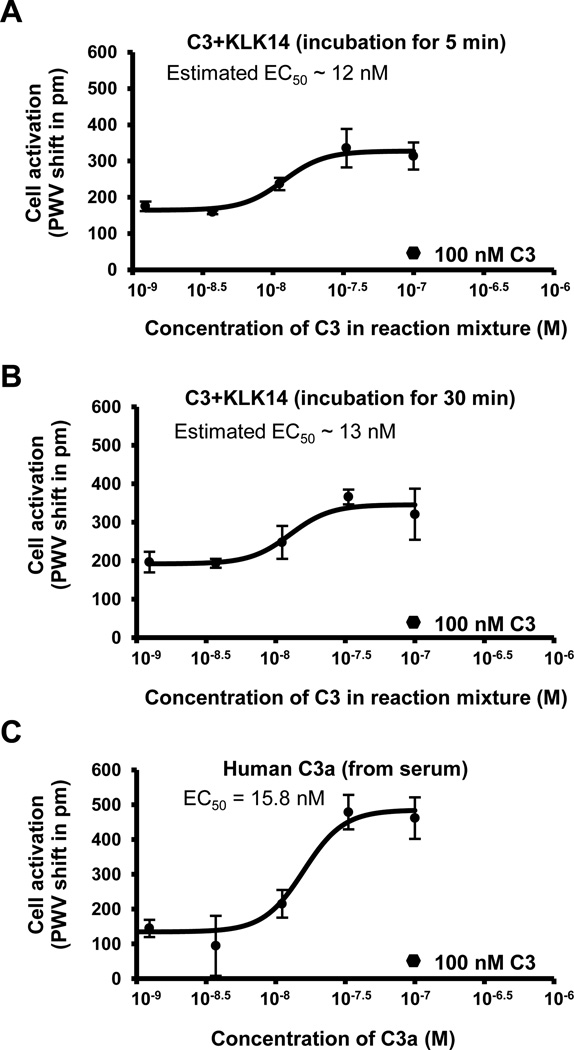
To investigate the biological function of the C3a-like fragments released from C3 by KLK14, we measured their signaling response in RBL-C3aR cells using the SRU BIND® photonic crystal sensor platform, which has been previously used to characterize anaphylatoxin signaling (26). Treatment of the RBL-C3aR cells with serial dilutions of C3 previously incubated with KLK14 for two time points, which were selected based on the MS-derived time course (Fig. 3), resulted in detectable cell activation with an estimated EC50 in the low (~12–13) nanomolar range (Fig. 4A, 4B). In contrast, pure C3 at 100 nM, which was used as a control, did not induce a significant response (Fig. 4; black hexagons). Importantly, the C3aR-mediated activity for the KLK14:C3 mixtures was in a similar range as for purified C3a (EC50 = 15.8 nM; Fig. 4C); C3a specifically acted on C3aR-RBL and did not stimulate RBL cells transfected with the C5a receptor (Supplemental Fig. S4A–B). Of note, the absolute concentration of biologically active C3a generated by KLK14 in our samples cannot be reliably determined and likely contains a mixture of intact and truncated C3a after 5 and 30 min of incubation with the enzyme (Fig. 3); given the importance of the arginated C-terminus for C3aR signaling (3,26), it is not expected that the truncated C3a fragments participate in active signaling. Indeed, incubation of C3a with KLK14 for 5 min led to visible decrease of the activation signal on C3aR-RBL cells when compared with time 0 (Supplemental Fig. S4C). Similarly, in the case of our transfected cell system, a role for other C3 species released during the proteolytic degradation of the remaining α-, α΄-, and/or β-chains is improbable. Since serum-derived proteinase inhibitors are known to restrict KLK14 enzyme activity (28), and the KLK14-mediated cell stimulation ability of C3 was abrogated in the presence of serum (Supplemental Fig. S4D), no further significant degradation of C3 fragments by KLK14 during this cell assay is expected after addition to serum-supplemented media. Similarly, the use of heat-inactivated serum renders significant desargination of C3a by carboxypeptidase N within the time frame of our experiment rather unlikely. The reason and importance of the observed lower maximal signal exhibited by the KLK14-mediated C3 fragments (Fig. 4A, 4B) as opposed to the C3a control (Fig. 4C) is not yet determined; besides experimental variability in cell seeding, a potential interference of components in the more complex matrix of the KLK14:C3 mixture cannot be excluded. Experiments with purified or recombinantly produced KLK14-induced C3 fragments may be performed in the future to better explain the C3aR-mediated signaling pattern more quantitatively. Still, the current semi-quantitative data on RBL-C3aR cells strongly indicate a biological activity for the C3 fragments that results from the proteolytic action of KLK14.
Figure 4. RBL-C3aR cell signaling mediated by the proteolytic action of KLK14.
A–B) Cell responses [peak wavelength value (PWV) shift in pm] triggered by C3aR signals were recorded in response to the KLK14-mediated C3 fragments after 5 min (A) or 30 min (B) of incubation (n=3). The x-axis represents the diluted concentration of C3 that was added to the reaction mixture with KLK14. C) Cell responses triggered by C3aR signals were recorded in response to C3a anaphylatoxin (n=2). The x-axis represents the C3a concentration. A typical cell response (average PWV shift of about 17 pm) to 100 nM of C3 (n=3) is also shown as a black hexagon in these graphs for comparison purposes.
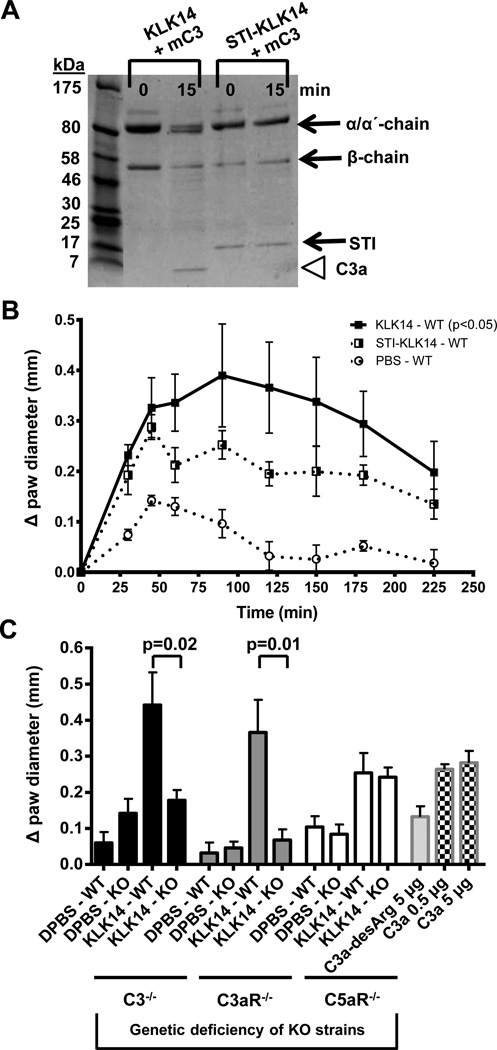
Administration of KLK14 results in complement-dependent paw edema in vivo
Based on our biochemical findings that KLK14 is a potential activator of human C3 (Figs. 1–3) that can induce C3aR-dependent cell signaling (Fig. 4), we hypothesized that the KLK14-released C3a-like fragments may exert pro-inflammatory roles. We therefore used a well-established mouse model of paw edema (34) in combination with different complement-deficient animals. As a first step, we confirmed that KLK14 was able to induce a similar cleavage pattern in vitro using mouse C3 as seen in human C3 incubations with KLK14 (above). Indeed, incubation of mouse C3 with KLK14 resulted in the cleavage of the α-chain with appearance of a C3a-sized band; this conversion was prevented in the presence of the STI-inhibited KLK14 (Fig. 5A). The intraplantar administration of KLK14 in paws of wild-type C57BL/6J mice or littermate controls of complement-deficient animals was able to induce a statistically significant edema response (Fig. 5B) comparable to that previously observed for trypsin (34) and formerly reported for KLK14 (35,36). The KLK14-generated response was attenuated by pre-incubation with STI (Fig. 5B). Further, this KLK14-mediated edema response was sustained at 120 min post-injection, in contrast to inactivated KLK14 (Fig. 5B) and vehicle alone (Fig. 5B and Fig. 5C: KLK14-WT bars). Similarly, intraplantar C3a injections in wild-type C57BL/6J mice resulted in a paw edema response at 120 min post-injection equivalent to that induced by KLK14 (Fig. 5C, square-dotted bars). As expected, the tissue edema response to C3a-desArg was significantly lower (Fig. 5C, light grey bar) when compared to C3a. The significance of the residual C3a-desArg-mediated response is not yet clear; while C3a-desArg does not bind to and activate C3aR (3,26), activities of this fragment potentially independent of C3aR have been reported in similar settings (37,38). In contrast, administration of KLK14 in C3-deficient animals resulted in a significantly decreased edema response 120 min post-injection and equivalent to edema caused by PBS alone (Fig. 5C, black bars). In agreement with these data, intraplantar KLK14 did not cause an edema response in the C3aR-deficient animals (Fig. 5C, grey bars), indicating that in this model the KLK14-mediated edema response is essentially C3a/C3aR-dependent. Notably, the swelling responses to vehicle alone were comparable in all animals. The same was true for the inactivated KLK14 responses in wild type (receptor-expressing) animals (not shown). Furthermore, since genetic deletion of the major skin proteinase-receptor, PAR2, cannot eliminate the murine paw edema response observed after intraplantar KLK14 administration [(36); K. Oikonomopoulou, unpublished data], the paw swelling observed in this model was not attributed to the previously reported KLK14-PAR2 signaling (25,35,39). Of importance, in the C5aR-deficient animals the paw swelling in response to KLK14 was equivalent to that in the wild-type littermate mice (Fig. 5C, white bars). This result renders a role for C5a in generating the KLK14-mediated inflammatory response very unlikely and further supports a C3b/C5-independent KLK14 edema pathway. Notably, these in vivo data are also in agreement with our in vitro findings of limited human C5 degradation by KLK14 that does not generate C5-derived effector fragments (Fig. 2C).
Figure 5. Edema response triggered by intraplantar injections of KLK14 in complement-deficient and wild-type mice.
A) Generation of C3a by cleavage of mouse C3 (mC3) is shown in the presence of active KLK14 enzyme (the C3a-like fragment is indicated with a white arrowhead); the absence of C3a release in the presence of STI-inhibited KLK14 is also demonstrated. B) Intraplantar administration of KLK14 resulted in a paw edema response, which was increased (p<0.05) compared with the PBS control and was partially inhibited by STI. C) A sustained response at 120 min for the animals injected with KLK14 alone, in contrast to vehicle, is depicted graphically (mean±SEM). The edema response was further shown to rely on the expression of C3 and C3aR, but not C5aR, and it was comparable to the response induced by 5 µg of C3a independently injected at the same site. The edema recorded upon administration of 0.5 µg and 5 µg of C3a and 5 µg of C3a-desArg are also shown for comparison purposes. In vivo treatments were performed in at least 4 mice per group and in three independently-organized experiments. Incubation of KLK14 with STI was performed for 30 min at 37°C using 1:5 molar ratio of enzyme to inhibitor.
Discussion
The main finding of our work is that KLK14, a prominent member of the kallikrein-related peptidase family, can generate the C3a anaphylatoxin from C3 in vitro and, presumably due to a comparable cleavage, can trigger an inflammatory edema response via activation of the C3aR in vivo. Work in the past decade has revealed the tissue expression patterns of members of the kallikrein-related peptidase family. Generally, KLKs share a wide range of expression in different tissues and settings (12,13) including the blood and skin, where complement components are also present (1,15,17). Our analyses presented in this study put a spotlight on C3 as a major target of KLKs. Even though we initially investigated the C3-processing activity of three members of the KLK family, we focused our subsequent studies on KLK14 as a prototype family member with high enzymatic activity and specificity for substrates like C3 that contain an Arg or Lys at their P1 position (40).
In our study we utilized an established murine paw edema model of intraplantar inflammation that has been extensively used to study the role of trypsin-like proteinases and PAR agonists in murine edema responses in peripheral tissues, including the skin (34,41). Most importantly, a rodent paw edema model has previously been used to study the role of the C3aR and its therapeutic modulation in adjuvant-induced arthritis (42), and to investigate the contribution of complement and C5aR to inflammatory reactions triggered after surgical incision (43). In agreement with our previous findings (35,36), which singled out PARs as potential targets of the KLK14 degradative activity that can potentially result in cell signaling, tissue inflammation, and edema (25,35,39), our current work confirmed that KLK14 can trigger swelling when injected intraplantarly in mice. In addition, the observations in this study critically extend this model by suggesting a role for KLK action via proteolytic mechanisms in addition to the ability to signal via PARs. The data reported here thereby implicate the targeting of the complement system by KLKs as an important regulator of inflammatory responses in peripheral tissues exemplified by our intraplantar model.
Specifically for the skin tissue, while several studies have investigated a role for the KLK cascade in skin homeostasis (i.e. desquamation, matrix remodelling) (13,14), their involvement in skin pathologies is not yet clear. Upregulation of KLKs in several skin diseases has been reported (13,14); for example, a role for proteinase-activated receptor (PAR) signaling by KLK14 in inflamed rosacea tissues (44) has been suggested. However, in a murine model of the Netherton syndrome, a pathological condition where activity of KLKs is highly increased, genetic deletion of the proteinase receptor was not able to abrogate cutaneous inflammation (45). Thus, our results offer a potential alternative mechanism of KLK-induced inflammation through complement activation that may contribute to skin pathologies.
The complement system, like the PARs (46), represents an important innate immune defense mechanism triggered by proteolytic activation of effector molecules (1,2). Complement activation in tissues in connection with local inflammatory processes is still poorly described and the absolute levels of its components in peripheral tissues, like the skin, are largely unknown. The inflammatory actions of complement, primarily a result of anaphylatoxin signaling, can, among other functions, induce cytokine release by immune and non-immune cells and regulate vascular permeability and cell adhesion (1,47), functions that can be of importance in the paw edema inflammation model used in this work and by other groups (42,43). Our study now shows that KLKs, and more specifically KLK14, can modulate C3a-mediated signaling in this established model of peripheral tissue inflammation.
More specifically, our data presented herein indicate that the C3aR can be a potential mediator of KLK-mediated localized inflammation in peripheral tissues including the skin. Based on the impaired reaction in C3aR−/− mice, the edema response is likely driven by the KLK-catalyzed generation of active complement C3a that stimulate C3aR. This is in agreement with previous work that suggested a role of C3aR in edema models of peripheral tissue inflammation (42). Although a role for secondary KLK14-mediated C3 fragmentation products in pro-inflammatory reactions cannot be fully excluded, a participation of the secondary C3a truncation fragment that was identified in our in vitro assays seems unlikely. In contrast to C5a, where the carboxypeptidase-processed product C5a-desArg still exerts signaling functions via C5aR, the corresponding truncation of C3a by even one C-terminal amino acid (i.e., to C3a-desArg) leads to complete loss of signaling activity (3,26). In the present study, we did not find evidence to support activation of C5aR signaling by KLK14 in the same setting, a finding which corresponds well with the observation that KLK14 exerts very limited activity towards C5. While the physiologically-relevant C3 and KLK14 concentrations in peripheral tissues are not known, the C3 levels utilized by our in vitro assays were close to typically observed plasma concentrations of C3 (~1–1.5 mg/ml) (48). Notably, as dictated by our in vitro analyses, the levels of KLK14 needed for activation of 1.5 mg/ml of C3 (about 8 µM) can be as low as 0.15 µg/ml (about 5 nM), an enzyme expression level that can be observed in tissue extracts and biological fluids (49).
Our new data indicating that KLK14 is able to stimulate C3aR-mediated cell/tissue responses adds C3aR, like PARs (25,39), to the group of G protein-coupled receptors involved in KLK-mediated inflammatory processes. Therefore, future work should focus on elucidating the mechanisms of the inflammatory action of KLK14 in peripheral tissues by activating the two receptor systems simultaneously (i.e., both PARs and C3aR). It will be of much interest to determine the role this process may play in the innate immune response in tissues like the skin. Moreover, it would be important to further investigate whether any other C3/C3a cleavage products generated by KLK14 may contribute to biological effects, including C3aR-independent signaling or antimicrobial functions. A similar hypothesis has been recently put forward for the processing of C5 by thrombin, leading to a unique C5b fragment with enhanced cell lytic activity (50).
In view of the recent literature that draws links between the KLK and thrombostasis proteolytic cascades (51), our new work suggests dual targets for these serine proteinases: (a) the PARs, which are activated by both coagulation enzymes and the KLKs (46), and (b) the complement system, which is affected by several coagulation factors (8,52) as well as by KLKs, as we now demonstrate. Thus, these three interconnected proteolytic cascades (thrombostasis, complement, and that of the KLKs) may well be considered as modulators and amplifiers of the innate immune response in the inflammatory microenvironment.
In conclusion, we provide the first evidence that kallikrein-related peptidases, apart from signaling via PARs and having degradative roles important for extracellular matrix remodeling and cell shedding, can exert inflammatory signaling in peripheral tissues including the skin by targeting complement component C3 and triggering release of C3a that, in turn, activates C3aR. This role of KLKs in modulating the availability of C3a anaphylatoxin adds to the recent literature that proposes KLKs as inflammatory modulators of cathelicidin activity and PAR signaling (13). In the future, it may therefore be interesting to extend these investigations to include other tissue-localized KLKs, PARs, and complement components in our models of peripheral tissue inflammation in order to arrive at a more complete picture of this intriguing crosstalk. Our data may also be of relevance for developing new therapeutic strategies for the treatment of inflammatory or cancerous skin conditions, where both KLKs and complement have been implicated.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Drs. E. Reis, I. Kourtzelis, A. Tzekou, G. Sfyroera, S. Rafail, and M. Neher, and A. Ulndreaj (all at the University of Pennsylvania, Philadelphia, PA, USA) for useful discussions during performance of this project, and Dr. R. Ramachandran and K. Chapman (from University of Calgary, Calgary, AB, Canada) for training on the paw edema assay.
These studies were supported by grants from the National Institutes of Health (NIH AI030040, AI068730, AI072106, AI097805, EY020633, GM097747: JDL, DR) and from the Canadian Institutes of Health Research (CIHR: EPD, MDH). KO is a recipient of an Alberta Heritage Foundation for Medical Research (AHFMR) and a National Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Fellowship.
Abbreviations used in this article
- ACN
acetonitrile
- AMC
7-amino-4-methylcoumarin
- C3aR
C3a receptor
- C5aR
C5a receptor
- C5L2
C5a receptor-like 2
- CID
collision-induced dissociation
- DPBS
Dulbecco's phosphate-buffered Saline
- ESI
electrospray ionization
- HEK
human embryonic kidney cells
- KLKs
kallikrein-related peptidases
- LC-MS
liquid chromatography-mass spectrometry
- PARs
proteinase-activated receptors
- PWV
peak wavelength values
- RBL-2H3
rat basophilic leukemia cells
- STI
soybean trypsin inhibitor
- UPLC
ultra performance liquid chromatography
Footnotes
Disclosures
The authors have no financial conflicts of interest.
Reference List
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Friec G, Kemper C. Complement: coming full circle. Arch. Immunol. Ther. Exp. (Warsz.) 2009;57:393–407. doi: 10.1007/s00005-009-0047-4. [DOI] [PubMed] [Google Scholar]
- 3.Klos A, Wende E, Wareham KJ, Monk PN. International Union of Pharmacology. LXXXVII. Complement peptide C5a, C4a, and C3a receptors. Pharmacol. Rev. 2013;65:500–543. doi: 10.1124/pr.111.005223. [DOI] [PubMed] [Google Scholar]
- 4.Eggertsen G, Hellman U, Lundwall A, Folkersen J, Sjoquist J. Characterization of tryptic fragments of human complement factor C3. Mol. Immunol. 1985;22:833–841. doi: 10.1016/0161-5890(85)90067-7. [DOI] [PubMed] [Google Scholar]
- 5.Wetsel RA, Kolb WP. Complement-independent activation of the fifth component (C5) of human complement: limited trypsin digestion resulting in the expression of biological activity. J. Immunol. 1982;128:2209–2216. [PubMed] [Google Scholar]
- 6.Wiggins RC, Giclas PC, Henson PM. Chemotactic activity generated from the fifth component of complement by plasma kallikrein of the rabbit. J. Exp. Med. 1981;153:1391–1404. doi: 10.1084/jem.153.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Discipio RG. The activation of the alternative pathway C3 convertase by human plasma kallikrein. Immunology. 1982;45:587–595. [PMC free article] [PubMed] [Google Scholar]
- 8.Amara U, Flierl MA, Rittirsch D, Klos A, Chen H, Acker B, Bruckner UB, Nilsson B, Gebhard F, Lambris JD, Huber-Lang M. Molecular intercommunication between the complement and coagulation systems. J. Immunol. 2010;185:5628–5636. doi: 10.4049/jimmunol.0903678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orr FW, Varani J, Kreutzer DL, Senior RM, Ward PA. Digestion of the fifth component of complement by leukocyte enzymes. Sequential generation of chemotactic activities for leukocytes and for tumor cells. Am. J. Pathol. 1979;94:75–83. [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuoka Y, Xia HZ, Sanchez-Munoz LB, Dellinger AL, Escribano L, Schwartz LB. Generation of anaphylatoxins by human beta-tryptase from C3, C4, and C5. J. Immunol. 2008;180:6307–6316. doi: 10.4049/jimmunol.180.9.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wingrove JA, Discipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- 12.Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer. 2004;4:876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 13.Sotiropoulou G, Pampalakis G. Kallikrein-related peptidases: bridges between immune functions and extracellular matrix degradation. Biol. Chem. 2010;391:321–331. doi: 10.1515/BC.2010.036. [DOI] [PubMed] [Google Scholar]
- 14.Eissa A, Diamandis EP. Human tissue kallikreins as promiscuous modulators of homeostatic skin barrier functions. Biol. Chem. 2008;389:669–680. doi: 10.1515/BC.2008.079. [DOI] [PubMed] [Google Scholar]
- 15.Klos A, Tenner AJ, Johswich KO, Ager RR, Reis ES, Kohl J. The role of the anaphylatoxins in health and disease. Mol. Immunol. 2009;46:2753–2766. doi: 10.1016/j.molimm.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nestle FO, Di MP, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 2009;9:679–691. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasch MC, van den Bosch NH, Daha MR, Bos JD, Asghar SS. Synthesis of complement components C3 and factor B in human keratinocytes is differentially regulated by cytokines. J. Invest Dermatol. 2000;114:78–82. doi: 10.1046/j.1523-1747.2000.00841.x. [DOI] [PubMed] [Google Scholar]
- 18.Dovezenski N, Billetta R, Gigli I. Expression and localization of proteins of the complement system in human skin. J. Clin. Invest. 1992;90:2000–2012. doi: 10.1172/JCI116080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purwar R, Wittmann M, Zwirner J, Oppermann M, Kracht M, ttrich-Breiholz O, Gutzmer R, Werfel T. Induction of C3 and CCL2 by C3a in keratinocytes: a novel autocrine amplification loop of inflammatory skin reactions. J. Immunol. 2006;177:4444–4450. doi: 10.4049/jimmunol.177.7.4444. [DOI] [PubMed] [Google Scholar]
- 20.Kotnik V. Complement in skin diseases. Acta Dermatovenerol. Alp Panonica. Adriat. 2011;20:3–11. [PubMed] [Google Scholar]
- 21.Markiewski MM, Lambris JD. Unwelcome complement. Cancer Res. 2009;69:6367–6370. doi: 10.1158/0008-5472.CAN-09-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jean D, Bar-Eli M, Huang S, Xie K, Rodrigues-Lima F, Hermann J, Frade R. A cysteine proteinase, which cleaves human C3, the third component of complement, is involved in tumorigenicity and metastasis of human melanoma. Cancer Res. 1996;56:254–258. [PubMed] [Google Scholar]
- 23.Basta M, Hammer CH. A rapid FPLC method for purification of the third component of human and guinea pig complement. J. Immunol Methods. 1991;142:39–44. doi: 10.1016/0022-1759(91)90290-v. [DOI] [PubMed] [Google Scholar]
- 24.Oikonomopoulou K, Hansen KK, Baruch A, Hollenberg MD, Diamandis EP. Immunofluorometric activity-based probe analysis of active KLK6 in biological fluids. Biol. Chem. 2008;389:747–756. doi: 10.1515/BC.2008.086. [DOI] [PubMed] [Google Scholar]
- 25.Oikonomopoulou K, Hansen KK, Saifeddine M, Tea I, Blaber M, Blaber SI, Scarisbrick I, ndrade-Gordon P, Cottrell GS, Bunnett NW, Diamandis EP, Hollenberg MD. Proteinase-activated receptors, targets for kallikrein signaling. J. Biol. Chem. 2006;281:32095–32112. doi: 10.1074/jbc.M513138200. [DOI] [PubMed] [Google Scholar]
- 26.Reis ES, Chen H, Sfyroera G, Monk PN, Kohl J, Ricklin D, Lambris JD. C5a receptor-dependent cell activation by physiological concentrations of desarginated C5a: insights from a novel label-free cellular assay. J. Immunol. 2012;189:4797–4805. doi: 10.4049/jimmunol.1200834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shamah SM, Cunningham BT. Label-free cell-based assays using photonic crystal optical biosensors. Analyst. 2011;136:1090–1102. doi: 10.1039/c0an00899k. [DOI] [PubMed] [Google Scholar]
- 28.Goettig P, Magdolen V, Brandstetter H. Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs) Biochimie. 2010;92:1546–1567. doi: 10.1016/j.biochi.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Circolo A, Garnier G, Fukuda W, Wang X, Hidvegi T, Szalai AJ, Briles DE, Volanakis JE, Wetsel RA, Colten HR. Genetic disruption of the murine complement C3 promoter region generates deficient mice with extrahepatic expression of C3 mRNA. Immunopharmacology. 1999;42:135–149. doi: 10.1016/s0162-3109(99)00021-1. [DOI] [PubMed] [Google Scholar]
- 30.Hopken UE, Lu B, Gerard NP, Gerard C. The C5a chemoattractant receptor mediates mucosal defence to infection. Nature. 1996;383:86–89. doi: 10.1038/383086a0. [DOI] [PubMed] [Google Scholar]
- 31.Kildsgaard J, Hollmann TJ, Matthews KW, Bian K, Murad F, Wetsel RA. Cutting edge: targeted disruption of the C3a receptor gene demonstrates a novel protective anti-inflammatory role for C3a in endotoxin-shock. J. Immunol. 2000;165:5406–5409. doi: 10.4049/jimmunol.165.10.5406. [DOI] [PubMed] [Google Scholar]
- 32.Brattsand M, Stefansson K, Lundh C, Haasum Y, Egelrud T. A proteolytic cascade of kallikreins in the stratum corneum. J. Invest Dermatol. 2005;124:198–203. doi: 10.1111/j.0022-202X.2004.23547.x. [DOI] [PubMed] [Google Scholar]
- 33.Bokisch VA, Dierich MP, Muller-Eberhard HJ. Third component of complement (C3): structural properties in relation to functions. Proc. Natl. Acad. Sci. U. S. A. 1975;72:1989–1993. doi: 10.1073/pnas.72.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paszcuk AF, Quintao NL, Fernandes ES, Juliano L, Chapman K, ndrade-Gordon P, Campos MM, Vergnolle N, Calixto JB. Mechanisms underlying the nociceptive and inflammatory responses induced by trypsin in the mouse paw. Eur. J. Pharmacol. 2008;581:204–215. doi: 10.1016/j.ejphar.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 35.Oikonomopoulou K, Hansen KK, Saifeddine M, Vergnolle N, Tea I, Blaber M, Blaber SI, Scarisbrick I, Diamandis EP, Hollenberg MD. Kallikrein-mediated cell signalling: targeting proteinase-activated receptors (PARs) Biol. Chem. 2006;387:817–824. doi: 10.1515/BC.2006.104. [DOI] [PubMed] [Google Scholar]
- 36.Oikonomopoulou K, Hansen KK, Chapman K, Vergnolle N, Diamandis EP, Hollenberg MD. Kallikrein-mediated activation of PARs in inflammation and nociception. Inflammation Research. 2007;56:S499–S502. [Google Scholar]
- 37.Fukuoka Y, Hugli TE. Anaphylatoxin binding and degradation by rat peritoneal mast cells. Mechanisms of degranulation and control. J. Immunol. 1990;145:1851–1858. [PubMed] [Google Scholar]
- 38.Mousli M, Hugli TE, Landry Y, Bronner C. A mechanism of action for anaphylatoxin C3a stimulation of mast cells. J. Immunol. 1992;148:2456–2461. [PubMed] [Google Scholar]
- 39.Gratio V, Loriot C, Virca GD, Oikonomopoulou K, Walker F, Diamandis EP, Hollenberg MD, Darmoul D. Kallikrein-related peptidase 14 acts on proteinase-activated receptor 2 to induce signaling pathway in colon cancer cells. Am. J. Pathol. 2011;179:2625–2636. doi: 10.1016/j.ajpath.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borgono CA, Michael IP, Shaw JL, Luo LY, Ghosh MC, Soosaipillai A, Grass L, Katsaros D, Diamandis EP. Expression and functional characterization of the cancer-related serine protease, human tissue kallikrein 14. J. Biol. Chem. 2007;282:2405–2422. doi: 10.1074/jbc.M608348200. [DOI] [PubMed] [Google Scholar]
- 41.Vergnolle N, Hollenberg MD, Sharkey KA, Wallace JL. Characterization of the inflammatory response to proteinase-activated receptor-2 (PAR2)-activating peptides in the rat paw. Br. J. Pharmacol. 1999;127:1083–1090. doi: 10.1038/sj.bjp.0702634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ames RS, Lee D, Foley JJ, Jurewicz AJ, Tornetta MA, Bautsch W, Settmacher B, Klos A, Erhard KF, Cousins RD, Sulpizio AC, Hieble JP, McCafferty G, Ward KW, Adams JL, Bondinell WE, Underwood DC, Osborn RR, Badger AM, Sarau HM. Identification of a selective nonpeptide antagonist of the anaphylatoxin C3a receptor that demonstrates antiinflammatory activity in animal models. J Immunol. 2001;166:6341–6348. doi: 10.4049/jimmunol.166.10.6341. [DOI] [PubMed] [Google Scholar]
- 43.Clark JD, Qiao Y, Li X, Shi X, Angst MS, Yeomans DC. Blockade of the complement C5a receptor reduces incisional allodynia, edema, and cytokine expression. Anesthesiology. 2006;104:1274–1282. doi: 10.1097/00000542-200606000-00024. [DOI] [PubMed] [Google Scholar]
- 44.Stefansson K, Brattsand M, Roosterman D, Kempkes C, Bocheva G, Steinhoff M, Egelrud T. Activation of proteinase-activated receptor-2 by human kallikrein-related peptidases. Journal of Investigative Dermatology. 2008;128:18–25. doi: 10.1038/sj.jid.5700965. [DOI] [PubMed] [Google Scholar]
- 45.Briot A, Lacroix M, Robin A, Steinhoff M, Deraison C, Hovnanian A. Par2 inactivation inhibits early production of TSLP, but not cutaneous inflammation, in Netherton syndrome adult mouse model. J. Invest Dermatol. 2010;130:2736–2742. doi: 10.1038/jid.2010.233. [DOI] [PubMed] [Google Scholar]
- 46.Ramachandran R, Noorbakhsh F, Defea K, Hollenberg MD. Targeting proteinase-activated receptors: therapeutic potential and challenges. Nat. Rev. Drug Discov. 2012;11:69–86. doi: 10.1038/nrd3615. [DOI] [PubMed] [Google Scholar]
- 47.Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J Immunol. 2013;190:3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reis ES, Falcao DA, Isaac L. Clinical aspects and molecular basis of primary deficiencies of complement component C3 and its regulatory proteins factor I and factor H. Scand. J Immunol. 2006;63:155–168. doi: 10.1111/j.1365-3083.2006.01729.x. [DOI] [PubMed] [Google Scholar]
- 49.Shaw JL, Diamandis EP. Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem. 2007;53:1423–1432. doi: 10.1373/clinchem.2007.088104. [DOI] [PubMed] [Google Scholar]
- 50.Krisinger MJ, Goebeler V, Lu Z, Meixner SC, Myles T, Pryzdial EL, Conway EM. Thrombin generates previously unidentified C5 products that support the terminal complement activation pathway. Blood. 2012;120:1717–1725. doi: 10.1182/blood-2012-02-412080. [DOI] [PubMed] [Google Scholar]
- 51.Blaber M, Yoon H, Juliano MA, Scarisbrick IA, Blaber SI. Functional intersection of the kallikrein-related peptidases (KLKs) and thrombostasis axis. Biol. Chem. 2010;391:311–320. doi: 10.1515/BC.2010.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oikonomopoulou K, Ricklin D, Ward PA, Lambris JD. Interactions between coagulation and complement--their role in inflammation. Semin. Immunopathol. 2012;34:151–165. doi: 10.1007/s00281-011-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.