Abstract
Background
KLF4 mediates inflammatory responses following vascular injury/disease; however, the role of KLF4 in abdominal aortic aneurysms [AAA] remains unknown. The goals of the present study were to: 1) determine the role of KLF4 in experimental AAA; and 2) determine the effect of KLF4 on smooth muscle cells in AAA.
Methods and Results
KLF4 expression progressively increased at day 3, 7, and 14 following aortic elastase perfusion in C57BL/6 mice. Separately, loss of a KLF4 allele conferred AAA protection using ERTCre+ KLF4 flx/wt mice in the elastase AAA model. In a third set of experiments, smooth muscle specific loss of 1 and 2 KLF4 alleles resulted in progressively greater protection using novel transgenic mice (MYHCre+ flx/flx, flx/wt and wt/wt) in the elastase AAA model compared to control. Elastin degradation, MAC2, and cytokine production (MCP1, TNFα, IL23) were significantly attenuated while α-actin staining was increased in KLF4 knock-out mice versus controls. Results were verified in global KLF4 and smooth muscle specific knock-out mice using an Angiotensin II model of aneurysm formation. KLF4 inhibition with siRNA attenuated down-regulation of smooth muscle gene expression in vitro, while in vivo studies demonstrated that KLF4 binds to promoters of smooth muscle genes by ChIP analysis. Finally, human aortic aneurysms demonstrated significantly higher KLF4 expression that was localized to smooth muscle cells (SMCs).
Conclusions
KLF4 plays a critical role in aortic aneurysm formation via effects on SMCs. These results suggest that KLF4 regulates SMC “phenotypic switching” and could be a potential therapeutic target for AAA disease.
Keywords: vascular biology, aneurysm, smooth muscle cell, transcription factors, KLF4, smooth muscle cells
Introduction
Abdominal aortic aneurysms (AAA) comprise the 10th leading cause of death among men over 65 and result in more than 15,000 surgical procedures annually in the United States 1, 2. Although prevalence of aneurysms is on the rise, the mechanisms that regulate aortic aneurysm formation remain unclear1, 2. Currently, aneurysm formation is normally accompanied by destruction of the intimal layer and loss of aortic smooth muscle cell contractile function3, 4. Furthermore, AAA formation is often accompanied by mural thrombus deposition, and infiltration by mesenchymal cells, processes influenced by immune regulation5, 6. Finally, collagen and elastin extracellular matrix (ECM) of the abdominal aorta is believed to undergo degradation and contribute to AAA formation via a myriad of mechanisms modulated by inflammatory cells 5, 6. The effects of smooth muscle cell apoptosis accompanied by ECM degradation of elastin and collagen leads to aneurysm formation; however, the mechanism that regulate this process and the cell-cell interactions are unknown.
Smooth muscle cells (SMC) are remarkably plastic and transition from a quiescent contractile state to a proliferative-migratory state during a process known as “phenotypic switching”7. This process is characterized by the coordinate down-regulation of markers of differentiated SMCs including SM22α, smooth muscle myosin heavy chain (SM-MHC), and SM α-actin, genes required for SMC contraction7. SMC phenotypic plasticity likely evolved for optimization of vascular repair following injury8, although it is also widely accepted that SMC phenotypic switching plays a key role in development and progression of atherosclerotic lesions9, and regulation of plaque stability. Our laboratory was the first to demonstrate that SMC phenotypic switching was an early event in aortic aneurysm formation4. However, there is currently little information to describe the mechanism of smooth muscle cell phenotypic switching in the context of aneurysm formation4.
Previously KLF4 has been shown to be a key factor required for SMC phenotypic switching following PDG-BB and POVPC treatments10–13. KLF4 has also been shown to play a critical role in the maintenance of pluripotency in embryonic stem cells (ESC)14, then declines as ESC differentiate into various cell types. KLF4 is transcriptionally induced in macrophages, SMCs, and other cell types with vascular injury-inflammation15,16. KLF4 along with Oct4, Sox2 and c-myc are also capable of reprogramming somatic cells into induced pluripotential stem (iPS) cells17. In addition, global conditional KLF4 knockout mice demonstrated a delay in SMC phenotypic switching following carotid artery endothelial injury18. Since SMC phenotypic switching occurs early in AAA formation, and the mechanisms by which this occurs in aneurysm formation are unknown, our overall goal was to investigate the effects of KLF4 on smooth muscle phenotype and aneurysm disease. In the present studies, we hypothesize that KLF4 deletion results in attenuated aneurysm formation and this occurs in part through phenotypic modulation of smooth muscle cells.
Materials and Methods
Defining KLF4 in the murine elastase aneurysm model
Two groups of 8–12 week old wild-type (WT) C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) underwent the murine aortic aneurysm model with either elastase or saline perfusion (WT elastase, WT saline) as described previously3, 4, 19–21. Infrarenal abdominal aortas from both groups were evaluated and harvested for tissue analysis at days 0, 3, 7, and 14.
Murine Elastase Model
Eight to 12-week male mice were injected with intraperitoneal (IP) ketamine solution, elastase perfused and harvest as previously described and carried out by Johnston et al.3, 4, 19–21. The aortas (or aneurysms, when present) were harvested, and either: 1) snap frozen in liquid nitrogen for analyses by real-time polymerase chain reaction (qPCR) or protein extraction, or 2) incubated overnight for histology or immunohistochemistry. Animal care and use were in accordance with the Guide for the Care and Use of Laboratory Animals. The animal protocol was approved by the University of Virginia Institutional Animal Care and Use Committee (#3634) in compliance with the Office of Laboratory Animal Welfare.
Creation of KLF4 transgenic mice
ERTCre+ KLF4 flx/wt mice and ERTCre+ KLF4 flx/wt mice were created by breeding ERT2 tamoxifen Cre mice with KLF4 flx/flx mice15, 18, 22. These mice are heterozygous knock-outs of KLF4 in all cell types and are necessary to use as ERTCre+ KLF4 flx/flx mice are lethal injections as they develop epithelial and gastrointestinal defects15, 23. When performing experiments ERTCre+ KLF4 flx/wt mice and their sibling age matched controls were used when performing elastase perfusion. These mice underwent a series of ten tamoxifen injections at 6–8 weeks over a 14 day period followed by 14 days of rest. These mice then underwent elastase perfusion followed by harvest 14 days later.
MYHCre+ KLF4 flx/flx, MYHCre+ flx/wt, and MYHCre+ wt/wt mice were created by breeding MYHCre+ mice[24] with KLF4 flx/flx mice15, 18, 22. These mice were then back-bred together to produce the experimental and control MYHCre+ KLF4 mice. The MYHCre+ transgene has been used successfully in the past to demonstrate specific smooth muscle specific knock-out of various genes following tamoxifen injections24, 25. These mice underwent the same series of injections and injuries as mentioned for the ERTCre+ mice above.
ERTCre+ KLF4 flx/wt ApoE−/− mice were created by breeding ERTCre+ KLF4 flx/flx onto an ApoE−/− background. Once ERTCre+ KLF4 flx/flx ApoE−/− mice were created, these mice were then bred with ApoE−/− mice to produce the experimental ERTCre+ KLF4 flx/wt ApoE−/− mice and their experimental controls. These mice underwent the same series of ten tamoxifen injections at 6–8 weeks of age followed by two weeks of rest and then angiotensin II infusion via osmotic pump.
MYHCre+ KLF4 flx/flx, flx/wt and wt/wt ApoE−/− mice were created by breeding MYHCre+ KLF4 flx/flx mice onto an ApoE−/− background. These mice were then back-bred together to create MYHCre+ KLF4 flx/wt ApoE−/− mice which were then bred together to produce the experimental MYHCre+ KLF4 flx/flx and flx/wt ApoE−/− mice and their WT controls. These mice also underwent the same series of injections, two weeks of rest and Angiotensin II infused osmotic pump insertion as mentioned previously.
Angiotensin II Infusion
Osmotic pumps (Alzet® 2004, Durect Corp., Cupertino, California) containing Ang II (1000 ng/kg/min, Sigma Aldrich Inc., St. Louis, Missouri) were introduced into 10-week-old ERTCre+ KLF4 flx/wt ApoE−/− and ERTCre- KLF4 flx/wt ApoE−/− male mice as previously described26, 27. Mice were housed and maintained at 70°F, 50% humidity, in 12-hour light-dark cycles per institutional animal protocols. All mice were fed ad libitum water and placed on high fat diet (TD 88137, Harlan Teklad Inc., Indianapolis, Indiana) with no restrictions on movement. Aneurysmal segments of the aortas (proximal to the renal arteries) were harvested after 28 days and processed for histology. At day 28, video micrometry measurements of the aortic wall diameter were performed in situ using a Q-Color3 Optical Camera (Olympus Corp., Center Valley, Pennsylvania) using QCapture Pro Software version 6.0 (QImaging Inc., Surrey, Canada). MYHCre+ KLF4 flx/flx, flx/wt and wt/wt ApoE−/− male mice underwent a similar procedure at 10 weeks of age as mentioned above. Kaplan-meier curves and log-rank (Mantel-Cox) tests tracked the percentage survival of mice over the 28 day period.
In a second model of Angiotensin II infusion, osmotic pumps containing Angiontensin II were introduced into ten-week-old ERTCre+ KLF4 flx/wt and ERTCre- KLF4 flx/wt male mice as previously described26, 27. Following Angiotensin II infusion, mice received biweekly injections of anti-TGFβ antibody(R and D systems) 28. After 28 days, aneurysmal segments were harvested as mentioned in the previous paragraph and percentage survival was tracked and log-rank(Mantel-Cox) tests were performed. The same procedure was performed for MYHCre+ KLF4 flx/flx, flx/wt and wt/wt mice at ten weeks.
mRNA and protein isolation
mRNA and protein were extracted from frozen aortic tissue samples with TRIzol reagent (Invitrogen, Life Technologies, Grand Island, NY) as previously described3,19,20.
Real time reverse transcription PCR analysis
Using isolated mRNA from the mouse aortas and aortic smooth muscle cells, cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-time reverse transcription PCR (RT-PCR) was performed using Sensifast SYBR Supermix (Bioline, Taunton, MA) with primers described previously10, 29–31. Target DNA was analyzed with Bio-Rad CFX Manager software (Bio-Rad, Hercules, CA) to obtain melt curves and takeoff values. Levels of mRNA were standardized either with GAPDH or 18s mRNA, which served as a housekeeping genes for comparison. All experiments were run three times in triplicate unless otherwise mentioned.
Histology
Murine aortas were harvested at sacrifice for histology analysis after undergoing left ventricular puncture and 4% paraformaldehyde (PFA) antegrade perfusion at physiologic pressure. Further fixation was achieved by overnight incubation in 4% PFA at 4°C followed by paraffin embedding and sectioning at 5 μm. After microwave antigen retrieval, antibodies were bound and detected using VectaStain Elite Kit (Vector Laboratories Inc., Burlingame, California). Antibodies for IHC staining were anti-rat Mac2 for macrophages (1:10,000; Cedarlane Laboratories, Burlington, Canada), anti-mouse anti-Neutrophil (Ly 6B.2) for neutrophils (1:10,000; AbD Serotec, Oxford, United Kingdom), anti-goat MMP2 for matrix metalloproteinase-2 (1:350; R&D Systems, Minneapolis, Minnesota), anti-goat MMP9 for matrix metalloproteinase-9 (1:400; R&D Systems, Minneapolis, Minnesota), and anti-mouse SMαA for smooth muscle α-actin (1:1000; Santa Cruz Biotechnology Inc., Santa Cruz, California). Visualization color development was completed using diaminobenzidine (Dako Corporation, Carpinteria, California) for SMαA, Mac2, anti-neutrophil, CD3ε, and MMP9.
Confocal immunohistochemistry was performed using immunoflourescent staining for KLF4 (1:200 dilution, R&D), macrophages with mac-2 (1:1000, Cedarlane Laboratories), smooth muscle cells with smooth muscle α actin (SMA) (1:1000, Abcam), and nuclei with DAPI (1:10,000, Invitrogen). Appropriate controls verified staining procedures. Human confocal immunoflourescent staining was performed for KLF4 (1:1000) dilution followed by TSA amplification using a TSA kit (Invitrogen), SMA for smooth muscle cells (1:1000, Cy3, Sigma Alrich Corporation), and nuclei using DAPI (1:10,000; Invitrogen).
Images were acquired using AxioCam Software version 4.6 via 10X, 40X, and 100X objectives and an AxioCam MRc camera (Carl Zeiss Inc., Thornwood, New York). Threshold gated positive signal was detected within the AOI and quantified using Image-Pro Plus version 7.0 (Media Cybernetics Inc., Bethesda, Maryland). Elastin degradation was quantified by counting the number of breaks per vessel and then averaged and graphed.
Cytokine Array
For the purpose of determining the effects of KLF4 total cell and smooth muscle cell deletion on pro-inflammatory cytokines in the aortic wall, mouse cytokine arrays (R&D Systems) were performed using isolated protein from mouse aortas at day 14 according to the manufacturer’s instructions (n=3 mice per group). Protein samples from each group were pooled for analysis, and all samples were run in duplicate19, 20.
Mouse aortic smooth muscle cell cultures
Mouse abdominal aortic smooth muscle were isolated and cultured as previously described5. For siRNA transfections, cells at passages 8–10 were plated in all 6 wells of a 6 well plate at 1×105 cells per well; 24 hours later cells were transfected with either a single siRNA to mouse KLF4 or and a non-targeting control11,12,32, 31. 24 hours later mouse aortic smooth muscle cells were stimulated by either elastin degradation products, phorbol ester, TGFbeta, Retinoic acid or IL1β (Upstate Biotechnology) or representative vehicle for 24 hours as described previously13. Cells were harvested and RNA was extracted using the TriZol method and RT-PCR was performed as described previously11, 12, 32.
ChIP Analysis
Quantitative chromatin immunoprecipitation assays (ChIP) were performed as described previously11, 33, 34. For each in vivo experiment, fifteen AAA samples were pooled into three sets of five samples each and ChIP analysis was performed. Experiments were carried out in triplicate and one representative experiment was shown. Antibodies include polycolonal KLF4 (R&D Systems). Real-time PCR primers were designed as previously described for SMA, SM22α, cFOS, and SM-MHC18, 34. For qPCR, 2 μl out of the 20 μl of extracted DNA was used in 50 cycles of amplification in 3 steps: 95°C for 15 s, 55°C for 30 s, and 68°C for 45 sec. At the end of the amplification cycles, dissociation curves were determined to rule out signal from primer dimers and other nonspecific dsDNA species. Data was normalized to IgG immunoprecipitated DNA levels. The size of the PCR products was confirmed on a 2% agarose gel stained with ethidium bromide.
Human Tissue Harvest
Normal abdominal aortas were obtained from transplant donors and aneurysmal aortas were taken from patients undergoing elective open abdominal aortic aneurysm repair. All aortic samples were harvested from patients without evidence of known collagen vascular disease. Aortic tissue specimens were explanted, placed on ice, and then immediately snap-frozen in liquid nitrogen. Collection of human tissue was approved by the patient’s written consent in compliance with the University of Virginia’s Human Subjects Review Committee (HSR #13178).
Statistical Methods
Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). Maximal aortic dilation (%) was calculated as [maximal aortic diameter − internal control diameter] ÷ internal control diameter * 100%. The internal control was a small segment of normal abdominal aorta just distal to the renal arteries that was above the proximal ligation. This section was not perfused with elastase, but it was susceptible to blood pressure changes from volume loss during the harvest as well as expected animal growth over time. Values are reported as mean ± standard error of the mean. Aortic dilation between groups was compared using Fisher’s exact test. Post hoc Tukey correction was applied to determine the significance of individual comparisons with α=0.05. When groups were compared, the Fisher’s exact test was used for significance. Pearson correlation coefficients (R) were used to determine strength of linear dependence and are reported as R value [95% confidence interval (CI)]. Differences between groups for histologic grading for macrophage infiltration, neutrophil infiltration, and elastin degradation were tested with the Fisher’s exact test. Angiotensin II experiment differences were measured using a log-rank(Mantel-Cox) test. All assays were performed in triplicate unless stated otherwise. Blinding to experimental groups was maintained as feasible. Data are presented as mean±SEM.
Results
KLF4 is elevated during Experimental Aneurysm Formation
KLF4 has previously been shown to be a key regulator of endothelial vascular injury and to be key in the down-regulation of smooth muscle marker gene expressio13,18. Therefore, we sought to determine if KLF4 expression increases during aneurysm formation. Aortic dilation in WT mice was significantly increased following elastase compared to saline perfusion20. Although no differences in aortic dilation were present at day 3, aortic dilation was apparent at day 7 (61.8 ± 4.0% with elastase vs. 47.6 ± 4.1% with saline, p < 0.05) and most prominent 14 days following perfusion (90.1 ± 3.7% with elastase vs. 45.8 ± 5.1% with saline, p<0.000120).
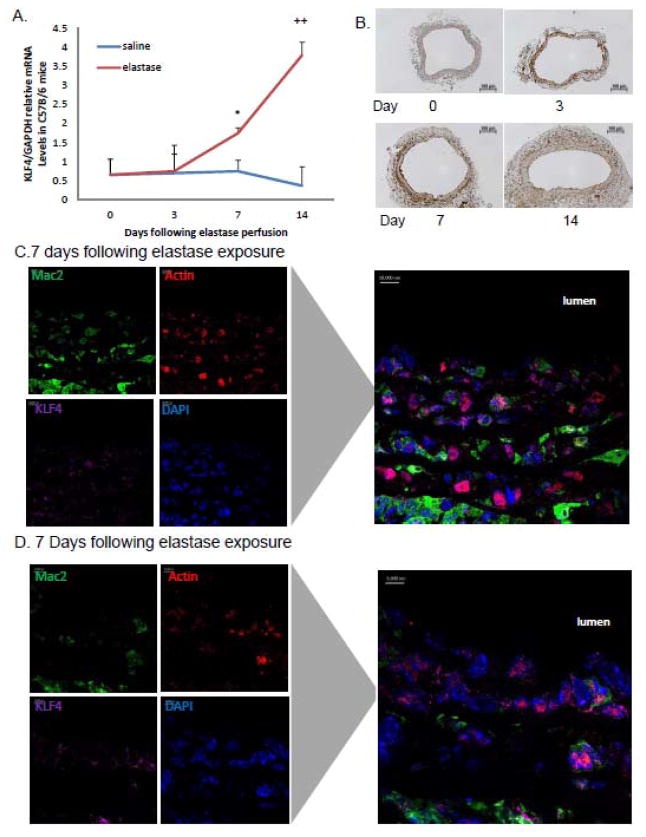
At baseline, murine aortas have low levels of KLF4 expression [Fig. 1A]. While there were no differences in KLF4 expression at 3 days, KLF4 expression at 7 and 14 days in elastase perfused aortas increased significantly compared to saline perfused aortas. Furthermore, by immunohistochemistry, KLF4 expression progressively increased following elastase perfusion as aortic diameter increased [Fig. 1B and Supplemental Fig. 1A]. To determine which cells types specifically expressed KLF4, confocal fluorescence microscopy was performed to evaluate macrophages, smooth muscle cells, and KLF4 7 days following elastase exposure. KLF4 co-localized primarily to intramural smooth muscle cells and macrophages [Fig. 1C and Supplemental Fig. 1B and C]. Aortic samples from 14 day elastase perfused aortas demonstrated decreased smooth muscle marker expression (data not shown), consistent with ours and others’ previous studies4,35,36.
Figure 1.
KLF4 is localized to smooth muscle cells and macrophages following elastase perfusion. A) C57Bl/6 mice underwent aortic perfusion with elastase or saline and animals were recovered. Aortas were harvested on days 0, 3, 7, and 14. RT-PCR for KLF4 gene expression relative to GAPDH from C57Bl/6 mice perfused with elastase or saline over time. N=6 mice per group. * = p<0.05 for WT elastase vs. saline while ++=p<0.005 for WT elastase vs saline via two way ANOVA. B) Aortic cross sectional histology with KLF4 antibody at day 0, 3, 7 and 14 days following elastase perfusion(n=6 mice per group). C) Confocal immunohistochemistry performed on aortic tissue from a WT elastase perfused mouse on day with macrophages (MAC2), smooth muscle cells (SMA), KLF4, and cell nuclei (DAPI). Confocal studies done at day 14 are not shown.
Heterozygous KLF4 deletion results in attenuated Aneurysm Formation
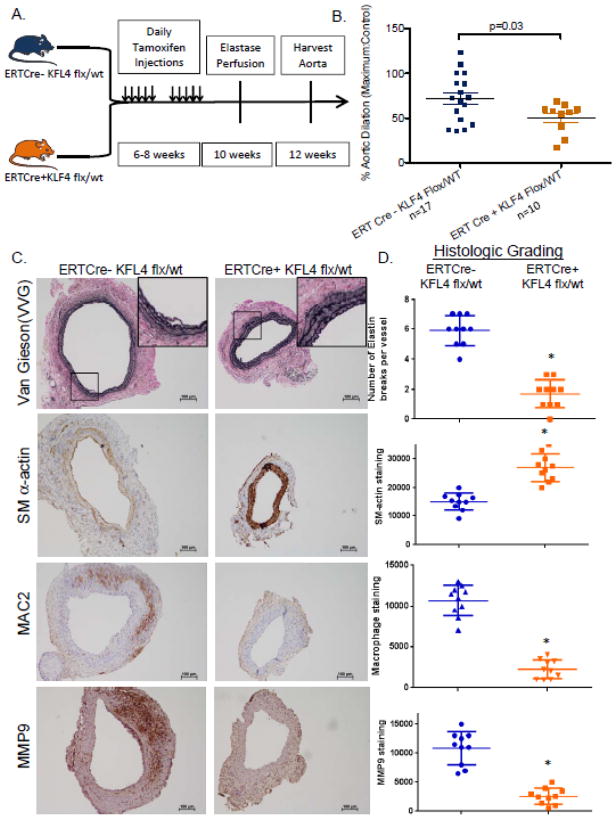
With KLF4 levels upregulated in AAA and known to alter vascular injury responses, we hypothesized that KLF4 plays a critical role in aneurysm formation. To test this, we required synthesis of unique transgenic mice since conventional KLF4 knock-out mice are embryonic lethal23. Tamoxifen inducible ERTCre+ KLF4 flx/wt mice and their controls were generated. These mice are total-cell tamoxifen dependent KLF4 heterozygous knock-outs and were necessary to use as inducible homozygous KLF4 knock-outs are lethal due to epithelial skin and gastrointestinal defects15, 23. Heterozygous KLF4 knockout mice received 10 tamoxifen injections to induce KLF4 loss. After a two week recovery period, elastase perfusion was performed and mice were harvested at 14 days [Fig. 2A]. Heterozygous KLF4 knock-out mice had significant reduction in maximal aortic dilation compared to WT mice [ERTCre+ KLF4 flx/wt:50.19+/−17.19% n=10 vs. ERTCre- KLF4 flx/wt: 72.27+/−27.13% n=17, p<0.003, Fig. 2B].
Figure 2.
Heterozygous deletion of KLF4 results in attenuated aneurysm formation. A) Model depicting the timeline of tamoxifen injections followed by recovery and subsequent elastase perfusion for the ERTCre +/− KLF4 Flx/wt (total-cell heterozygous KLF4 deleted mice). B) Maximal % aortic dilation is shown for the two groups of mice at day 14 [ERTCre+ KLF4 flx/wt:50.19+/−17.19% n=10 vs. ERTCre- KLF4 flx/wt: 72.27+/−27.13% n=17, p<0.003 by Fisher’s exact test, Fig. 2B]. C) Elastin fibers shown with Verhoeff-van Gieson (VVG) staining. Smooth muscle cell staining shown by SM α-actin staining. Macrophage and MMP9 staining were visualized with MAC2 and anti-MMP9 respectively. D) Quantification of number of elastin breaks per vessel. * indicates significantly less elastin breaks per vessel with a p-value <0.05 compared with WT controls. Quantification of SM α-actin, MAC2, and MMP9 staining using imagePro. * indicates significantly less staining with a p-value <0.05 by Fisher’s exact test as compared with WT elastase perfused controls.
Next, given the significant difference in aneurysm formation between KLF4 flx/wt ERT Cre+ mice and their controls, we examined elastin degradation and expression of SM α-actin and MAC2 into the medial cell layer at 14 days. Vernhoff’s Van Gieson staining demonstrated significantly decreased elastin fragmentation in KLF4 heterozygous knock-out mice [Fig 2C and 2D], while smooth muscle cell α-actin staining was significantly increased in ERTCre+ KLF4 flx/wt knock-outs compared with WT controls [Fig. 2C and 2D]. Moreover, KLF4 heterozygous deletion resulted in decreased macrophage and neutrophil staining compared with WT controls [Fig. 2C and Supplementary Fig 2]. Finally, MMP9 but not MMP2 had significantly less staining in heterozygous KLF4 knockouts compared to controls at 14 days [Fig 2C, 2D and Supplementary Fig. 2]. The preceding data suggests that deletion of even 1 allele of KLF4 in the entire animal attenuates aneurysm formation and demonstrates that KLF4 is important for SMC marker gene expression as well as macrophage and neutrophil recruitment into the aorta.
Smooth muscle specific deletion of KLF4 attenuates aneurysm formation
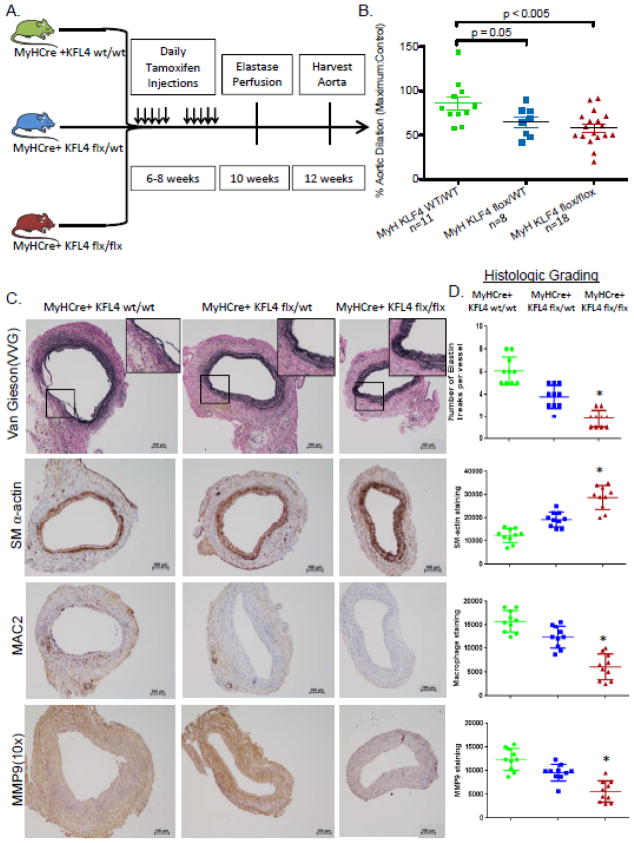
As there are both smooth muscle and leukocyte sources of KLF4 [Fig 1C], we next sought to determine the contribution of smooth muscle derived KLF4 in AAA. To investigate this, smooth muscle specific myosin heavy chain (MYH) Cre mice24 were bred with KLF4 flx/flx mice to produce novel MYHCre+ KLF4 Flx/Flx, MYHCre+ KLF4 Flx/wt, and MYHCre+ KLF4 WT/WT mice[Supplemental Fig 3]. Using a tamoxifen-elastase experimental design [Fig 3A], deletion of 1 or 2 alleles of smooth muscle specific KLF4 showed significant step-wise protection from AAA formation compared to controls [MYHCre+ KLF4 Flx/Flx: 57.97 +/−18.64% n=18 vs. KLF4 flx/wt SM-MHC 64.83+/− 16.84 n=8 vs. MYHCre+ KLF4 WT/WT:85.89+/−24.45 n=11; Fig 3B]. Elastin degradation was significantly less in heterozygous mice [MYHCre+ KLF4 flx/wt] and even greater protection in homozygous smooth muscle deleted KLF4 mice [MYHCre+ KLF4 flx/flx, Fig. 3C]. Similarly, there was evidence of increasing protection from macrophage and neutrophil inflammation in the MYHCre+ KLF4 flx/wt and flx/flx animals compared to WT animals [Fig. 3C and supplemental Fig 4]. Smooth muscle cell deletion of KLF4 resulted in significantly less MMP2 and MMP9 staining [Fig 3C, D and Supplemental Fig. 4].
Figure 3.
Smooth muscle specific knock-out of KLF4 results in attenuated aneurysm formation. A) Model depicting the timeline of tamoxifen injections followed by recovery and subsequent elastase perfusion for the MYHCre+/− KLF4 mice. B) Maximal % aortic dilation is shown for the three groups of mice at day 14. Sample images are shown [MYHCre+ KLF4 Flx/Flx: 57.97 +/−18.64% n=18 vs. KLF4 flx/wt SM-MHC 64.83+/− 16.84 n=8 vs. MYHCre+ KLF4 WT/WT:85.89+/−24.45 n=11, p<0.05 by Fisher’s exact test]. C) Elastin fibers shown with Verhoeff-van Gieson (VVG) staining. Smooth muscle cell staining shown by SM α-actin staining. Macrophage and MMP9 staining were visualized with MAC2 and anti-MMP9 respectively. D) Quantification of number of elastin breaks per vessel. * indicates significantly less elastin breaks per vessel with a p-value <0.05 compared with WT controls. Quantification of SM α-actin, MAC2, and MMP9 staining using imagePro. * indicates significantly less staining with a p-value <0.05 by Fisher’s exact test as compared with WT elastase perfused controls.
KLF4 modulates cytokine expression and smooth muscle marker genes during aneurysm formation
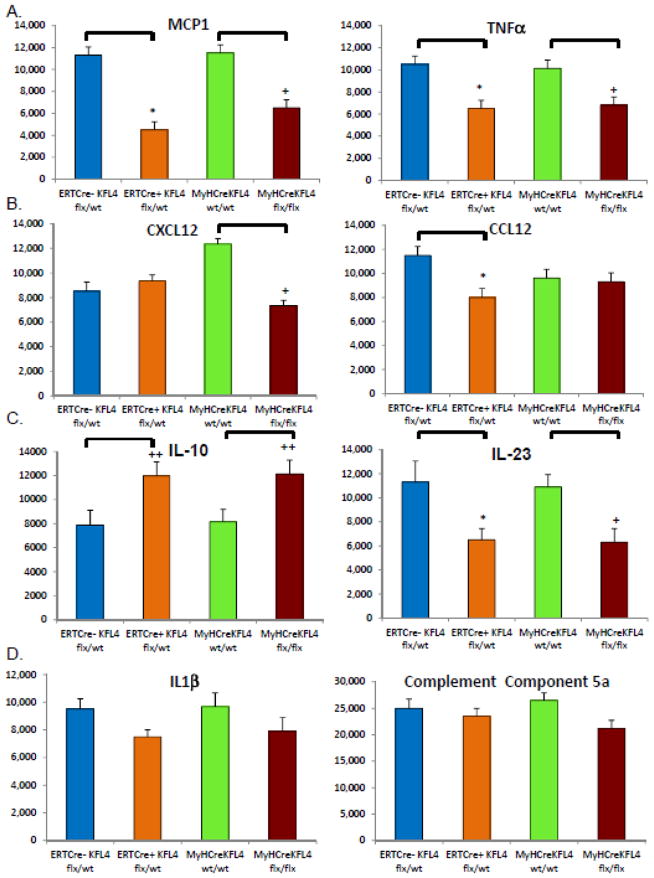
In order to determine cytokines involved in AAA formation that may be modulated by KLF4, cytokine protein array analysis was performed on purified protein extracts from elastase perfused aortic tissue harvested 14 days following perfusion. Aortic tissue from total cell heterozygous deletion had significantly less MCP1, TNFα, CCL12, and IL-23 [Fig 4]. Conversely, there were increased levels of IL-10, a known anti-inflammatory cytokine [Fig. 4]. Interestingly, KLF4 did not modulate IL-1β, CXCL12 or Complement Component 5A cytokine levels which have been linked previously to experimental AAA20,37.
Figure 4.
KLF4 deletion alters cytokines important to aneurysm formation following elastase perfusion. Protein array for ERTCre+ KLF4 flx/wt (Global KLF4 heterozygous knock-out), ERTCre- KLF4 flw/wt (Global control), MYHCre+ wt/wt (Smooth muscle control) and MYHCre+ KLF4 flx/flx (Smooth muscle homozygous KLF4 KO) elastase perfused and harvested at day 14 evaluating CXCL12, CCL12, Complement component 5a, MCP1, TNFα, TL-23, IL1β, IL-10. Three mouse aortas were pooled for each array. Results are the average of two independent experiments19,20. * indicates significant down-regulation of ERTCre+ KLF4 flx/wt mice in comparison to their WT controls with p-value <0.05 by Fisher’s exact test. + indicates significant down-regulation of MYHCre+ KLF4 flx/flx mice in comparison to their WT controls with p-value <0.05 compared to WT control by Fisher’s exact test. ++ indicates significant up-regulation of ERTCre+ KLF4 flx/wt mice as compared to their WT controls with p-value <0.005 by Fisher’s exact test. +++ indicates significant up-regulation of MYHCre+ KLF4 flx/flx mice in comparison with their controls by Fisher’s exact test.
Cytokine array analysis was also performed on aortic tissue harvested on day 14 from smooth muscle specific homozygous KLF4 deleted mice [MYHCre+ KLF4 flx/flx] and WT control mice [MYHCre+ KLF4 wt/wt]. Cytokine levels of MCP1, TNFα, CXCL12, and IL-23 were decreased in KLF4 deleted mice [MYHCre+ KLF4 flx/flx mice, p<0.05 per Fisher’s exact test for all, Fig 4]. Moreover, smooth muscle specific knock-out of KLF4 increased IL-10 levels. KLF4 knock-out in smooth muscle cells did not modulate IL-1β, CCL12 or Complement Component 5A cytokine levels [Fig. 4]. Of interest, CXCL12 was not modulated in global heterozygous KLF4 knockout mice (ERTCre+ KLF4 flx/wt) while smooth muscle specific knock-out of KLF4 modulated CXCL12. Conversely, CCL12 was modulated with total cell heterozygous knock-out of KLF4 while KLF4 was not modulated in the smooth muscle specific knock-out of KLF4.
To determine if these protein cytokine levels are reflected in changes in staining patterns, we stained total cell heterozygous and smooth muscle specific KLF4 knock-out mice with an antibody to MCP-1, a cytokine we previously documented to be highly critical in experimental AAA3. Heterozygous and smooth muscle specific knock-out of KLF4 resulted in decreased MCP-1 staining [Supplemental Fig. 5]. Taken together, these data indicate that KLF4 deletion decreased protein cytokine levels of a number of relevant inflammatory cytokines in aortic aneurysms.
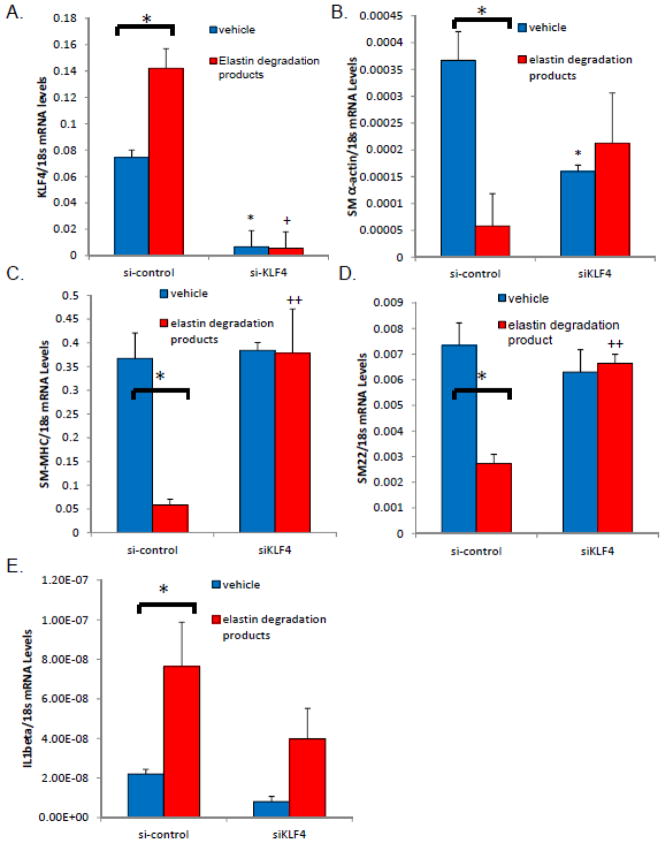
siRNA to KLF4 inhibits downregulation of SMC marker genes in vitro
Previously, KLF4 has been shown to repress SMC marker gene expression in vivo and in cultured SMC through multiple mechanisms10, 34, 38 and most recently via directly binding to smooth muscle marker genes in vivo via the G/C Repressor element to mediate transcriptional silencing following injury13. Currently, there is no known role for KLF4 modulation of smooth muscle marker genes during aneurysm formation. To determine if KLF4 could mediate SMC marker gene down-regulation during aneurysm formation, we isolated primary mouse abdominal aortic smooth muscle cells. We next treated these cells with siRNAs to KLF4 or control siRNAs followed by novel treatment with elastin degradation products [10 ng/mL]. We verified that KLF4 mRNA expression was down-regulated via RT-PCR with siRNA treatment [Fig. 5A]. Second, we examined the levels of SM α-actin, SM22α, and SM-MHC levels following elastin degradation product treatment (EDP)[Fig. 5B–D]. All three smooth muscle marker genes were down-regulated with treatment of elastin degradation product treatment for 24 hours, similar to what has been seen previously with PDGF-BB or POVPC treatments10,12. Importantly, transfection of siRNAs to KLF4 resulted in a lack of down-regulation of smooth muscle marker genes following EDP treatment [Fig. 5B–D]. IL1β did not demonstrate de-regulation in response to siKLF4 [Fig. 5E]. These findings are consistent with our in vivo studies and suggest that KLF4 is a key modulator of smooth muscle gene expression in vitro following treatment with elastin degradation products.
Figure 5.
Knock-down of KLF4 modulates smooth muscle marker genes following elastin degradation product treatment. A) Mouse abdominal aortas were isolated, the adventia and endothelial layer stripped and the abdominal cells were placed in culture. Cells were cultured until passage 6 and smooth muscle marker genes were verified at passage six and then again at passage 11 for expression (Data not shown). From these cultures, cells were plated at 1×105 cells per well and treated with siKLF4 or control, scrambled siRNA in serum free media13. 24 hours following transfection cells were treated with elastin degradation products for 24 hours and then harvested, RNA extracted, and RT-PCR performed. Brackets indicate a significant response (p>0.05) to treatment with elastin degradation products. * indicates significant down-regulation of vehicle treated cells in response to siKLF4 expression as compared with control treated siRNA by Fisher’s exact test. Results are the average of three independent experiments performed in triplicate.
In separate experiments, we demonstrated that KLF4 appears to be important for smooth muscle marker gene down-regulation in abdominal aortic smooth muscle cells following IL1β treatment, since we have shown previously IL1β and Receptor deleted mice are highly protected from aneurysm formation20[Supplemental Fig. 6]. In additional subsequent experiments, we demonstrated that KLF4 over-expression alone appears to inhibit smooth muscle marker gene expression and that this effect is enhanced with cytokine treatment [Supplemental Fig. 7]. However, KLF4 did not negatively affect the expression of smooth muscle marker genes following retinoic acid, phorbol ester or TGF-β treatment [Supplemental Fig. 8–10].
KLF4 binds to promoters of SMCs in vitro and in vivo during aneurysm formation
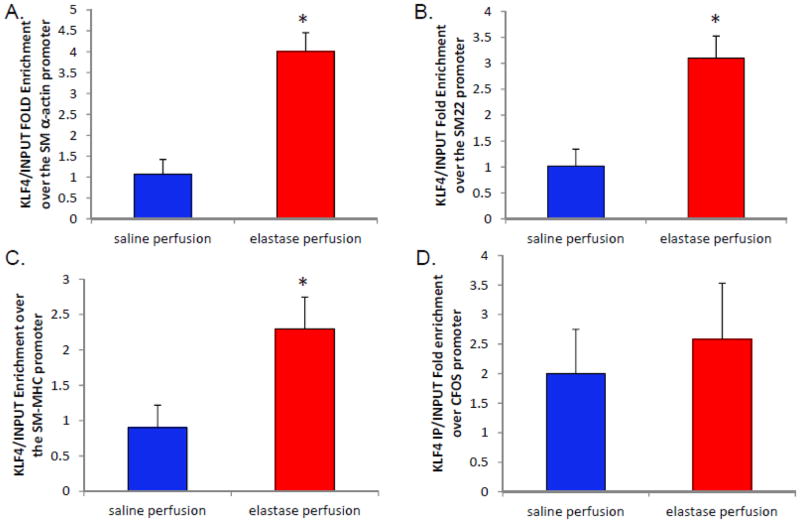
Previously, it has been demonstrated that KLF4 binds directly to smooth muscle marker genes following PDGF-BB and POVPC treatment13; however, there is no evidence to date that KLF4 modulates smooth muscle marker genes during aneurysm formation. We treated abdominal aortic mouse smooth muscle cells with elastin degradation products for 24 hours following serum starvation and then performed chromatin immunoprecipitation (ChIP) analysis to determine if KLF4 binds to smooth muscle marker genes following EDP treatment [Supplemental Fig. 11]. KLF4 significantly binds to the SM α-actin, SM22α, and SM-MHC promoters following EDP treatment [Supplemental Fig. 11]. These data indicate that KLF4 regulates smooth muscle marker gene down-regulation in vitro following EDP treatment.
Importantly, KLF4 binding to SMC marker genes has never been evaluated in vivo during aneurysm formation. Thus, ChIP assays were performed on aortas from WT mice 14 days following elastase perfusion. KLF4 was found to bind specifically to the promoters of smooth muscle marker genes SM α-actin, SM22α and SM-MHC, while KLF4 did not bind the C-FOS promoter either before or after elastase perfusion [Fig. 6]. Collectively, these data suggest a new model where KLF4 plays a role in the phenotypic modulation of smooth muscle cells and suggest that KLF4 plays a role in aneurysm formation via smooth muscle cell modulation.
Figure 6.
KLF4 bind smooth muscle cell marker genes in vivo during aneurysm formation. A–D) 14 day elastase and saline perfused C57/B6 mice underwent ChIP analysis for KLF4 binding in vivo during aneurysm formation13. Aortas were ground and subsequently underwent formalin crosslinking and sonication. Following sonication and pre-clearage, the homogenate underwent immunoprecipitation for either KLF4 or IgG. The precipitate was then washed and the crosslinking was reversed and proteins denatured. RT-PCR was then performed for SM α-actin, SM22α, SM-MHC and c-FOS. Results are the average of three independent pooled experiments of 15 animals each performed in triplicate. *indicates significant binding over controls with a p-value <0.05 by Fisher’s exact test.
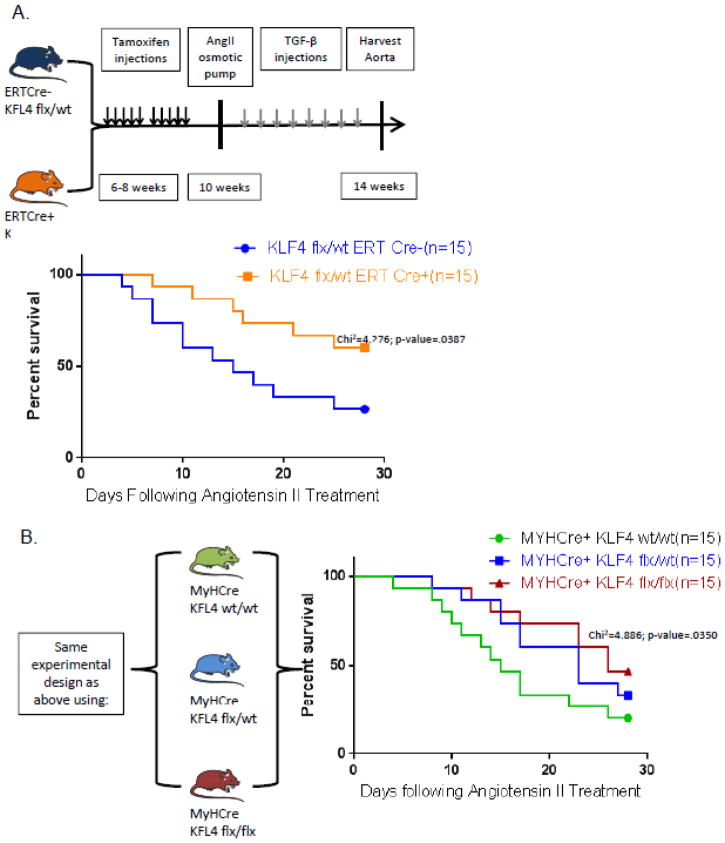
KLF4 deletion attenuates aneurysm formation following Angiotensin II treatment
Since the elastase perfusion model is criticized as an acute artificial (surgical) model of AAA, we sought to determine if KLF4 could also attenuate aneurysm formation in other established models of aneurysm formation. To determine if KLF4 is protective of aneurysm formation we used: 1) ERTCre+ KLF4 flx/wt mice bred on an ApoE−/− background infused with Angiotensin II for 28 days26[Supplemental Fig. 12A]; and 2) ERTCre+ KLF4 flx/wt mice infused with Angiotensin II with a series of TGFβ injections[Fig. 7A]28. Kaplan-meier curves of ERTCre+ KLF4 flx/wt deleted mice demonstrated that KLF4 protects from death due to aneurysm rupture in both Angiotensin II models of aneurysm formation [Fig. 7A, B and Supplemental Fig. 6A, Chi2=4.276, p-value= .0387 by log-rank test].
Figure 7.
KLF4 affects aneurysm formation following Ang II pump insertion. A) Model depicting the timeline of tamoxifen injections followed by recovery and subsequent Angiotensin II pump insertion for ERTCre +/− KLF4 Flx/wt mice (Global heterozygous KLF4 deleted mice). Kaplan-meier survival curves of ERTCre+/− KLF4 flx/wt mice following Angiotensin II pump insertion with log-rank tests as indicated28. B) Model for the MYHCre+ KLF4 flx/flx, flx/wt, and wt/wt mice and pump insertion.
To further study the role of KLF4 in smooth muscle cells specifically, we also utilized these two models of Angiotensin II aneurysm formation in MYHCre+ KLF4 flx/flx, flx/wt, wt/wt. Smooth muscle specific knock-out of KLF4 was also protective of mouse survival over the 28 day course of aneurysm formation following Angiotensin II pump insertion [Fig. 7B and Supplemental Fig. 12B, Chi2=4.886 p-value=0.035 by log-rank test]. These data further demonstrate the critical importance of smooth muscle derived KLF4 in two additional models of aneurysm formation enhancing our findings within the elastase model of aneurysm formation.
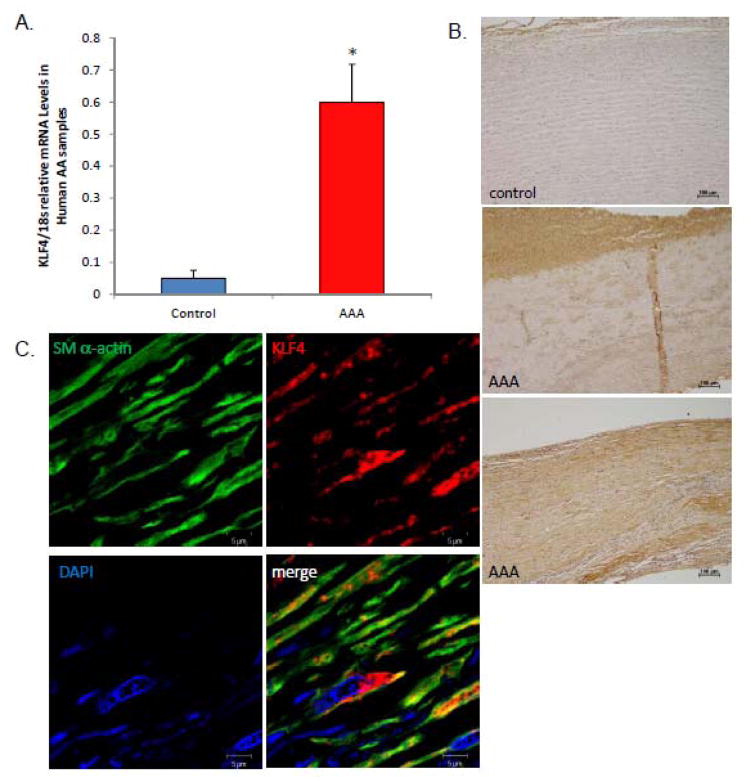
KLF4 is up-regulated in human aneurysms and is located in smooth muscle cells
We have seen that KLF4 is important in multiple models of murine aneurysm formation; however, we sought to determine if KLF4 could be relevant in human aortic aneurysms. KLF4 levels were increased in human AAA compared to healthy donor controls via RT-PCR [Fig. 8A]. KLF4 staining was markedly increased in human aneurysm tissue versus controls via immunohistochemistry [Fig. 8B]. Finally, confocal immunofluorescence of SM α-actin and KLF4 demonstrated that KLF4 (red) and SM α-actin (green) are localized in the same cells within human tissue as in our experimental model [Fig. 8C]. These data suggest that KLF4 may be relevant in human aneurysm formation and, in conjunction with our experimental models, suggest that KLF4 could be considered as a novel therapeutic target in human aneurysm formation.
Figure 8.
KLF4 is elevated in human AAAs and localizes to smooth muscle cells. A) qPCR of KLF4 in human AAA tissue versus healthy donor abdominal aorta. * indicates significant binding over human controls. B) Immunohistochemical staining of KLF4 in control versus aneurysm tissue. C) Confocal immunohistochemistry performed on human aneurysm tissue stained with SM α-actin, KLF4, and cell nuclei.
Discussion
Deletion of KLF4 results in decreased aortic aneurysm formation in both a heterozygous and a smooth muscle specific knock-out mouse elastase AAA model. KLF4 deletion also affected mouse survival from aneurysm rupture in two angiotensin II models of aneurysm formation. More importantly, we demonstrated KLF4 attenuates smooth muscle marker gene expression and that KLF4 binds to smooth muscle marker genes in vivo during aneurysm formation. These studies document KLF4 binding to smooth muscle marker gene promoters in vivo in the context of aneurysm formation. Moreover, we verified KLF4 is upregulated in humans during aneurysm formation and that KLF4 is localized to smooth muscle cells. The present studies suggest that KLF4 contributes to suppression of SMC marker genes in response to an ever-growing range of agents in vitro and in vivo. Taken together, our results identify KLF4 as a molecular regulator of aneurysm formation that mediates SMC phenotypic switching during aneurysm formation.
A key unresolved question is the mechanism of activation of KLF4 expression in SMC, as the cytokines that up-regulate this gene are most likely key to the initiating progression of the aneurysm phenotype. Furthermore, KLF4 activation is of great interest given this gene is normally epigenetically silenced in virtually all differentiated somatic cells39, 40. The KLF4 promoter contains a number of conserved regulatory elements for AP-1, GATA-1, Sp1, NFκB, and HLH factors22; however, few studies have directly assessed activation of the KLF4 promoter under different stimuli30, 41. There are a number of relevant unresolved issues regarding mechanisms responsible for activation of KLF4 during aneurysm formation. First, few studies have been published that have determined mechanisms that activate KLF4 upon vascular injury or disease. These mechanisms may be key to blocking up-regulation of KLF4 and their inhibition could represent a reasonable drug target against aneurysm formation. Second, the mechanisms responsible for controlling the different roles of KLF4 in various cell types during vascular injury and/or disease remain unclear and at times contradictory. It is possible that post-translational modifications, such as differential mRNA splicing, or chemical modifications, such as sumoylation or acetylation, regulate effects of KLF4 in different cell types. It has been shown that TGFβ induced expression of SM α-actin in cultured SMC is mediated in part through inactivation of KLF4 through protein sumoylation42. Thus, selectively targeting these modifications of KLF4 to produce a net decline in aneurysm formation may pose a challenge. Alternatively, targeting the down-stream effects of KLF4 activation could also be considered. Third, other Kruppel-like factors may be involved in regulating aneurysm formation. For example, KLF2 has been shown to have a profound role in atherosclerosis43 and future investigation will merit studying both KLF2’s role during aneurysm formation as well as the interplay among multiple KLFs. In summary, further studies are needed to determine if these factors regulate activation of KLF4 in SMC in vivo and if post-translational modifications regulate KLF4 function.
These studies suggest that KLF4 could be a potential therapeutic target against aneurysm formation. However, there are a number of potential challenges associated with targeting a transcription factor as a potential drug target. Since KLF4 is a transcription factor and can have both positive and negative interactions on many different promoters in many different cell types, the challenge will be to specifically design a drug target that can modulate KLF4’s negative effects on aneurysm formation. Interestingly, our use of a KLF4 heterozygous tamoxifen-dependent total knock-out of KLF4 [Fig. 2] reflects decreases we would see with pharmacologic inhibition of KLF4 and suggests that inhibition of KLF4 in all cell types would be overall beneficial for preventing aneurysm formation. However, it is unknown if inhibition of KLF4 in humans would ultimately revert SMCs to a non-inflammatory phenotype in AAA. Another additional issue that should be considered is KLF4 is one of the transcription factors required for stem cell function. Our lab has demonstrated previously that stem cells appear to be pro-active when added exogenously following aneurysm treatments19; however, KLF4 expression during aneurysm formation appears to be detrimental for aneurysm formation. How are these two issues resolved within the cellular context and what sort of modulated cell type does KLF4 cause smooth muscle cells to become? These issues have to be addressed and sufficiently understood in order for KLF4 to be utilized as a therapeutic target for aortic aneurysms.
In summary, the present studies provide the first direct evidence that KLF4 is an important regulator of aneurysm formation specifically in smooth muscle cells. Moreover, our results support a model whereby KLF4 mediates phenotypic switching of SMCs in vivo during aneurysm formation via binding to smooth muscle marker gene promoters. Although the present studies have focused on phenotypic modulation of smooth muscle cells and KLF4 during aneurysm formation, KLF4 also may have broad significance for the overall aneurysm phenotype given that KLF4 is already known to play key roles in mediating cellular function and proliferation in other cell types16, 23, 39, 44. However, further studies are needed to directly test if KLF4 play similar roles in these other cell types during aneurysm formation.
Supplementary Material
Acknowledgments
We thank Anthony Herring, Rupande Tripathi, Sandi Walton, Melissa Bevard, and Laura Shankman for their knowledge and technical expertise.
Sources of Funding: This work was supported by NIH K08 HL098560 (G.A.) and RO1 HL081629 (G.R.U.) grants. This project was supported by Award Number T32HL007849 from the National Heart, Lung, and Blood Institute(NHLBI) (W. Johnston, PI: Irving L. Kron, MD), and by the Thoracic Surgery Foundation for Research and Education (TSFRE) Research Grant (PI: G. Ailawadi). The content is solely the responsibility of the authors and does not necessarily represent the views of the NHLBI or the TSFRE.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Lederle FA, Johnson GR, Wilson SE. The aneurysm detection and management study screening program: Validation cohort and final results. Archives of Internal Medicine. 2000;160:1425–30. doi: 10.1001/archinte.160.10.1425. [DOI] [PubMed] [Google Scholar]
- 2.Lederle FA, Johnson GR, Wilson SE. Yield of repeated screening for abdominal aortic aneurysm after a 4-year interval. Archives of Internal Medicine. 2000;160:1117–21. doi: 10.1001/archinte.160.8.1117. [DOI] [PubMed] [Google Scholar]
- 3.Moehle CW, Bhamidipati CM, Alexander MR, Mehta GS, Irvine JN, Salmon M, Upchurch GR, Kron IL, Owens GK, Ailawadi G. Bone marrow derived MCP1 required for experimental aortic aneurysm formation and smooth muscle phenotypic modulation. J Thorac Cardiovasc Surg. 2011;142:1567–74. doi: 10.1016/j.jtcvs.2011.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ailawadi G, Moehle CW, Pei H, Walton SP, Yang Z, Kron IL, Lau CL, Owens GK. Smooth muscle phenotypic modulation is an early event in aortic aneurysms. J Thorac Cardiovasc Surg. 2009;138:1392–9. doi: 10.1016/j.jtcvs.2009.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu K, Mitchell RN, Libby P. Inflammation and Cellular Immune Responses in Abdominal Aortic Aneurysms. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26:987–94. doi: 10.1161/01.ATV.0000214999.12921.4f. [DOI] [PubMed] [Google Scholar]
- 6.Freestone T, Turner RJ, Coady A, Higman DJ, Greenhalgh RM, Powell JT. Inflammation and Matrix Metalloproteinases in the Enlarging Abdominal Aortic Aneurysm. Arteriosclerosis, Thrombosis, and Vascular Biology. 1995;15:1145–51. doi: 10.1161/01.atv.15.8.1145. [DOI] [PubMed] [Google Scholar]
- 7.Chamley-Campbell J, Campbell G, Ross R. The smooth muscle cell in culture. Physiol Reviews. 1979;59:1–6. doi: 10.1152/physrev.1979.59.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Owens GK. Regulation of Differentiation of Vascular Smooth Muscle Cells. Physiol Reviews. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 9.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. American Journal of Physiology - Cell Physiology. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 10.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, Leitinger N, Owens GK. Oxidized Phospholipids Induce Phenotypic Switching of Vascular Smooth Muscle Cells In Vivo and In Vitro. Circ Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. American Journal of Physiology - Cell Physiology. 2008;295:C1175–C1182. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. American Journal of Physiology - Cell Physiology. 2007;292:C886–C895. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 13.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative Binding of KLF4, pELK-1, and HDAC2 to a G/C Repressor Element in the SM22α Promoter Mediates Transcriptional Silencing During SMC Phenotypic Switching In Vivo / Novelty and Significance. Circ Res. 2012;111:685–96. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–60. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 15.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–45. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, SenBanerjee S, Jain MK. Kruppel-like Factor 4 Is a Mediator of Proinflammatory Signaling in Macrophages. Journal of Biological Chemistry. 2005;280:38247–58. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, Kaestner KH, Owens GK. Conditional Deletion of Kruppel-Like Factor 4 Delays Downregulation of Smooth Muscle Cell Differentiation Markers but Accelerates Neointimal Formation Following Vascular Injury. Circ Res. 2008;102:1548–57. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma AK, Lu G, Jester A, Johnston WF, Zhao Y, Hajzus VA, Saadatzadeh MR, Su G, Bhamidipati CM, Mehta GS, Kron IL, Laubach VE, Murphy MP, Ailawadi G, Upchurch GR. Experimental Abdominal Aortic Aneurysm Formation Is Mediated by IL-17 and Attenuated by Mesenchymal Stem Cell Treatment. Circulation. 2012;126:S38–S45. doi: 10.1161/CIRCULATIONAHA.111.083451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston WF, Salmon M, Su G, Lu G, Stone M, Zhao Y, Owens GK, Upchurch GR, Ailawadi G. Interleukin-1β Neutralization Prevents Experimental Aortic Aneurysm Formation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33:294–304. doi: 10.1161/ATVBAHA.112.300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eliason JL, Hannawa KK, Ailawadi G, Sinha I, Ford JW, Deogracias MP, Roelofs KJ, Woodrum DT, Ennis TL, Henke PK, Stanley JC, Thompson RW, Upchurch GR. Neutrophil Depletion Inhibits Experimental Abdominal Aortic Aneurysm Formation. Circulation. 2005;112:232–40. doi: 10.1161/CIRCULATIONAHA.104.517391. [DOI] [PubMed] [Google Scholar]
- 22.Mahatan C, Kaestner K, Geiman D, Yang VW. Characterization of the structure and regulation of the murine encoding gut-enriched Kruppel-like factor. Nucleic Acid Research. 1999;27:4562–9. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segre J, Bauer C, Fuchs E. KLF4 is a transcription factor required for establishing the barrier function of the skin. Nature Genetics. 1999;22:356–60. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 24.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind S, Offermanns S. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008:64–8. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 25.Nemenoff RA, Horita H, Ostriker AC, Furgeson SB, Simpson PA, VanPutten V, Crossno J, Offermanns S, Weiser-Evans MC. SDF-1 Induction in Mature Smooth Muscle Cells by Inactivation of PTEN Is a Critical Mediator of Exacerbated Injury-Induced Neointima Formation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31:1300–8. doi: 10.1161/ATVBAHA.111.223701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saraff K, Babamusta F, Cassis LA, Daugherty A. Aortic Dissection Precedes Formation of Aneurysms and Atherosclerosis in Angiotensin II-Infused, Apolipoprotein E-Deficient Mice. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003;23:1621–6. doi: 10.1161/01.ATV.0000085631.76095.64. [DOI] [PubMed] [Google Scholar]
- 27.Daugherty A, Cassis LA. Mouse Models of Abdominal Aortic Aneurysms. Arteriosclerosis, Thrombosis, and Vascular Biology. 2004;24:429–34. doi: 10.1161/01.ATV.0000118013.72016.ea. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Ait-Oufella H, Herbin O, Bonnin P, Ramkhelawon B, Taleb S, Huang J, Offenstadt G, Combadiere C, Renia L, Johnson JL, Tharaux PL, Tedgui A, Mallat Z. TGF-β activity protects against inflammatory aortic aneurysm progression and complications in angiotensin II infused mice. J Clin Invest. 2010;120:422–32. doi: 10.1172/JCI38136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Sinha S, Shang Y, Hoofnagle MH, Owens GK. PDGF bb induced suppression of SM α-actin in SMC is mediated by KLF4/GKLF dependent repression of myocardin-SRF dependent transcription. Journal of Biological Chemistry. 2005;280:9719–27. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 30.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. American Journal of Physiology - Heart and Circulatory Physiology. 2009;296:H1027–H1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherepanova O, Pidkovka N, Yoshida T, Gan Q, Adiguzel E, Bendick MP, Berliner J, Leitinger N, Owens GK. Oxidized Phospholipids induce type VIII collagen expression and vascular smooth muscle cell migration. Circ Res. 2009;104:609–18. doi: 10.1161/CIRCRESAHA.108.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1 and histone deacetylases cooperatively suppress smooth muscle differentiation markers in reponse to oxidized phospholipids. Am J Physiology cell physiology. 2008;295:C1175–82. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. A G/C Element Mediates Repression of the SM22a Promoter Within Phenotypically Modulated Smooth Muscle Cells in Experimental Atherosclerosis. Circ Res. 2004;95:981–8. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 34.McDonald O, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF Binding to CARG-box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Candales A, Holmes DR, Liao S, Scott MJ, Wickline SA, Thompson RW. Decreased vascular smooth muscle cell density in medial degeneration of human abdominal aortic aneurysms. Am J Pathology. 1997:993–1007. [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson RW, Liao S, Curci JA. Vascular smooth muscle cell apoptosis in abdominal aortic aneurysms. Coron Artery Dis. 1997;8:623–31. doi: 10.1097/00019501-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Harkin DW, Romaschin A, Taylor SM, Rubin BB, Lindsay TF. Complement c5a receptor antagonist attenuates multiple organ injury in a model of ruptured abdominal aortic aneurysm. J Vasc Surg. 2004;39:196–206. doi: 10.1016/j.jvs.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like Factor 4 Abrogates Myocardin-induced Activation of Smooth Muscle Gene Expression. Journal of Biological Chemistry. 2005;280:9719–27. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional Profiling of Kruppel-like Factor 4 Reveals a Function in Cell Cycle Regulation and Epithelial Differentiation. Journal of Molecular Biology. 2003;326:665–77. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shields JM, Christy RJ, Yang VW. Identification and Characterization of a Gene Encoding a Gut-enriched Kruppel-like Factor Expressed during Growth Arrest. Journal of Biological Chemistry. 1996;271:20009–17. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. opposing Effects of kruppel-like factor 4 and Kruppel-like factor 5 on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Research. 2002;30:2736–41. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai-Kowase K, Ohshima T, Matsui H, Tanaka T, Shimizu T, Iso T, Arai M, Owens GK, Kurabayashi M. PIAS1 Mediates TGFβ-Induced SM α-Actin Gene Expression Through Inhibition of KLF4 Function-Expression by Protein Sumoylation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2009;29:99–106. doi: 10.1161/ATVBAHA.108.172700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atkins GB, Wang Y, Mahabeleshwar GH, Shi H, Gao H, Kawanami D, Natesan V, Lin Z, Simon DI, Jain MK. Hemizygous deficiency of Kruppel-like factor 2 augments experimental atherosclerosis. Circ Res. 2008;103:690. doi: 10.1161/CIRCRESAHA.108.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like Factor 4 Regulates Endothelial Inflammation. Journal of Biological Chemistry. 2007;282:13769–79. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.