Abstract
To understand the emergence of human higher cognition, we must understand its biological substrate—the cerebral cortex, which considers itself the crowning achievement of evolution. Here, we describe how advances in developmental neurobiology, coupled with those in genetics, including adaptive protein evolution via gene duplications and the emergence of novel regulatory elements, can provide insights into the evolutionary mechanisms culminating in the human cerebrum. Given that the massive expansion of the cortical surface and elaboration of its connections in humans originates from developmental events, understanding the genetic regulation of cell number, neuronal migration to proper layers, columns, and regions, and ultimately their differentiation into specific phenotypes, is critical. The pre- and postnatal environment also interacts with the cellular substrate to yield a basic network that is refined via selection and elimination of synaptic connections, a process that is prolonged in humans. This knowledge provides essential insight into the pathogenesis of human-specific neuropsychiatric disorders.
Introduction
Since the time of Darwin's The Origin of Species about 200 years ago, there has been little disagreement among scientists that the brain, and more specifically its covering, cerebral cortex, is the organ that enables human extraordinary cognitive capacity that includes abstract thinking, language, and other higher cognitive functions. Thus, it is surprising that relatively little attention has been given to the study of how the human brain has evolved and become different from other mammals or even other primates (Clowry et al., 2010). Yet, the study of human brain evolution is essential for understanding causes and to possibly develop cures for diseases in which some of the purely human behaviors may be disrupted, as in dyslexia, intellectual disability (ID), attention deficit hyperactivity disorder (ADHD), autism spectrum disorder (ASD), and schizophrenia, as well as a number of human-specific neurodegenerative conditions including Alzheimer's disease (e.g., Casanova and Tillquist, 2008; Geschwind and Konopka, 2009; Knowles and McLysaght, 2009; Li et al., 2010; Miller et al., 2010; Preuss et al., 2004; Xu et al., 2010).
Traditionally, it is comparative anatomy that has informed our understanding of how our brain may have evolved over 300 million years of mammalian evolution (Kaas, 2013; Preuss, 1995). These studies left no doubt that the human cerebral cortex has expanded significantly relative to other hominids, including introduction of new regions in the frontal and parieto-temporal lobes in humans (Dunbar, 1993; Fjell et al., 2013; Preuss, 1995; Rakic, 2009; Teffer and Semendeferi, 2012). It also became evident that although the basic principles of brain development in all mammals may be conserved, the modifications of developmental events during evolution produce not only quantitative but qualitative changes as well (Table 1).
Table 1.
Differences between Developing Human and Mouse Neocortex
| Quantitative Differences* | Qualitative Differences |
|---|---|
| • Number of neurons and surface area ~1000 × larger | • Introduction of new genes, gene variants, regulatory elements, and expression patterns |
| • Number or ontogenetic (radial) columns ~1000× higher | • Introduction of new neuronal types (e.g. Predecessors, VEN -Spindle cells; ISN) |
| • Length of cell cycle length (~ 3- 4 × longer) | • Distinct upper stratum of the outer subventricular zone (OSVZ) |
| • Duration of cortical neurogenesis ~ 20 × longer | |
| • Subplate Zone occupies several-fold larger portion of the embryonic cerebral wall | • Subset of GABAergic interneurons originate from the dorsal VZ/SVZ |
| • Configuration gyrencephalic rather than lissencephalic | • Neuronal migration to thalamus from the GE absent in all examined mammals (CGT) |
| • Birth occurs during later stages of cortical development | • Transient Subpial Granular Layer (SGL, absent in rodents and carnivores) |
| • Lower density of neurons, larger neuropil, higher dendritic and axonal branching | • Distinct Radial Glial Cells (early GFAP+ and differentiated and genetically distinct) |
| • Modification of common cytoarchitectonic areas (e.g. layer IV in A17) | |
| • Tempo of cortical maturation in relation to onset of reproduction very different | • Introduction of new cytoarchitectonic areas (e.g. A22, 28, 44, 45, 46) |
| • Very prolonged neoteny with cortical maturation surpassing puberty and adolescence | • Absence of neuronal turnover and resistance to regeneration |
The quantitative statements above are only an approximation of the level of an order of magnitude to highlight how large the differences are. The inclusion of all the references and precise numbers and measurements that may differ due to the individual variations within species as well as the methods used are not practical or appropriate in this short Perspective article.
Due to the limits of the space, we cannot provide a comprehensive review of this wide-ranging topic. Instead, we will focus on the expansion and elaboration of the human cerebral neocortex and provide our own personal perspective on some of the key advances in this area, including the high promise, as well as enormous challenges ahead. We organize our thoughts into two major areas—the phenotype-driven and genome-driven approaches, which, unfortunately, only rarely meet in the middle. Our hope is that in the near future, it will be possible to connect some of the known human genetic adaptations to the developmental and maturational features that underlie uniquely human cognitive abilities.
The Phenotype-Based Approach
Cortical Expansion
It is well established that the expansion of the cortex occurs primarily in surface area rather than in thickness. This is most pronounced in anthropoid primates, including humans, in which the neocortex comprises up to 80% of the brain mass. We have also known for a long time that the neocortex is subdivided into distinct cytoarchitectonic areas with neurons organized in horizontal layers or laminae, and vertical (radial) columns or modules, which have increased in number, size, and complexity during cortical evolution (Mountcastle, 1995; Goldman-Rakic, 1987). Of course, brain size is not simply a matter of cell number; it also reflects cell density arrangements and connectivity (Herculano-Houzel et al., 2008), which is relevant here, as the distance between cell bodies in the cerebral cortex, especially prefrontal regions of humans, is greater than in other primates (Semendeferi et al., 2011). Thus, three essential features account for the changes in cerebral size over mammalian evolution: large changes in cell number, morphology, and composition.
However, it is not sufficient to enlarge the entire brain, as Neanderthals had large brains, and modern human brain size may differ by 2-fold among individuals. From this perspective, many genes that modify cell cycle can increase or decrease brain size but not necessarily in a manner that is relevant to cerebral evolution. A salient recent example worth discussing is the sophisticated analysis of the function of BAF-170 in mouse brain development (Tuoc et al., 2013). This study shows that BAF-170 controls cortical neurogenesis via modulating the intermediate progenitor pool and therefore suggests that this could be related to enhanced intellectual capacities of primates via cortical enlargement. However, we posit that the overall expansion of the entire volume of the cortex, including its width, as well as potentially other brain regions, suggests that this particular gene is actually unlikely to be involved in the specific and selective expansion of cortical surface area occurring in primate and human brain evolution.
The cellular mechanism for the enormous cortical expansion in the surface area without a comparable increase in thickness has been first explained by the radial unit hypothesis (RUH) (Rakic, 1988). According to the RUH, tangential (horizontal) coordinates of cortical neurons are determined by the relative position of their precursor cells in the proliferative zone lining the cerebral ventricles, while their radial (vertical) position is determined by the time of their origin. Thus, the number of the radial ontogenetic columns determines the size of the cortical surface, whereas the number of cells within the columns determines the thickness. This model frames the issue of the evolution of cerebral cortical size and its thickness in the context of understanding the mechanisms governing genetic regulation of cell number and their allocation to different regions (Casanova and Tillquist, 2008; Elsen et al., 2013; Hevner and Haydar, 2012; Molnár, 2011). Furthermore, according to the RUH, the initial increase in the number of neural stem cells occurs by symmetrical divisions in the ventricular zone (VZ) before the onset of neurogenesis and the formation of the subventricular zone (SVZ) (Bystron et al., 2008; Rakic, 1988, 2009; Stancik et al., 2010). This highlights genes involved in the control of the duration and mode of cell division (symmetric/asymmetric) as important factors for cerebral expansion in evolution (Huttner and Kosodo, 2005; Rakic, 2009). Finally, the manner by which a larger number of postmitotic cells migrate radially from the proliferative VZ/SVZ to become deployed in the cortical plate as a relatively thin sheet is a biological necessity that enables cortical expansion during evolution (Heng et al., 2008; Noctor et al., 2001; Rakic, 1988, 1995; Takahashi et al., 1999; Yu et al., 2009). More recently, electroporation and transgenic technologies show intermixing of the ontogenetic columns in the SVA that is necessary for the formation of functional columns with different compositions and constellations of cell types (Figure 1A; Torii et al., 2009). However, the relation of ontogenetic columns to functional columns of the adult cortex remains to be defined (e.g., Mountcastle, 1995).
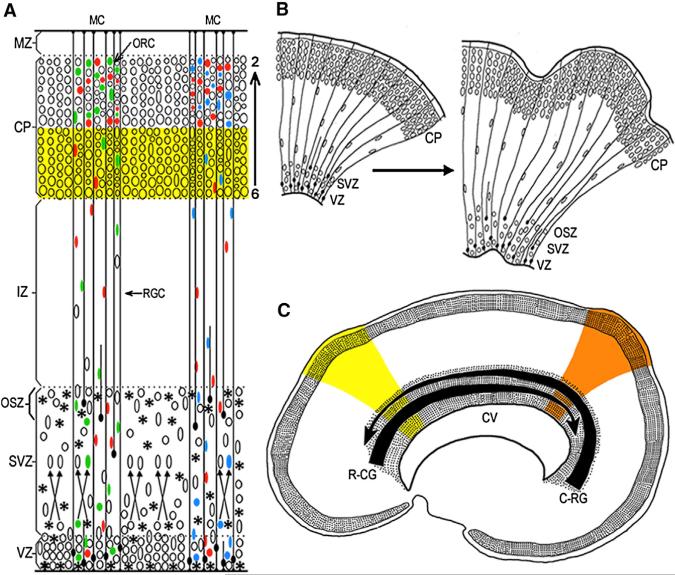
Figure 1. Radial Unit Model of the Deployment of Postmitotic Migratory Neurons and Their Settling Pattern into the Horizontal-Laminar, Inside-Out, and Vertical-Columnar Organization.
(A) Neuronal progenitors in the proliferative ventricular and subventricular zones (VZ/SVZ/OSZ) and their progenies exhibit clonal heterogeneity (indicated by the differently colored ellipses). Several clones become intermixed in the SVZ, before migrating across the intermediate zone (IZ) along elongated shafts of the radial glial cells (RGC) into the cortical plate (CP). Newborn neurons bypass previously generated cells of the deeper layers (yellow stripe) in the inside-out sequence (layers 6 to 2) to participate in pheno-typically and functionally heterogeneous mini-columns (MC) consisting of several ontogenetic radial columns (ORC) (e.g., Rakic, 1988; Torii et al., 2009).
(B) Graphic explanation of the Radial Unit Hypothesis of cortical expansion, by either preventing programmed cell death or increasing the rate of proliferation in mice, can produce a larger number of radial units that, constrained by the radial glial scaffolding, generate an expanded cellular sheet, which begins to buckle and transforms a lissencephalic (on the left) to the gyrencephalic (on the right) cerebrum. Based on studies in primates and experiments in mice (e.g., Kuida et al., 1998; Rakic, 2009; Haydar et al., 2003; Chenn and Walsh, 2003).
(C) Illustration of the concept, how opposing rostro-caudal (R-CG) and caudo-rostral (C-RG) molecular gradients, that form the protomap in embryonic VZ/SVZ lining cerebral ventricle (CV) can introduce new subtypes of neurons that migrate to the overlying CP and establish new cortical areas in the superjacent CP, indicated by yellow and orange color stripes. Based on experimental data in mice (e.g., Grove and Fukuchi-Shimogori, 2003; O'Leary and Borngasser, 2006; Cholfin and Rubenstein, 2008).
Since the length of the cell cycle is a major determinant of the number of cells produced, it is paradoxical that the duration of the cell cycle in primates is about five times longer than that in mouse (Kornack and Rakic, 1998; Lukaszewicz et al., 2006). Nevertheless, due to the greatly extended duration of cortical neurogenesis in primates (~100 days in human, 60 days in macaque monkeys—compared to 6 days in mice), the number of successive cell-division cycles that generate cortical cells can account for the enormous expansion of cortical surface (Caviness et al., 2003; Rakic, 1995). In contrast, the hypercellularity of upper layers can be attributed to the enlargement of the SVZ in human (Bystron et al., 2008). Its outer portion, termed OSVZ, has massively expanded in human and nonhuman primates, which is likely important for human brain evolution (Kennedy and Dehay, 2012; LaMonica et al., 2012). Furthermore, the 4-fold increase in the width of layer IV in primates, but not rodents, is in part a result of an increased production of cells destined for these areas in the VZ/SVZ subjacent to area 17 compared to 18 at the time of genesis of the upper layers (Kornack and Rakic, 1998; Lukaszewicz et al., 2006; Polleux et al., 1997). Thus, an explanation of genetic regulation of the length of progenitor cell divisions in the VZ/SVZ may provide clues to how these changes may have occurred during evolution (Kennedy and Dehay, 1993; Rakic, 1995; Tarui et al., 2005; Xuan et al., 1995). Finally, delay in the switch between symmetric and asymmetric divisions in the VZ/SVZ could indirectly cause the enlarged cortical surface of the cerebral cortex (Rakic, 1995). Indeed, the decrease of programmed cell death (Haydar et al., 2003; Kuida et al., 1998), or increase in number of cell cycles (Chenn and Walsh, 2002, 2003), can expand the mouse neocortical surface without an increase in its width, consistent with what may have occurred during mammalian brain evolution (Figure 1B). The elimination of the isochronously dividing cells by low doses of ionizing radiation in monkey embryos at early stages of development results in a decrease in cortical surface with little effect on its thickness, whereas later irradiation deletes individual layers and reduces cortical thickness without overall decrease in surface (Selemon et al., 2013).
The mitotic activity in the VZ can be divided into the stage before and after onset of neurogenesis that is followed by neuronal migration (Rakic, 1988). The duration of the first phase, and of the cell cycle, determines the number of radial units and, indirectly, the size of cortical areas, while duration of the second phase determines the number of neurons within each ontogenetic column. It is also during this second phase that the time of neuron origin determines laminar phenotype of generated neurons (Caviness and Rakic, 1978; McConnell, 1995; Rakic, 1974). More recent studies indicate that the switch between the two phases of cortical development may be triggered by the activation of numerous putative regulatory genes that control the mode of mitotic division and cell polarity in the VZ/SVZ including Notch, Numb, Cadherin, and AMP-activated protein kinase (e.g., Amato et al., 2011; Kwan et al., 2008; Liu et al., 2008, 2012b; Rash et al., 2013; Rasin et al., 2007; Solecki et al., 2006). Several lines of evidence from experimental manipulation as well as the pathogenesis of particular cortical malformations in humans suggest that these two phases can be separately affected: a deficit occurring during the first phase produces a cortex with a small surface area but normal or enlarged thickness (lissencephaly), whereas a defect during the phase of ontogenetic column formation produces polymicrogyria with thinner cortex and relatively normal or larger surface (e.g., Reiner et al., 1993). Although the developmental mechanisms underlying the natural occurrence and patterning of cortical gyri in other species remain largely unknown, several theories have been proposed (e.g., Van Essen, 1997).
The relevance of the process of progenitor proliferation to human brain evolution is also supported by evidence of positive selection of genes involved in regulating brain size via cell-cycle control in humans, notably ASPM and MCPH1, in which mutations cause intellectual disability in humans (Bond et al., 2002; Evans et al., 2004; Jackson et al., 2002; Kouprina et al., 2004; Zhang, 2003). Thus, although the RUH provides a framework for understanding of cortical expansion during evolution, this process requires activity of many genes. Given the high level of amino acid conservation of many of the key neurodevelopmental proteins across mammalian evolution, introduction of small modifications in the timing of developmental events via the adaptive evolution of new regulatory elements, as has occurred in limb evolution, is also likely to play a role (Cotney et al., 2013; Prabhakar et al., 2008).
Introduction of New Areas and Cells
In addition to dramatically expanding surface area, the neocortex also divided into more complex and more distinct cytoarchitectonic maps by both the differential growth of existing, as well as the introduction of novel, areas (e.g., (Krubitzer and Kaas, 2005; Preuss, 2000). The final pattern and relative size of cytoarchitectonic subdivisions of the neocortex are probably regulated by a different set of genes than those regulating neuronal number and, in addition, must be coordinated through reciprocal cell-cell interactions with various afferent systems.
The protomap hypothesis (PMH) of cortical parcellation (Rakic, 1988) postulates that intersecting gradients of molecules might be expressed across the embryonic cerebral wall that guides and attracts specific afferent systems to appropriate position in the cortex, where they can interact with a responsive set of cells. The prefix “proto” indicates the malleable character of this primordial map, as opposed to the concept of equipotential cortical plate consisting of the undifferentiated cells that is eventually shaped and subdivided entirely by the instructions from those afferents (Creutzfeldt, 1977). There has been increasing evidence of differential gene expression across the embryonic cerebral wall that indicates prospective subdivisions of the neocortex. Some of these molecules may be expressed as opposing gradients or in a region-specific manner before and/ or independently, since dilation of input does not prevent formation of at least some basic region-specific cytoarchitectonic features (Figure 1C; Arimatsu et al., 1992; Cohen-Tannoudji et al., 1994; Grove and Fukuchi-Shimogori, 2003; Miyashita-Lin et al., 1999; O'Leary and Borngasser, 2006; Rakic et al., 1991; Rubenstein and Rakic, 1999). An instructive example of how new regions could emerge via differential expansion of the cortical surface is the demonstration that frontal cortex can be enlarged in surface area without change in the size of other areas via manipulation of the transcription factors Fgf8, Fgh17, and Emx2 (Cholfin and Rubenstein, 2008). Opposing molecular gradients during development can, at some points of their intersection, provide instructions and coordinates for the creation of the new neocortical areas (Figure 1C). For example, prospective Broca and Wernicke areas, which are formed as islands in the frontal and temporal lobes display a distinct temporarily enriched gene expression pattern that is distinct from the mice or macaque cerebrum at the comparable prenatal stages (e.g. Abrahams et al., 2007; Johnson et al., 2009; Pletikos et al., 2013; Figure 2).
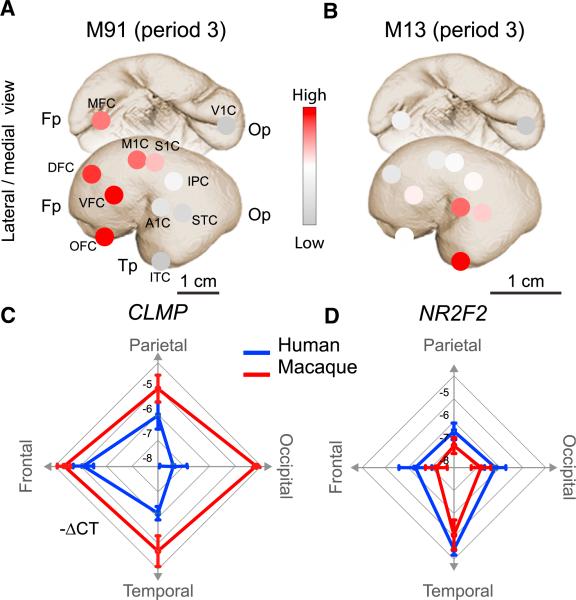
Figure 2. Gene Coexpression Modules and Hub Genes in the Developing Human and Macaque Monkey Cerebral Cortex.
(A and B) The average scaled expression of all genes in two coexpression modules with gradient-like patterns. (A) Genes in M91 exhibit a pattern with graded expression along the anterior-posterior axis. (B) Genes in M13 show a gradient in the neocortical areas of the temporal lobe. (C and D) Radar charts with qRT-PCR data of hub genes in modules 91 and 13.
(C) Areal expression of CLMP (hub gene of M91) demonstrates a clear gradient-like expression pattern in humans (blue), with a graded expression from the frontal lobe to the occipital lobe. This gradient-like expression is not present in the macaque monkey (red). (D) Areal expression of NR2F2 (hub gene of M13) exhibits a gradient-like expression pattern that is conserved between the human (blue) and macaque monkey (red) temporal cortices. Based on Pletikos et al. (2013).
The complex process of radial glia-guided neuronal migration of projection neurons was probably introduced during evolution to enable translation of the protomap at the VZ to the overlying cerebral cortex and preservation of neuronal positional information (Rakic, 1988). The cellular and molecular mechanisms underlying this complex event involve cooperation of multiple genes and molecules including astrotactin, doublecortin, glial growth factor, erbB, Reelin, Notch, NJPA1, Integrins, Sparc-like1, Ephs, MEKK4, various calcium channels, receptors, and many others (e.g., Anton et al., 1999; Gleeson and Walsh, 2000; Hatten, 2002; Gongidi et al., 2004; Hashimoto-Torii et al., 2008; Hatten, 2002; Komuro and Rakic, 1998; Nadarajah and Parnavelas, 2002; Reiner et al., 1993; Sarkisian et al., 2006; Torii et al., 2009). Radial migration is particularly elaborate in the convoluted primate cerebrum, requiring modification of the radial glia (Rakic, 2003). This process is extremely relevant to human cerebral function, as neuronal migrational abnormalities are a major cause of human neurodevelopmental conditions (Gleeson and Walsh, 2000; Lewis and Levitt, 2002). Yet mutations that cause severe abnormalities in human brain may cause far more subtle phenotypes in mouse, consistent with the exigencies of much longer migration in humans (e.g., Gleeson and Walsh, 2000; Lewis and Levitt, 2002).
There are also several examples of new types of neurons in the human cerebrum, including von Economo neurons, which are also observed in the brains of other large mammals and yet still may play an important role in human cerebral cortex (Butti et al., 2013). The more elaborate dendritic trees in humans, greatest in prefrontal cortex, coupled with overall larger neuropil and more spacing between pyramidal neurons in human (Bianchi et al., 2013a; Teffer et al., 2013) suggests differences in local circuit organization. Relevant to this notion is that one characteristic of human cerebral cortex is the high number and variety of local circuits neurons, most of which are inhibitory GABAergic interneurons (e.g., DeFelipe et al., 2002). One recent finding is that, unlike in the rodent, where GABA interneurons are generated in the ganglionic eminence (GE) of the ventral telencephalon (e.g., Anderson et al., 2001; de Carlos et al., 1996), a subclass of these cells in the human embryos are generated in the VZ/SVZ of the dorsal telencephalon (e.g., Jakovcevski et al., 2011; Letinic et al., 2002; Petanjek et al., 2009). Genes involved in the development of the GABAergic cells, including Nkx2.1, that are present in the mouse GE are present in both GE as well as dorsal VZ/SVZ in human embryos (Allan Brain Institute). A deeper discussion of all of the human-specific quantitative and qualitative differences, including cell types and morphology, are beyond the scope of this chapter, but some key phenotypic differences are highlighted in Table 1. Clearly more phenotype discovery is needed (Preuss, 2012).
Effect of the Environment, Neoteny, and Heterochrony
The increase of cortical surface during evolution by the introduction of new radial units, as well as the expansion and elaboration of the cytoarchitectonic areas, provide an opportunity for evolution to create novel input/target/output relationships with other structures via natural selection. Thus, we emphasize that the primordial protomap provides only a blueprint with a specific biological potential that can fully differentiate into a species-specific archetype of neural connections through reciprocal interactions between interconnected levels (e.g., Molnár and Blakemore, 1995; O'Leary and Stanfield, 1989; Rakic, 1981; Rakic et al., 1991, 2009; Selemon et al., 2013). The reciprocal interaction with subcortical structures and other cortical areas is essential but not a sufficient determinant of regional specification and circuit formation, which are subsequently modified by coordinated electrical activity (Katz and Crowley, 2002; Katz and Shatz, 1996).
The essential role of the environment and extended postnatal development in human brain is further emphasized by the observation that relative to all other primates, the human brain comprises a far smaller percentage of its eventual adult mass at birth (Bianchi et al., 2013b; Robson and Wood, 2008). Yet, in adulthood, human cortical neuropil is significantly expanded in comparison with chimpanzee, especially in prefrontal cortex (Spocter et al., 2012). Additionally, in humans, the processes of dendritic and synaptic maturation, as well as synaptic elimination, are prolonged relative to other mammals and primates (Bianchi et al., 2013b; Huttenlocher et al., 1982). In nonhuman primates, synaptic overproduction continues and elimination starts only after puberty (Bourgeois and Rakic, 1993; Rakic et al., 1986). The extraordinary scale of axon overproduction and elimination via competition present in primates (LaMantia and Rakic, 1990; Rakic and Riley, 1983a, 1983b) has not been observed in rodents. Remarkably, in humans, the period of synaptic elimination in the prefrontal association cortex lasts until the third decade of life (Petanjek et al., 2011) a remarkable level of neoteny. Similarly, the process of cortical myelination, which ends in puberty in chimpanzees, continues into the third decade in humans (Miller et al., 2012). This heterochronic development of cortical regions in humans is supported by a wide range of other methods (e.g., Chugani et al., 1987; Khundrakpam et al., 2013; Shaw et al., 2008), all of which indicate delayed development (neoteny) of cortical regions that have been most related to human higher cognition. Interestingly, a recent study suggests that although synaptic elimination may be synchronous across cortical regions in chimpanzee, the maturation of dendritic arborization is delayed in frontal cortex versus sensory and motor cortex, similar to what is observed in humans (Bianchi et al., 2013b).
Thus, there appears to be a gradient of neoteny and heterochrony in cortical circuit development that is most pronounced in humans in tertiary association regions. However, this process has parallels in chimpanzee, and, to a lesser extent, monkeys. These observations are consistent with the gradual, stepwise emergence of the delayed and heterochronic development of cortical regions over primate evolution, such as the prefrontal lobe, that are crucial for the development of human higher cognition. The significant conceptual (Changeux and Danchin, 1976), as well as biomedical, implications of these observations is indicated by the number of hypotheses that link inappropriate synaptic pruning and the prolonged development of human prefrontal cortex to various neuropsychiatric disorders and intellectual abilities (Paus et al., 2008; Selemon et al., 2013). However, understanding the role of heterochrony in the phylogenetic development of the brain presents special problems because of the complex interplay among multiple epigenetic factors that regulate gene expression during development (Changeux and Chavaillon, 1995; Rakic, 1995). During the genesis of the cerebral cortex, such cellular interactions probably play a more significant role than in any other organ, and this, as well as the paucity of crucial comparative developmental studies, is perhaps why progress in this field has been slow. A first step is to identify key differences in the adult and work backward to understand their ontogeny. But such differences are likely to be many; evidence of significant evolutionary adaptations at the molecular level of even a primary sensory region of the visual system in humans is evident in adults (Preuss, 2000; Preuss et al., 2004), but their genesis is unknown. Connecting such morphological phenotypes, as well as the basic developmental mechanisms controlling production, migration, and areal allocation of neurons, to genetic adaptations that have occurred in the anthropoid primate and human lineages is the next critical step if we are to understand human cortical evolution. It is clearly not a one-way process, as genetic distinctions can be used to guide phenotype discovery. These genetic factors are addressed in the following sections.
The Genomic-Driven Approach
Comparative Genomics in the Postgenome Era
Comparative genomics provides a powerful platform for identifying the genes and adaptive regulatory changes involved in cerebral cortical expansion, arealization, and other human-specific cellular or connectivity phenotypes (e.g., Table 1; Li et al., 2013; Rilling et al., 2008). The basic assumption underlying this paradigm is that changes in the genome on the human line-age, whether individual nucleotides, insertion-deletions (indels), or larger structural chromosomal variation, underlie the basic developmental processes described above. By comparing the human sequence to other mammals, one can infer that common DNA sequences represent those of the common ancestor and that those that differ between the two represent changes occurring in either species. Critical to interpretation of these data is comparison to another species that is a common but more distantly related ancestor, called an outgroup, without which understanding whether the observed differences occur on the human lineage is not possible (reviewed in Preuss et al., 2004; Varki and Altheide, 2005). Many forms of genetic variation that distinguish human from other species have been identified (reviewed in O'Bleness et al., 2012; Scally et al., 2012; Varki et al., 2008). The process of identifying variation is framed by the daunting prospect of sifting through tens of millions of base pairs that differ between humans and their closest relatives to identify those that are most divergent. Once such variants are found, connecting them to specific tissues, such as the brain, and, within the brain, to specific phenotypes, poses additional challenges. Thus, it should not be surprising that few clear smoking guns have been identified that distinguish the human brain from that of other species, including anthropoid primates.
Identifying Multiple Forms of Genetic Drivers
It is estimated that single-nucleotide differences, indels, and structural chromosomal changes comprising about 4% of the genome differ between humans and chimpanzees, providing a finite space for exploring the differences between ourselves and our closest living ancestor (Cheng et al., 2005; Prado-Martinez et al., 2013; Prüfer et al., 2012; Sudmant et al., 2013). Until the last decade, identifying key functional elements was practically restricted to evolutionary comparisons focused primarily on known coding regions, despite the fact that the importance of regulatory variation outside of coding sequences was well appreciated (King and Wilson, 1975). As our ability to annotate function has increased, so has the appreciation that there is a great deal of our functional genome outside of that accounting for protein-coding genes, ranging from multiple classes of non-coding RNA (Mercer et al., 2009) to known and cryptic regulatory elements (Bernstein et al., 2012).
Evolution of the Coding Genome at the Single-Nucleotide Level
As there are only about two dozen genes estimated to be present in human (derived; Table 2) and not in chimpanzee, most analyses of the protein-coding genome focus on differences between proteins shared between humans and other primates. In this case, changes that alter amino acids (missense or nonsense) between several species are compared to background changes—those that do not alter coding sequence, such as silent polymorphisms within protein-coding regions, or variants within introns, or those entirely outside of genic regions. The key issue here is that in the case of modern humans, neutral changes and genetic drift predominate due to small initial population sizes and population bottlenecks. The usual metrics used compare two species on a gene-wide basis, for example Ka/Ki (number of amino acid changing variants/number of noncoding variant background) or Ka/Ks (number of amino acid changing variants/number of synonymous variants). As genomics have continued to expand our notion of the functional genome, one must ask what is reasonable to use as neutral background (Bernstein et al., 2012; Mercer et al., 2009; Varki et al., 2008). Furthermore, it is clear that not all protein-coding domains are equivalent when it comes to conservation of their functional role.
Table 2.
Classes of Genetic Features that Differentiate Humans from Other Species, Some of which May Contribute to Human-Specific Features
| Class | Number | Reference |
|---|---|---|
| Accelerated genes in humans with respect to chimpanzees and gorillas | 663 genes | Column1; Scalley et al. 2012 |
| Brain-expressed genes in humans with dN/dS > 1 comparing human to chimp, gorilla, macaque, marmoset, and mouse lemur | 342 genesa | http://ensembl.org, http://brainspan.org |
| Human-specific deletions in otherwise highly conserved regions (hCONDELs) | 510 deletions | McLean et al., 2011 |
| Regions of differential H3K4 trimethylation compared to chimps and macaques | 410 peaks | Shulha et al., 2012 |
| Human-specific gene duplications compared to gorilla, chimpanzee, and orangutan | 23 gene families, of which 8 are fixed in humansb | Sudmant et al., 2010; Dennis et al., 2012 |
| Copy number increases | 15c | O'Bleness et al., 2012 |
| Purely de novo human genes | 3d | Knowles and McLysaght, 2009 |
| Purely de novo human genes | 1 | Li et al., 2010 |
| Human-accelerated regions | 202 regionse | Pollard et al., 2006 |
| Human-specific intergenic highly transcribed regions | 24f | Xu et al., 2010 |
| Human-specific miRNAs | 2-22g | Berezikov et al., 2006 |
| Human-specific miRNAs | 10 | Hu et al., 2012 |
| miRNAs with human-specific cortical expression patterns | 5 | Hu et al., 2011 |
| Telencephalic enhancers | 307 peaks in human genome nonalignable to mouseh | Visel et al., 2013 |
| Human-specific genes upregulated in PFC compared to cerebellum, thalamus, striatum, and hippocampus | 54i | Zhang et al., 2011 |
Some of these will be neutrally evolving on the human lineage.
Uniformly diploid in other great apes.
Many listed as having “likely” phenotypic consequences.
Uniprot currently lists the existence of one of these proteins as “unknown” and the other two as only having evidence at the transcript level.
The acceleration in a minority of these regions is consistent with biased gene conversion.
Observed in both pools of humans but not in the chimp pool or macaque pool. This will include lincRNAs and poorly annotated UTRs and was based on pooling a handful of representatives from each species, which may complicate inference of species specificity.
Many of these were only observed once in human, suggesting that depth may pose a challenge to infer species specificity.
As this is with respect to mouse, these peaks will reflect 80–100 million years of divergence along each lineage.
Species specificity was based on automated alignments, which may overcall lineage-specific genes.
Another issue is the timescale. Intraspecies comparisons of sequence depend on having sufficient number of events to have power to detect significant deviations from neutral expectations. This means that comparisons between the hominid line-ages, or even old-world primates and other mammals such as rodents, have significantly more power to detect primate-specific changes than comparisons of human and chimpanzee have to detect human-specific changes. However, the vastly different population sizes and histories of these mammals, for example, mice and men, can undermine many of the standard assumptions made in these analyses (e.g., Oldham and Geschwind, 2005). These issues highlight some of the key limitations of purely statistical approaches when assessing natural selection at the protein-coding level and, conversely, highlight the need to develop experimental systems for testing such hypotheses.
Realizing these limitations, it is still of interest to know whether protein-coding genes are under positive selection in humans or in anthropoid primates relative to other mammals. Although some studies have suggested that brain genes are under positive selection with respect to the rest of the genome (Dorus et al., 2004), the weight of the accumulating evidence suggests that this is not the case (Wang et al., 2007). Rather, protein-coding genes in the nervous system have on average fewer nonneutral changes and thus are under increased purifying selection. This is consistent with the notion that human CNS complexity yields evolutionary constraint.
However, the increased evolutionary constraint on brain-expressed genes overall does not preclude adaptive evolution of individual genes; rather, it puts them into stronger relief. Genome-wide comparisons reveal that approximately 500–1,000 genes are likely under strong positive selection in humans based on changes in their coding sequence (Clark et al., 2003; Chimpanzee Sequencing and Analysis Consortium, 2005; Scally et al., 2012). Although brain genes are not overall under positive selection, there is an enrichment for brain-related functions among those that are (Kamada et al., 2011; Liu et al., 2012a). One particularly salient example is the transcription factor FoxP2, which was originally identified for its role in a rare speech and language disorder, and more recently with developmental dyspraxia in humans (Noonan et al., 2006). Remarkably, sequencing of the Neanderthal and Denisovan genomes revealed that they share the human-derived form of FoxP2 (Meyer et al., 2012; Noonan et al., 2006). One of the human-derived changes was present in carnivores, reducing the statistical evidence for the adaptive evolution of FoxP2. Here, the power of modern molecular genetics and neuroscience was brought to bear in two studies, one in vitro and one in vivo, which tested the functional impact of the two amino acid changes. In the first, mouse FoxP2 was humanized (hFoxP2) and compared with the endogenous mouse form, revealing functional changes in striatal circuitry coupled with cellular alterations, including increased dendritic length in the mouse with hFoxP2, consistent with previous analyses of FoxP2 targets (Spiteri et al., 2007; Vernes et al., 2011). In the second study (Konopka et al., 2009), overexpression of the human and chimpanzee FoxP2 in human cells was performed to compare its transcriptional targets, revealing striking differences between the two species’ FoxP2 forms, many of which reflected in vivo gene expression differences observed between human and chimpanzee brain. In addition to genes important for neurodevelopment and synaptic function, human differential FoxP2 targets also included genes involved in branchial arch formation and craniofacial development, which suggests potential coevolution of both the CNS and articulatory structures necessary for spoken language (Konopka et al., 2009). Additional layers of complexity exist; recent studies comparing the hFoxp2 to the mouse version suggest that FoxP2 function may extend beyond circuit formation and plasticity to directing neural progenitor proliferation (Tsui et al., 2013), thus providing another window for directing cortical evolution. The work in mouse, as well as other research in songbirds, by showing that FoxP2 directs fundamental aspects of sensori-motor integration rather than being a language gene per se (e.g., auditory-guided vocal and other forms of motor learning), also demonstrates how cross-species comparative studies can inform our mechanistic understanding of language through identifying shared and derived elements (Fisher and Ridley, 2013; Konopka et al., 2012).
Gene Duplication and Deletion
From a purely quantitative perspective, gene duplications and deletions comprise more of the genetic landscape relevant to interspecies comparisons than do single base pair changes (Conrad and Antonarakis, 2007). Genome duplication played a major role in the development of the vertebrate lineage, yet connecting these changes to function has proven difficult (Van de Peer et al., 2009). Work from Eichler and colleagues also shows that the rate of accumulation of duplications has increased in African Great Apes relative to all other primates and that because of the repetitive elements surrounding these regions, many are the source of disease-related copy number variation in humans (Conrad and Antonarakis, 2007; Marques-Bonet et al., 2009). In humans, there are several hundred identified regions of interspersed segmental duplications. Since duplicated genes are likely to be under less initial constraint than the ancestral form, they also provide a fertile platform for adaptive evolution. Less clear is the role of genic deletions (Prado-Martinez et al., 2013). One clearly important example of duplication is the Duff1220 protein domain, whose role in cerebral development and function remains under investigation (Dumas et al., 2012; Popesco et al., 2006).
It was experimentally demonstrated recently that gene duplication influences vertebrate cognitive evolution via investigation of the role of paralogues of the DLG family of synaptic signaling molecules and two NMDA receptor subunits derived from genome duplications in the vertebrate radiation (Nithianantharajah et al., 2013; Ryan et al., 2013). These are challenging studies to perform from many perspectives (Belgard and Geschwind, 2013); one particularly innovative aspect of the work by Nithianantharajah et al. (2013) is the cross-species investigation of cognitive phenotypes in mouse and human using the CANTAB, which reveals parallel deficits in attention, memory, and visuo-spatial discrimination in knockout mice and human subjects with DLG2 mutations, three of whom suffer from schizophrenia. Ryan et al. (2013) perform domain swapping in particularly divergent regions of the NMDA receptor subunits GluN2a and GluN2b that enables them to relate different subunit components to distinct aspects of learning including executive function, which is related to the expansion of the frontal lobes in primates.
Rather than focusing on conserved features of the mammalian synapse, Charrier et al. (2012) and Dennis et al. (2012) extensively characterize a complex set of remarkable gene duplication events occurring over the course of the last three and a half million years that yielded a truncated form of the protein SRGAP2A protein, SRGAP2C in humans. They find that SRGAP2C is also expressed in H. neanderthalensis and Deniso-vans (Dennis et al., 2012) but not in any of the great apes, suggesting that it arose approximately one million years ago, consistent with a new role in human brain function. Through elegant in vitro and in vivo experiments in mouse (Charrier et al., 2012), they show that SRGAP2 leads to a higher density of dendritic spines, as well as longer dendritic shafts, which are known to be more human-like phenotypes when compared with other mammals (Benavides-Piccione et al., 2002). Thus, SRGAP2C may at least partially underlie the neoteny in synaptic refinement observed in humans described in the section on phenotypes. The next step will be connecting these phenotypes to circuits and behavior: for example, how do more human-like spines affect mouse behavior and cognition?
Nonprotein-Coding Elements: Regulatory Regions and Noncoding RNA
The protein-coding genome accounts for about 2% of the human genome, but it is estimated that at least another 10%–15% is also functional, including presumed and cryptic regulatory elements and thousands of transcribed noncoding RNAs (Ponting and Hardison, 2011). This nonprotein-coding regulatory portion was emphasized by the now classic study by King and Wilson (1975). Yet assigning function to these regions has only recently become practical (Pollard et al., 2006; Prabhakar et al., 2006).
Identifying Regulatory Elements
As a complement to genome sequence, the ENCODE project has the laudable goal of providing an “encyclopedia” of functionally annotated DNA and a foundational regulatory map of the human genome across tissue and cell types (Gerstein et al., 2012). A key issue is that since chromatin structure, DNA methylation, and subsequently promoter binding vary across cell types and tissues, we need to have this information in specific neural cell types and in human cerebral cortex across development, which has not yet been completed (hence the call for a “psychENCODE”; http://grants.nih.gov/grants/guide/rfa-files/RFA-MH-14-020.html). Still, focused genome-wide studies have yielded important advances, including the study of highly conserved, yet rapidly evolving regions of the genome (in primates and humans) that have revealed more than a hundred new putative enhancers. Elegant in vivo reporter assays show that most have tissue-specific early developmental functions, most frequently in the CNS (Visel et al., 2008). In fact, other forms of presumed human-specific gene regulation are also enriched near genes involved in CNS function and development (McLean et al., 2011). Currently, over 500 human accelerated regions (but otherwise highly conserved) (HARs) and a similar number of primate accelerated regions (PARs) have been identified based on comprehensive analysis of human-constrained genome sequence in 29 mammals (Jones et al., 2012). However, a remarkable 40% of human-constrained sequence (nearly 2% of the genome!) identified in this conservative manner remains essentially uncharacterized, indicating room for a remarkable amount of future discovery. Similarly, there are over 500 regions that are highly conserved in mammals through chimpanzees but deleted in humans (suggestive of function) that may regulate more than 1,000 genes (McLean et al., 2011). To complicate matters further, mobile repeats such as Alu elements (Cordaux and Batzer, 2009) have rapidly evolved in African great apes, with the greatest number occurring in humans. Such transposable elements have been shown to regulate gene expression and thus represent another layer of regulatory complexity introduced in primates and accelerated in humans. Furthermore, to understand the role of these genomic events in human brain evolution, their function must be interrogated in a tissue- and stage-specific manner in cerebral cortex.
In a recent tour de force, Rubenstein and colleagues combined computational analysis of sequence conservation with DNA binding assays in mouse and humans (chromatin IP, etc.) and in vivo validation in developing mouse to provide a catalog of human telencephalic enhancers (Visel et al., 2013). This includes several that may be associated with human neuropsychiatric diseases and a significant proportion that are presumed human or primate specific (Visel et al., 2013). Future studies querying laminar and cell-type-specific regulation in high resolution at multiple stages will be necessary to complete a map of human cortical regulatory elements as a crucial foundational resource. Like the functional work on gene duplications, this work again demonstrates how combination of cross-species bioinformatics and mouse experimentation can provide mechanistic insight into brain evolution.
Noncoding RNA
Noncoding RNAs provide another layer of regulatory complexity that needs to be considered in understanding human brain evolution. Some have proposed that noncoding RNA and RNA editing mechanisms may serve as a major driver of brain evolution (Barry and Mattick, 2012). Unfortunately, little is known about the roles of various forms of noncoding transcripts, from miRNAs through lincRNAs in human brain (Ulitsky and Bartel, 2013). Complicating their identification and study is the very rapid sequence divergence in many noncoding regions, whether purely regulatory or coding for transcripts, such as lincRNAs.
One notable example of a noncoding RNA that is involved in human brain evolution is HAR-1, a long noncoding RNA originally identified as the most accelerated noncoding transcribed genomic region in humans (Pollard et al., 2006). HAR-1 shows strikingly restricted expression in Cajal-Retzius neurons in the marginal zone during the time of neuronal migration in the cerebral cortex, consistent with a fundamental role in human cerebral cortical development and evolution. Precisely what this role is remains to be determined, perhaps by adapting the experimental approaches pioneered in the study of duplicated genes.
In contrast with protein-coding genes, miRNAs are extremely divergent between humans and other mammals. There are nearly twice the number of miRNAs in humans as in mice (and six times the number in Drosophila [Berezikov, 2011]). The organization and diversity of human miRNAs is consistent with the model that gene duplication and transposon insertion lead to reduced constraint early in the emergence of paralogues and is a major driver of mammalian evolution. Although potentially confounded by the different stages compared, sequencing of human fetal and adult chimpanzee brain miRNAs identified about 20 human-specific, and over 100 primate-specific, miRNAs when compared with other vertebrates (Berezikov, 2011). These provide a fertile ground for understanding complex gene regulation in human cerebral development, for example, how these miRNAs relate to the expansion of specific neural progenitor pools predicated by the protomap hypothesis, as well as unique cellular and synaptic features of human cortical architecture.
Transcriptomics
One weakness of isolated interspecies sequence comparisons is that most genes expressed in the cerebral cortex are also expressed in other tissues, so it is not possible to unequivocally assign organ-specific function to human-specific DNA changes without further experimental evidence (Prabhakar et al., 2008; Visel et al., 2013). A complement to sequence analysis is the analysis of gene expression, which can help in understanding the particular role of genetic variation at the level of the specific tissue. Analysis of gene expression at the RNA or protein level also provides a phenotype in between the structural or cognitive phenotypes in question and DNA variation (Geschwind and Konopka, 2009). Several studies have now shown that there are significant differences between the species, identifying hundreds of genes changing on the human lineage (Khaitovich et al., 2006; Preuss et al., 2004). However, there are many caveats in interpreting these differences, including the role of the environment and the challenge in distinguishing which changes in expression are adaptive changes, rather than the expected neutral changes due to genetic drift (Khaitovich neutral model). These confounders have been reviewed in detail (Khaitovich et al., 2006; Preuss et al., 2004).
Increased Molecular Complexity in Human Frontal Lobe and Neoteny
By organizing genes into coexpression modules, network analysis provides a functional context from which to assess the significance of expression changes and can further help to prioritize individual genes from long lists of differential expressed genes (Konopka et al., 2012; Oldham et al., 2006; Oldham et al., 2008). This approach has highlighted accelerated changes in the cerebral cortex, most specifically the frontal lobe on the human lineage (Konopka et al., 2012). Konopka et al. (2012) found that even with frontal lobe, there was significantly more transcriptional network complexity in humans compared with chimpanzee and macaque, consistent with an increase in cellular and molecular complexity within a single brain region in humans. Remarkably, these human-specific networks are comprised of genes involved in neuronal morphology and synaptic function, as well as genes related to FoxP2 (Konopka et al., 2012). The next step is to understand to what extent these networks reflect differences in cell types themselves or molecular signaling within cells in the human frontal lobe (Konopka et al., 2012; Ponting and Oliver, 2012). Furthermore, understanding how the specific genes identified here relate to specific human-derived pheno-types related to local circuit organization in the prefrontal cortex, such as elaborated dendritic branching, or increased inhibitory neuron density, can now be experimentally approached.
Marked acceleration of human-specific changes in the frontal lobe has also been observed specifically in the class of genes with developmental trajectories that differed between the species (Somel et al., 2011). Support for this contention is the observation that humans and other primates differ in terms of the delay in the upregulation of gene expression related to synaptic function in human frontal cortex (Liu et al., 2012a). Coupled with in vitro experimental validation, Somel and colleagues’ work represents one of the first studies to begin to use transcriptional phenotypes to identify potential causal drivers of adaptive evolution and connect these to specific brain regions and functional processes (Somel et al., 2011). These data and the effect of the human-specific SRGAP2c on dendritic development (Charrier and Polleux, 2012) may provide the first known molecular signatures of neoteny that characterizes human cognitive and behavioral development.
Other recent work, showing that one of the most human accelerated classes of genes involves those that are expressed during brain development, provides additional evidence that characterizing human or primate-derived developmental mechanisms will be critical to understanding human evolution (Zhang et al., 2011). Understanding whether these “new” genes are involved in human and or primate-specific neural progenitor cell-cycle regulation enriched in the OSVZ (Lui et al., 2011; Molnár et al., 2006) or alternatively overlap with those involved in frontal cortex dendritic/synaptic development or maturation presents a clear means for connecting evolutionary genomic findings with human cerebral cortical phenotypes underlying the evolution of human cognition.
Finally, the origin of transcriptional changes in the cerebral cortex on the human lineage is not known in most cases (Oldham et al., 2006), but they may be related to the evolution of the human-specific regulatory or noncoding elements discussed above. Integration of many of the data sets cited here, coupled to experimental manipulations, now permits making such causal connections. Additionally, environmental or genetic factors may also mediate changes in gene expression via DNA methylation. A recent tour de force analysis of human and mouse frontal cortex methylation throughout postnatal development reveals dynamic, age-dependent changes (Lister et al., 2013), consistent with marked epigenetic remodeling during neural development and maturation. These investigators also identify multiple regions of differential methylation between mouse and human cortex, which not surprisingly are significantly associated with regulatory regions (Lister et al., 2013). In additional to providing a genomic methylation roadmap in a tissue that is key for human evolution for the first time, this work indicates that careful matching for developmental stage is necessary for comparative analysis across species.
Although epigenetic comparisons of primate and human brain are in their early stages, some interesting findings have emerged (Shulha et al., 2012; Zeng et al., 2012b). Integration of cross-species methylation and expression data have revealed significant correlations between the two in humans and primates, suggesting that differential methylation may drive the evolution of specific gene expression patterns in humans (Zeng et al., 2012b). Comparative analysis of major histone marks related to open chromatin (transcriptionally active regions) in sorted neuronal nuclei revealed adaptive evolution in humans relative to chimpanzee at several loci, including DPP10 (Shulha et al., 2012). Chromosome confirmation-capture and analysis of DPP10 expression supports a human-specific regulatory network at this neuropsychiatric disease-relevant locus (Shulha et al., 2012). The next steps that will afford us a more holistic understanding of genome and regulatory evolution, by connecting changes in genome sequence, chromatin structure, and transcriptional regulation, are now within reach.
Human Cortical Lamination
No genome-wide comparison of human and primate cortical laminae has yet been conducted. However, laminar comparisons of the expression of about 1,000 curated genes between human and mouse by in situ hybridization reveal that, overall, there is widespread conservation of patterns, with a 20% overall divergence in expression patterns in primary visual cortex, and an approximately 25% divergence in temporal lobe (Zeng et al., 2012a). Remarkably, the most divergent class of genes is cortical neuronal markers, including laminar markers (59% divergence) and putative interneuron markers (41%). The authors emphasize that almost half of the genes that were layer V markers in mouse either were not expressed differentially or were layer III markers in human, consistent with the expansive long-range intercortical connections in primate relative to rodent. These investigators also explored the patterns of a subset of genes evolving on the human lineage, observing widespread cortical expression for most of these, consistent with a trans-neuronal, rather than a subclass-specific, role. Now, work in this area must move to the level of specific cell types, including glia, as unbiased, genome-wide comparisons of mouse and human gene expression networks suggest more divergence in glia than in most neurons (Miller et al., 2010).
Conclusions
The evolution of the human brain is a vast subject. We argue that although we are at a stage where large-scale genomic data collection is clearly useful and already has provided a key foundation, it is not sufficient. A theoretical framework founded on understanding the key processes of neurodevelopment and cortical neural function that distinguish primates and humans from other mammals is essential. The radial unit and protomap hypotheses provide structures on which to explore specific early developmental events’ role in human cerebral cortical evolution. However, understanding differences in both the pace and final state and diversity of cortical neuronal phenotypes in humans will require further comparative cellular, behavioral, and anatomical studies to provide a true catalog of human differences. Comparisons with our closest living ancestors, the chimpanzee, will be critical to define human specificity, but broader phylogenetic comparisons including widely used experimental models such as invertebrates, mice, and other primates are also fundamental. But even that may not guarantee success. One example of a well-described anatomical human adaptation that has been particularly vexing to connect to developmental or molecular mechanisms is the genesis of human cerebral asymmetry, which is fundamental to the emergence of human language. Its anatomical basis has been appreciated for nearly a half century, yet, despite more than a decade of significant progress in defining the molecular pathways involved in visceral asymmetry, relatively little is understood about how this might connect to cerebral cortex asymmetry.
It is also clear that gene regulation has played a key role in human cerebral evolution. Integration of the multiple types of functional genes, from those coding proteins to multiple forms of noncoding RNAs, as well as mechanisms of gene regulation, will require innovative systems biology methods. Nevertheless, we are now at a place where we can connect differentially expressed genes to biological processes and understand the regulatory elements that may drive these processes, moving from an era of genomic and molecular description to functional testing in model systems. Many challenges remain, including the tradeoffs between matching the intricacies of in vivo development often only approachable in nonprimates, such as mouse, and the vast species differences that warrant adopting in vitro human models. Technological advances, including three-dimensional organoid cultures (Lancaster et al., 2013) or mouse and mouse-human chimeras (Goldman et al., 2012), will soon improve this situation. The confluence of advances in comparative genomics and modern neurobiology has made what in the past may have seemed like an experimentally intractable problem readily addressable. It is imperative that we continue to take on this challenge, as understanding human brain evolution is likely to be important for understanding many human neuro-psychiatric diseases.
ACKNOWLEDGMENTS
P.R. is supported by the NIH grants R01 DA023999 and R01 NS014841 and the Kavli Institute for Neuroscience at Yale. P.R. also thanks members of his lab for helpful discussions. D.H.G. is supported by NIH/NIMH grants R01 MH100027 (ACE Network Award), R37 MH060233 (MERiT Award), P50 HD055784 (ACE Center Award), and R01 MH094714, and Simons SFARI Award 206744. D.H.G. thanks T. Grant Belgard, PhD, for very helpful discussions and construction of Table 2 and Lauren Kawaguchi for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abrahams B, Tentler D, Perederiy J, Oldham M, Coppola G, Geschwind D. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl. Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato S, Liu X, Zheng B, Cantley L, Rakic P, Man HY. AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science. 2011;332:247–251. doi: 10.1126/science.1201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Anton ES, Kreidberg J, Rakic P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminarorganization of the cerebral cortex. Neuron. 1999;22:227–289. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- Arimatsu Y, Miyamoto M, Nihonmatsu I, Hirata K, Uratani Y, Hatanaka Y, Takiguchi-Hayashi K. Early regional specification for a molecular neuronal phenotype in the rat neocortex. Proc. Natl. Acad. Sci. USA. 1992;89:8879–8883. doi: 10.1073/pnas.89.19.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G, Mattick JS. The role of regulatory RNA in cognitive evolution. Trends Cogn. Sci. 2012;16:497–503. doi: 10.1016/j.tics.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Belgard TG, Geschwind DH. Retooling spare parts: gene duplication and cognition. Nat. Neurosci. 2013;16:6–8. doi: 10.1038/nn.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides-Piccione R, Ballesteros-Yáñez I, DeFelipe J, Yuste R. Cortical area and species differences in dendritic spine morphology. J. Neurocytol. 2002;31:337–346. doi: 10.1023/a:1024134312173. [DOI] [PubMed] [Google Scholar]
- Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi S, Stimpson CD, Bauernfeind AL, Schapiro SJ, Baze WB, McArthur MJ, Bronson E, Hopkins WD, Semendeferi K, Jacobs B, et al. Dendritic morphology of pyramidal neurons in the chimpanzee neocortex: regional specializations and comparison to humans. Cereb. Cortex. 2013a;23:2429–2436. doi: 10.1093/cercor/bhs239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi S, Stimpson CD, Duka T, Larsen MD, Janssen WG, Collins Z, Bauernfeind AL, Schapiro SJ, Baze WB, McArthur MJ, et al. Synaptogenesis and development of pyramidal neuron dendritic morphology in the chimpanzee neocortex resembles humans. Proc. Natl. Acad. Sci. USA. 2013b;110(Suppl 2):10395–10401. doi: 10.1073/pnas.1301224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Bourgeois JP, Rakic P. Changes of synaptic density in the primary visual cortex of the macaque monkey from fetal to adult stage. J. Neurosci. 1993;13:2801–2820. doi: 10.1523/JNEUROSCI.13-07-02801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butti C, Santos M, Uppal N, Hof PR. Von Economo neurons: clinical and evolutionary perspectives. Cortex. 2013;49:312–326. doi: 10.1016/j.cortex.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Tillquist CR. Encephalization, emergent properties, and psychiatry: a minicolumnar perspective. Neuroscientist. 2008;14:101–118. doi: 10.1177/1073858407309091. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr., Rakic P. Mechanisms of cortical development: a view from mutations in mice. Annu. Rev. Neurosci. 1978;1:297–326. doi: 10.1146/annurev.ne.01.030178.001501. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr., Goto T, Tarui T, Takahashi T, Bhide PG, Nowakowski RS. Cell output, cell cycle duration and neuronal specification: a model of integrated mechanisms of the neocortical proliferative process. Cereb. Cortex. 2003;13:592–598. doi: 10.1093/cercor/13.6.592. [DOI] [PubMed] [Google Scholar]
- Changeux JP, Chavaillon J. Origins of the Human Brain. Clarendon Press; Oxford: 1995. [Google Scholar]
- Changeux JP, Danchin A. Selective stabilisation of developing synapses as a mechanism for the specification of neuronal networks. Nature. 1976;264:705–712. doi: 10.1038/264705a0. [DOI] [PubMed] [Google Scholar]
- Charrier C, Polleux F. [How human-specific SRGAP2 gene duplications control human brain development]. Med. Sci. (Paris) 2012;28:911–914. doi: 10.1051/medsci/20122811003. [DOI] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, Polleux F. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Pääbo S, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb. Cortex. 2003;13:599–606. doi: 10.1093/cercor/13.6.599. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J. Comp. Neurol. 2008;509:144–155. doi: 10.1002/cne.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani HT, Phelps ME, Mazziotta JC. Positron emission tomography study of human brain functional development. Ann. Neurol. 1987;22:487–497. doi: 10.1002/ana.410220408. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Clowry G, Molnár Z, Rakic P. Renewed focus on the developing human neocortex. J. Anat. 2010;217:276–288. doi: 10.1111/j.1469-7580.2010.01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Tannoudji M, Babinet C, Wassef M. Early determination of a mouse somatosensory cortex marker. Nature. 1994;368:460–463. doi: 10.1038/368460a0. [DOI] [PubMed] [Google Scholar]
- Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutzfeldt OD. Generality of the functional structure of the neocortex. Naturwissenschaften. 1977;64:507–517. doi: 10.1007/BF00483547. [DOI] [PubMed] [Google Scholar]
- de Carlos JA, López-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J. Neurosci. 1996;16:6146–6156. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J, Alonso-Nanclares L, Arellano JI. Microstructure of the neocortex: comparative aspects. J. Neurocytol. 2002;31:299–316. doi: 10.1023/a:1024130211265. [DOI] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Dumas LJ, O'Bleness MS, Davis JM, Dickens CM, Anderson N, Keeney JG, Jackson J, Sikela M, Raznahan A, Giedd J, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am. J. Hum. Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar RIM. Coevolution of neocortical size, group size, and language in humans. Behav. Brain Sci. 1993;16:681–694. [Google Scholar]
- Elsen GE, Hodge RD, Bedogni F, Daza RA, Nelson BR, Shiba N, Reiner SL, Hevner RF. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc. Natl. Acad. Sci. USA. 2013;110:4081–4086. doi: 10.1073/pnas.1209076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Ridley M. Evolution. Culture, genes, and the human revolution. Science. 2013;340:929–930. doi: 10.1126/science.1236171. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Amlien I, Tamnes CK, Grydeland H, Engvig A, Espeseth T, Reinvang I, Lundervold AJ, Lundervold A, Walhovd KB. High-expanding cortical regions in human development and evolution are related to higher intellectual abilities. Cereb. Cortex. 2013 doi: 10.1093/cercor/bht201. Published online August 19, 2013. http://dx.doi.org/10.1093/cercor/bht201. [DOI] [PubMed]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Walsh CA. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci. 2000;23:352–359. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- Goldman SA, Nedergaard M, Windrem MS. Glial progenitor cell-based treatment and modeling of neurological disease. Science. 2012;338:491–495. doi: 10.1126/science.1218071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Development of cortical circuitry and cognitive function. Child Dev. 1987;58:601–622. [PubMed] [Google Scholar]
- Gongidi V, Ring C, Moody M, Brekken R, Sage EH, Rakic P, Anton ES. SPARC-like 1 regulates the terminal phase of radial glia-guided migration in the cerebral cortex. Neuron. 2004;41:57–69. doi: 10.1016/s0896-6273(03)00818-3. [DOI] [PubMed] [Google Scholar]
- Grove EA, Fukuchi-Shimogori T. Generating the cerebral cortical area map. Annu. Rev. Neurosci. 2003;26:355–380. doi: 10.1146/annurev.neuro.26.041002.131137. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Torii M, Sarkisian MR, Bartley CM, Shen J, Radtke F, Gridley T, Sestan N, Rakic P. Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron. 2008;60:273–284. doi: 10.1016/j.neuron.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- Haydar TF, Ang E, Jr., Rakic P. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc. Natl. Acad. Sci. USA. 2003;100:2890–2895. doi: 10.1073/pnas.0437969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, Guillemot F. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S, Collins CE, Wong P, Kaas JH, Lent R. The basic nonuniformity of the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2008;105:12593–12598. doi: 10.1073/pnas.0805417105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Haydar TF. The (not necessarily) convoluted role of basal radial glia in cortical neurogenesis. Cereb. Cortex. 2012;22:465–468. doi: 10.1093/cercor/bhr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, Guo S, Xi J, Yan Z, Fu N, Zhang X, Menzel C, Liang H, Yang H, Zhao M, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, Taylor MS, Tang L, Li J, Liu J, et al. Evolution of the human-specific microRNA miR-941. Nat Commun. 2012;3:1145. doi: 10.1038/ncomms2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR, de Courten C, Garey LJ, Van der Loos H. Synaptogenesis in human visual cortex—evidence for synapse elimination during normal development. Neurosci. Lett. 1982;33:247–252. doi: 10.1016/0304-3940(82)90379-2. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr. Opin. Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I, Mayer N, Zecevic N. Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cereb. Cortex. 2011;21:1771–1782. doi: 10.1093/cercor/bhq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanović D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. The evolution of brains from early mammals to humans. Wiley Interdiscip. Rev. Cogn. Sci. 2013;4:33–45. doi: 10.1002/wcs.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada F, Aoki Y, Narisawa A, Abe Y, Komatsuzaki S, Kikuchi A, Kanno J, Niihori T, Ono M, Ishii N, et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J. Hum. Genet. 2011;56:34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- Katz LC, Crowley JC. Development of cortical circuits: lessons from ocular dominance columns. Nat. Rev. Neurosci. 2002;3:34–42. doi: 10.1038/nrn703. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kennedy H, Dehay C. Cortical specification of mice and men. Cereb. Cortex. 1993;3:171–186. doi: 10.1093/cercor/3.3.171. [DOI] [PubMed] [Google Scholar]
- Kennedy H, Dehay C. Self-organization and interareal networks in the primate cortex. Prog. Brain Res. 2012;195:341–360. doi: 10.1016/B978-0-444-53860-4.00016-7. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khundrakpam BS, Reid A, Brauer J, Carbonell F, Lewis J, Ameis S, Karama S, Lee J, Chen Z, Das S, Evans AC, Brain Development Cooperative Group Developmental changes in organization of structural brain networks. Cereb. Cortex. 2013;23:2072–2085. doi: 10.1093/cercor/bhs187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Orchestration of neuronal migration by activity of ion channels, neurotransmitter receptors, and intracellular Ca2+ fluctuations. J. Neurobiol. 1998;37:110–130. [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM, Geschwind DH. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc. Natl. Acad. Sci. USA. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon YH, Collura R, Ruvolo M, Barrett JC, Woods CG, et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krubitzer L, Kaas J. The evolution of the neocortex in mammals: how is phenotypic diversity generated? Curr. Opin. Neurobiol. 2005;15:444–453. doi: 10.1016/j.conb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia AS, Rakic P. Axon overproduction and elimination in the corpus callosum of the developing rhesus monkey. J. Neurosci. 1990;10:2156–2175. doi: 10.1523/JNEUROSCI.10-07-02156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr. Opin. Neurobiol. 2012;22:747–753. doi: 10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annu. Rev. Neurosci. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- Li CY, Zhang Y, Wang Z, Zhang Y, Cao C, Zhang PW, Lu SJ, Li XM, Yu Q, Zheng X, et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput. Biol. 2010;6:e1000734. doi: 10.1371/journal.pcbi.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hu X, Preuss TM, Glasser MF, Damen FW, Qiu Y, Rilling J. Mapping putative hubs in human, chimpanzee and rhesus macaque connectomes via diffusion tractography. Neuroimage. 2013;80:462–474. doi: 10.1016/j.neuroimage.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hashimoto-Torii K, Torii M, Haydar TF, Rakic P. The role of ATP signaling in the migration of intermediate neuronal progenitors to the neocortical subventricular zone. Proc. Natl. Acad. Sci. USA. 2008;105:11802–11807. doi: 10.1073/pnas.0805180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, Yuan Y, He L, Oleksiak A, Zhang Y, et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012a;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun L, Torii M, Rakic P. Connexin 43 controls the multipolar phase of neuronal migration to the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2012b;109:8280–8285. doi: 10.1073/pnas.1205880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Cortay V, Giroud P, Berland M, Smart I, Kennedy H, Dehay C. The concerted modulation of proliferation and migration contributes to the specification of the cytoarchitecture and dimensions of cortical areas. Cereb. Cortex. 2006;16(Suppl 1):i26–i34. doi: 10.1093/cercor/bhk011. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Girirajan S, Eichler EE. The origins and impact of primate segmental duplications. Trends Genet. 2009;25:443–454. doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK. Strategies for the generation of neuronal diversity in the developing central nervous system. J. Neurosci. 1995;15:6987–6998. doi: 10.1523/JNEUROSCI.15-11-06987.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AM, Sestan N, Wildman DE, et al. Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci. USA. 2012;109:16480–16485. doi: 10.1073/pnas.1117943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, Rubenstein JL. Early neocortical regionalization in the absence of thalamic innervation. Science. 1999;285:906–909. doi: 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- Molnár Z. Evolution of cerebral cortical development. Brain Behav. Evol. 2011;78:94–107. doi: 10.1159/000327325. [DOI] [PubMed] [Google Scholar]
- Molnár Z, Blakemore C. How do thalamic axons find their way to the cortex? Trends Neurosci. 1995;18:389–397. doi: 10.1016/0166-2236(95)93935-q. [DOI] [PubMed] [Google Scholar]
- Molnár Z, Métin C, Stoykova A, Tarabykin V, Price DJ, Francis F, Meyer G, Dehay C, Kennedy H. Comparative aspects of cerebral cortical development. Eur. J. Neurosci. 2006;23:921–934. doi: 10.1111/j.1460-9568.2006.04611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle V. The evolution of ideas concerning the function of the neocortex. Cereb. Cortex. 1995;5:289–295. doi: 10.1093/cercor/5.4.289. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nat. Rev. Neurosci. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Nithianantharajah J, Komiyama NH, McKechanie A, Johnstone M, Blackwood DH, St Clair D, Emes RD, van de Lagemaat LN, Saksida LM, Bussey TJ, Grant SG. Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat. Neurosci. 2013;16:16–24. doi: 10.1038/nn.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noonan JP, Coop G, Kudaravalli S, Smith D, Krause J, Alessi J, Chen F, Platt D, Pääbo S, Pritchard JK, Rubin EM. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat. Rev. Genet. 2012;13:853–866. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Borngasser D. Cortical ventricular zone progenitors and their progeny maintain spatial relationships and radial patterning during preplate development indicating an early protomap. Cereb. Cortex. 2006;16(Suppl 1):i46–i56. doi: 10.1093/cercor/bhk019. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Stanfield BB. Selective elimination of axons extended by developing cortical neurons is dependent on regional locale: experiments utilizing fetal cortical transplants. J. Neurosci. 1989;9:2230–2246. doi: 10.1523/JNEUROSCI.09-07-02230.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Geschwind DH. Evolutionary genetics: the human brain — adaptation at many levels. Eur. J. Hum. Genet. 2005;13:520–522. doi: 10.1038/sj.ejhg.5201401. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl. Acad. Sci. USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN. Why do many psychiatric disorders emerge during adolescence? Nat. Rev. Neurosci. 2008;9:947–957. doi: 10.1038/nrn2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Berger B, Esclapez M. Origins of cortical GABAergic neurons in the cynomolgus monkey. Cereb. Cortex. 2009;19:249–262. doi: 10.1093/cercor/bhn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Šimic G, Rasin MR, Uylings HB, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. USA. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletikos M, Sousa AMM, Sedmak S, Meyer KA, Zhu Y, Cheng F, Li M, Kawasawa YI, Šestan N. Temporal specification and bilaterality of human neocortical topographic dene expression. Neuron. 2013 doi: 10.1016/j.neuron.2013.11.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Polleux F, Dehay C, Kennedy H. The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. J. Comp. Neurol. 1997;385:95–116. [PubMed] [Google Scholar]
- Ponting CP, Hardison RC. What fraction of the human genome is functional? Genome Res. 2011;21:1769–1776. doi: 10.1101/gr.116814.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL. Brain, know thy transcriptome, know thyself. Neuron. 2012;75:543–545. doi: 10.1016/j.neuron.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O'Connor TD, Santpere G, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM. The argument from animals to humans in cognitive neuroscience. In: Gazzaniga MS, editor. The Cognitive Neurosciences. MIT Press; Cambridge: 1995. pp. 1227–1241. [Google Scholar]
- Preuss TM. Taking the measure of diversity: comparative alternatives to the model-animal paradigm in cortical neuroscience. Brain Behav. Evol. 2000;55:287–299. doi: 10.1159/000006664. [DOI] [PubMed] [Google Scholar]
- Preuss TM. Human brain evolution: from gene discovery to phenotype discovery. Proc. Natl. Acad. Sci. USA. 2012;109(Suppl 1):10709–10716. doi: 10.1073/pnas.1201894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM, Cáceres M, Oldham MC, Geschwind DH. Human brain evolution: insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, et al. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Rakic P. Development of visual centers in the primate brain depends on binocular competition before birth. Science. 1981;214:928–931. doi: 10.1126/science.7302569. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- Rakic P. Elusive radial glial cells: historical and evolutionary perspective. Glia. 2003;43:19–32. doi: 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Riley KP. Overproduction and elimination of retinal axons in the fetal rhesus monkey. Science. 1983a;219:1441–1444. doi: 10.1126/science.6828871. [DOI] [PubMed] [Google Scholar]
- Rakic P, Riley KP. Regulation of axon number in primate optic nerve by prenatal binocular competition. Nature. 1983b;305:135–137. doi: 10.1038/305135a0. [DOI] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232:232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- Rakic P, Suñer I, Williams RW. A novel cytoarchitectonic area induced experimentally within the primate visual cortex. Proc. Natl. Acad. Sci. USA. 1991;88:2083–2087. doi: 10.1073/pnas.88.6.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Ayoub AE, Breunig JJ, Dominguez MH. Decision by division: making cortical maps. Trends Neurosci. 2009;32:291–301. doi: 10.1016/j.tins.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash BG, Tomasi S, Lim HD, Suh CY, Vaccarino FM. Cortical gyrification induced by fibroblast growth factor 2 in the mouse brain. J. Neurosci. 2013;33:10802–10814. doi: 10.1523/JNEUROSCI.3621-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat. Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat. Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Robson SL, Wood B. Hominin life history: reconstruction and evolution. J. Anat. 2008;212:394–425. doi: 10.1111/j.1469-7580.2008.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Rakic P. Genetic control of cortical development. Cereb. Cortex. 1999;9:521–523. doi: 10.1093/cercor/9.6.521. [DOI] [PubMed] [Google Scholar]
- Ryan TJ, Kopanitsa MV, Indersmitten T, Nithianantharajah J, Afinowi NO, Pettit C, Stanford LE, Sprengel R, Saksida LM, Bussey TJ, et al. Evolution of GluN2A/B cytoplasmic domains diversified vertebrate synaptic plasticity and behavior. Nat. Neurosci. 2013;16:25–32. doi: 10.1038/nn.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian MR, Bartley CM, Chi H, Nakamura F, Hashimoto-Torii K, Torii M, Flavell RA, Rakic P. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon LD, Ceritoglu C, Ratnanather JT, Wang L, Harms MP, Aldridge K, Begović A, Csernansky JG, Miller MI, Rakic P. Distinct abnormalities of the primate prefrontal cortex caused by ionizing radiation in early or midgestation. J. Comp. Neurol. 2013;521:1040–1053. doi: 10.1002/cne.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, Travis K, Buckwalter J. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb. Cortex. 2011;21:1485–1497. doi: 10.1093/cercor/bhq191. [DOI] [PubMed] [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, Greenstein D, Clasen L, Evans A, Rapoport JL, et al. Neurodevelopmental trajectories of the human cerebral cortex. J. Neurosci. 2008;28:3586–3594. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Crisci JL, Reshetov D, Tushir JS, Cheung I, Bharadwaj R, Chou HJ, Houston IB, Peter CJ, Mitchell AC, et al. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol. 2012;10:e1001427. doi: 10.1371/journal.pbio.1001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Govek EE, Tomoda T, Hatten ME. Neuronal polarity in CNS development. Genes Dev. 2006;20:2639–2647. doi: 10.1101/gad.1462506. [DOI] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am. J. Hum. Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spocter MA, Hopkins WD, Barks SK, Bianchi S, Hehmeyer AE, Anderson SM, Stimpson CD, Fobbs AJ, Hof PR, Sherwood CC. Neuropil distribution in the cerebral cortex differs between humans and chimpanzees. J. Comp. Neurol. 2012;520:2917–2929. doi: 10.1002/cne.23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J. Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE, 1000 Genomes Project Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, Mohajeri K, Kondova I, Bontrop RE, Persengiev S, et al. Great Ape Genome Project Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–1382. doi: 10.1101/gr.158543.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS., Jr. Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J. Neurosci. 1999;19:10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarui T, Takahashi T, Nowakowski RS, Hayes NL, Bhide PG, Caviness VS. Overexpression of p27 Kip 1, probability of cell cycle exit, and laminar destination of neocortical neurons. Cereb. Cortex. 2005;15:1343–1355. doi: 10.1093/cercor/bhi017. [DOI] [PubMed] [Google Scholar]
- Teffer K, Semendeferi K. Human prefrontal cortex: evolution, development, and pathology. Prog. Brain Res. 2012;195:191–218. doi: 10.1016/B978-0-444-53860-4.00009-X. [DOI] [PubMed] [Google Scholar]
- Teffer K, Buxhoeveden DP, Stimpson CD, Fobbs AJ, Schapiro SJ, Baze WB, McArthur MJ, Hopkins WD, Hof PR, Sherwood CC, Semendeferi K. Developmental changes in the spatial organization of neurons in the neocortex of humans and common chimpanzees. J. Comp. Neurol. 2013 doi: 10.1002/cne.23412. Published online July 10, 2013. http://dx.doi.org/10.1002/cne.23412. [DOI] [PMC free article] [PubMed]
- Torii M, Hashimoto-Torii K, Levitt P, Rakic P. Integration of neuronal clones in the radial cortical columns by EphA and ephrin-A signalling. Nature. 2009;461:524–528. doi: 10.1038/nature08362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulates neurogenesis during embryonic cortical development. J. Neurosci. 2013;33:244–258. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Varki A, Altheide TK. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat. Rev. Genet. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Chien HC, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou CK, Tsai SF, Wu CI, Shen CK. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu AG, He L, Li Z, Xu Y, Li M, Fu X, Yan Z, Yuan Y, Menzel C, Li N, et al. Intergenic and repeat transcription in human, chimpanzee and macaque brains measured by RNA-Seq. PLoS Comput. Biol. 2010;6:e1000843. doi: 10.1371/journal.pcbi.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012a;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am. J. Hum. Genet. 2012b;91:455–465. doi: 10.1016/j.ajhg.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Landback P, Vibranovski MD, Long M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]