My interest in gene therapy began in the early 1980s as a fresh associate professor at the University of Michigan. The University of Michigan had just recruited a new wave of young scientists who went on to become important leaders in the field of molecular medicine, including Francis Collins (current NIH director), Gary Nabel (chief scientific officer at Sanofi), Betsy Nabel (president of Brigham and Women's Hospital), Graig Thompson (CEO of Sloan Kettering Memorial), Jeffrey Leiden (CEO and president of Vertex Pharmaceuticals), and James Wilson (director of the Penn Gene Therapy Program). Most were appointed in medicine, where William Kelley (former CEO of the Penn Health System and dean of the School of Medicine) was chair. I had the good fortune to work with and be influenced by these talented scientists and particularly by Bill Kelley, who was a visionary leader and a huge proponent of gene therapy, including the development of herpes simplex viral (HSV) vectors for treatment of Lesch Nyhan disease (Palella et al., 1989). I was also blessed by my association with Myron (Mike) Levine in the Department of Human Genetics. Mike took me under his wing and taught me how to use genetics to study virus biology, whereupon his insights influenced me for the rest of my career.
The potential for genetic intervention in the treatment of human disease began essentially with the advent of genetic mapping of disease genes and determination of their resulting pathologies. It appeared that gene therapy offered unlimited potential for “curing” at least recessive inborn errors of metabolism by gene “replacement” therapy. The most important question was how to get the therapeutic gene into the correct tissue with appropriate level and duration of expression. Because viruses could efficiently enter and replicate in cells of the body reflecting their unique biology and host cell preferences, it appeared that recombinant viruses could be engineered as effective gene delivery vehicles by replacing pathogenic genes with therapeutic sequences. I became intrigued with the concept of viruses as universal gene delivery tools. On the basis of the fact that herpes viruses persisted in humans in a latent state for life with preference for specific cell types, it seemed feasible to exploit this group of human viruses for gene therapy. Since some therapeutic gene cassettes were likely to be large, especially in combination with complex regulatory elements, large payloads would require large vectors. I became convinced that HSV could fill this niche, particularly for gene delivery to the nervous system, the natural site of HSV latency. However, when we began these studies, there was relatively little known about the molecular biology of HSV replication and pathogenesis, requiring us to peel back many layers of vector-engineering complexities to create a useful gene delivery platform. This quest has taken 30 years of work, beginning at the University of Michigan and continuing at the University of Pittsburgh, and only recently do I think we have succeeded in creating broadly useful vector designs. Our headway in vector engineering also addressed other pressing vector problems that included the need for low effective dosing with minimal inflammatory responses, targeted infection and long-term gene expression in specific cell types, and the ability to rapidly produce and manufacture vectors for clinical applications.
The Natural Biology of HSV
HSV is transmitted by direct contact. For HSV type 1 (HSV-1), direct contact most often occurs on the face around the lips, and the most common disease, fever blisters, is rather benign but still unpleasant. There are, nevertheless, rare cases of HSV-1 migrating to the brain and causing encephalitis, and infections of the cornea are a leading cause of blindness. HSV-2 is more often associated with genital lesions, is generally more pathogenic, and can produce a devastating infection of newborns if acquired during childbirth. HSV-1 was selected for vector development since limited viral pathogenesis was considered important.
HSV replicates in epithelial cells of the skin or mucosa where it gains access to sensory nerve terminals. Remarkably, the de-enveloped virus particle is efficiently delivered by retrograde axonal transport to the nerve cell nucleus, where the viral genome persists as a latent circular episome for life. A second remarkable feature of HSV is the persistent expression of an unstable, noncoding 8.3 kb latency active transcript (LAT) but with the accumulation of a highly stable LAT-derived 2 kb intron in most latently infected cells (Stevens, 1987); the presence of this intron is a hallmark of latency. Of interest was the finding that LAT expression is not essential for the establishment or maintenance of latency at least in animal models, suggesting that this locus can be altered and its regulatory sequences exploited for ectopic long-term gene expression. The molecular mechanisms that underlie the establishment of latency are still under investigation, but natural virus latency is limited to neurons accounting for HSV's neurotropic status. Latent virus can be reactivated to reestablish an active infection of the skin at the original site of infection, but the virus does not spread in the body of individuals with normal immune function. Thus, HSV is a localized infection dependent on the site of entry into the host. These features, skin delivery and long-term LAT expression, provided the drivers for development of HSV vectors for the treatment of peripheral nervous system diseases. Our task was to create an HSV vector that would serve as a platform for transgene expression in a latent-like state without the ability to reactivate.
HSV Molecular Biology
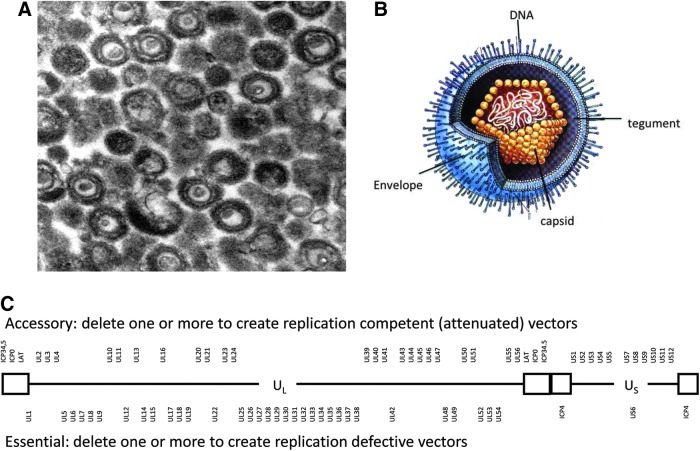
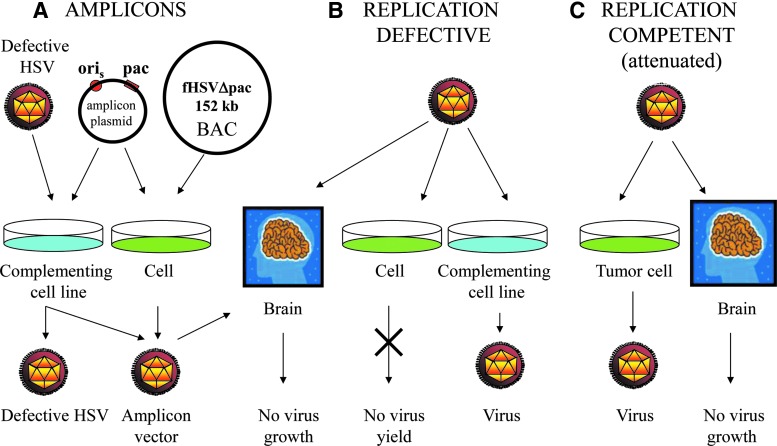
Herpes simplex virus is among the largest DNA viruses developed for gene transfer. The viral genome is 152 kb in length and approximately 84 genes are encoded. The virus has an envelope, a subenvelope structure referred to as the tegument, and a regular icosahedral capsid (Fig. 1A and B). The linear double-stranded genome is composed of a long (UL) and short (US) genomic segment each flanked by inverted repeats creating an internal region referred to as the joint. The genes of HSV are classified as accessory and essential (Fig. 1C). In general, accessory genes can be individually deleted without substantially compromising virus replication in standard cell cultures, although multiple gene deletions can have additive effects on virus growth. Nevertheless, accessory genes play an important role in completing the virus life cycle in the host, and thus deletion of certain genes can limit virus replication in vivo. By contrast, deletion of any essential gene completely blocks productive virus infection. Figure 2 identifies the important elements of the HSV genome that are important to HSV vector engineering. Several key genes in regulating virus replication are located in repeat regions and are therefore diploid. Consequently, the approximately 19 kb joint region can be deleted without substantially compromising virus replication, creating a large space for addition of foreign sequences. The manipulation of these classes of viral genes has led to the creation of three types of HSV vectors: amplicon, replication-defective, and replication-competent vectors (Fig. 3).
FIG. 1.
HSV-1 particle and genome structure. (A) Electron micrograph of purified HSV particles reveals the envelope, capsid, and condensed DNA. (B) Representation of the particle locates the envelope with spike glycoproteins, tegument, regular icosahedral capsid, and internal DNA (reproduced from the thesis of Paola Grandi, University of Ferrara, 2002). (C) The linear double-stranded DNA is shown as a diagram. Boxes represent the inverted terminal repeats that flank the unique long (UL) and unique short (US) sequences. Accessory viral genes (upper) can be manipulated to create attenuated viruses, and essential genes (lower) can be deleted to make replication-defective viruses. HSV-1, herpes simplex virus type 1.
FIG. 2.
Genome structure: location of gene classes. (A) Diagram of the linear viral genome. Blue boxes, inverted repeats. Terminal repeat (TR) and internal repeat (IR) flanking the long (L) and short (S) unique segments. (B) The viral transcription units are color coded as immediate early (IE, green), early (E, yellow), early-late (L1, red), late (L2, brown), and latency (LAT) gene (purple). The blue boxes represent the repeat regions that can be manipulated to construct replication-defective vectors. The internal joint region contains genes that are duplicated in the terminal repeats and can be deleted for insertion of foreign sequences. The IE genes are deleted to create completely silent vectors. The LAT region contains elements that can be used for insertion of transgenes that remain active in the absence of the IE genes.
FIG. 3.
HSV vector design strategies. (A) Amplicon designs. Amplicons are genome-length plasmid-like vectors that contain a viral origin (ori) and packaging (pac) sequences that allow amplification of transfected amplicon DNA and incorporation into virus particles for amplicon delivery to cells. Replication-defective helper virus supplies viral functions needed to replicate amplicon DNA and to make helper and amplicon particles in complementing (blue) cells. Cotransfection of noncomplementing cells (green) with amplicon DNA and helper DNA lacking packaging sequences will produce helper-free amplicon particles. (B) Replication-defective vector designs. Replication-defective viruses are deleted for one or more essential virus genes and must be grown on cell lines that complement these deleted functions in trans (blue). Infection of noncomplementing cells in culture will not produce virus (green), and infection of other tissues (e.g., brain) will not allow virus replication. (C) Replication-competent (attenuated) vector designs. These vectors can be propagated on noncomplementing cells (green) and attenuated by removal of neuropathogenic accessory genes for targeting of brain tumors without harming normal brain tissue.
Replication-defective vectors
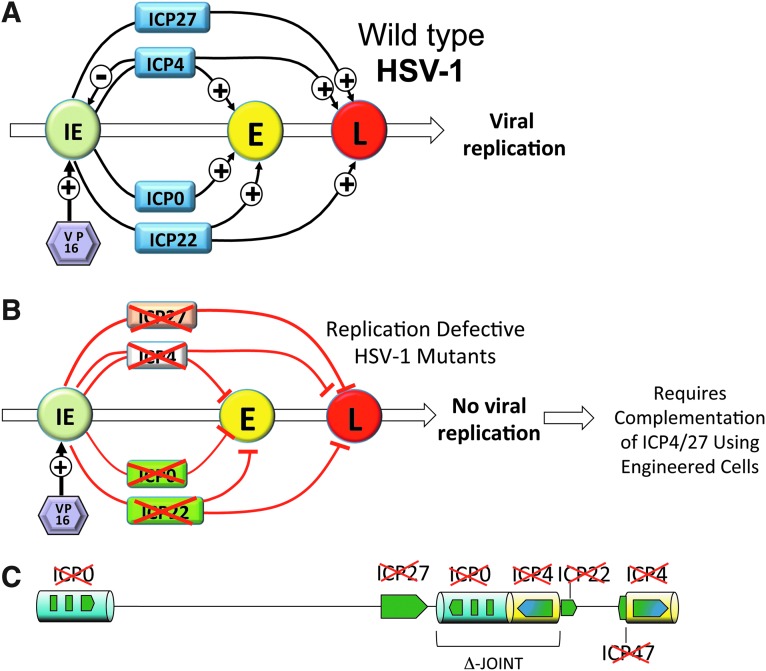
In the late 1980s and early 1990s, our research group sought to create HSV vectors that were completely nontoxic to host cells and incapable of virus growth but capable of expressing transgenes in subpopulations of neurons. While this was a daunting task since the virus had only recently been cloned (Goldin et al., 1981) and sequenced (McGeoch, 1987; McGeoch et al., 1988; Perry and McGeoch, 1988; Wu et al., 1988), we were nevertheless able to take advantage of an important aspect of HSV biology, cascade gene regulation (Fig. 4A). Together, the first wave of genes (immediate early [IE] functions) provided activities to overcome innate immune responses, hide infected cells from immune surveillance, block cell division, prevent cell suicide, prevent repression of viral gene expression, and activate the expression of early (E) and late (L) genes during progression of the virus replication cycle. Two of the IE genes, ICP4 and ICP27, were known to be essential for virus replication and thus deletion of either of these genes completely interrupted virus growth at an early stage and provided the platform for the first- and second-generation vectors (Fig. 4B). Because these genes are essential, a method for growing such viruses was critical to vector engineering and production. By the mid-1990s, cell lines capable of complementing both ICP4 and ICP27 had been produced, thus allowing production of a vector backbone that only expressed the remaining IE genes, ICP0, ICP22, and ICP47 (Shepard and DeLuca, 1991; Marconi et al., 1996). The deletion of both essential IE genes prevented the occurrence of replication-competent recombinants during complementation, an important vector safety feature. Fortunately, we found that the 4/27 deletion vector was able to infect sensory nerves after intradermal delivery (Goins et al., 1994). The vector was transported to the nerve cell body and the mutant virus genome became established long-term but without the ability to reactivate. Moreover, it was possible to achieve transient gene expression in neurons using strong promoters such as the human cytomegalovirus (HCMV) IE gene promoter. The remaining viral IE genes were rapidly silenced and the neurons appeared to be unharmed. Although the HCMV promoter was only transiently active in the ganglia, the viral latency gene promoter remained active, as evidenced by the expression of LAT, suggesting that these vectors achieved a latent-like state. These HSV mutants represented the first vectors useful for gene transfer to peripheral nerves, opening up the door for gene therapy.
FIG. 4.
Cascade expression of HSV genes. (A) The viral tegument protein VP16 (purple) activates the IE gene promoters (green) to express the IE gene products (blue) that in turn activate the promoters of the E genes (yellow). The E gene products replicate the viral DNA, releasing expression of the L gene products (red) that package viral genomes de novo and form the virus particle structure. (B) ICP4 (diploid) and ICP27 are essential IE genes. Expression of E and L genes is blocked in replication-defective HSV deleted for ICP4 and/or ICP27. Vector production requires the use of engineered complementing cells. First-generation replication-defective HSV vectors are deleted for ICP4 (orange). Second-generation vectors are deleted for both ICP4 (orange) and ICP27 (white). Third-generation vectors are functionally deleted for all of the IE genes ICP4, ICP27, ICP0 (diploid) (green), ICP22 (green), and ICP47 (not shown) and are noncytotoxic or interfere with cell division. (C) Genomic structure of the third-generation vector: deletion of the joint sequence and all remaining IE genes blocks the expression of any viral genes requiring complementation of ICP4, ICP27, and ICP0 in engineered U2OS cells.
A second breakthrough came from Bill Goins in our group, who discovered a sequence in the latency gene locus downstream of the latency promoter that possessed independent promoter activity. Bill referred to this element as the second latency active promoter (LAP2). LAP2 was moveable and capable of expressing transgenes at genomic locations outside of the latency locus and for extended times (Goins et al., 1994). The discovery of LAP2 and the capability of constructing ICP4/27 deletion vectors together provided a vector system useful for long-term gene therapy of the brain and sensory nerve conditions (Glorioso et al., 2003).
Despite these advances, our vectors were still cytotoxic to cells in culture and failed to persist in nonneuronal tissues, thereby limiting the broad utility of HSV vectors. ICP0 in particular caused cell cycle arrest and apoptosis (Samaniego et al., 1997). While removal of the remaining HSV IE genes was possible, two roadblocks were encountered. First, in the absence of ICP0, vector transgene expression was silenced and, second, ICP0-deleted virus production was poor in 4/27 complementing cells. The toxicity of ICP0 made the engineering of cell lines that complemented all three deleted genes challenging. Fortuitously, it was discovered that certain tumor lines, in particular the human osteosarcoma line U2OS, naturally complemented ICP0 (Yao and Schaffer, 1995), suggesting the possibility of exploiting an ICP4- and ICP27-complementing U2OS line for production of virus mutants lacking expression of the complete set of IE genes. Only recently have we been able to create such cell lines allowing the propagation of vectors that are functionally deleted for all of the IE genes. Because removal of the IE genes renders the rest of the viral genome transcriptionally inactive (Fig. 4C), these new vectors (Fig. 4D) fail to express any viral genes, are completely devoid of toxicity, and can persist in any nondividing cell type both in vitro and in vivo.
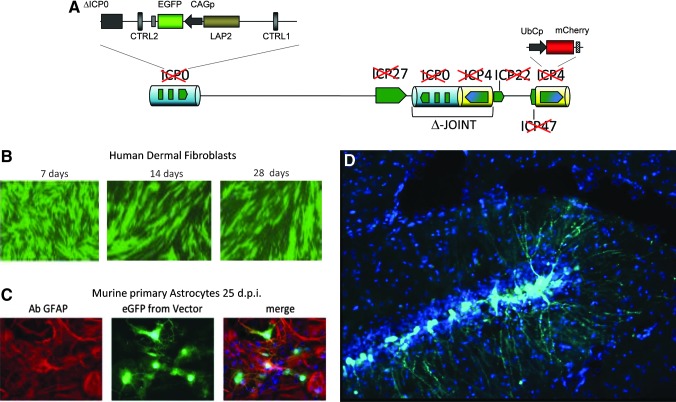
There remained the problem of how to overcome transgene shutdown, a problem associated with the rapid formation of heterochromatin. Moving to bacterial artificial chromosomes for rapid HSV vector engineering, a talented researcher in our group, Yoshitaka Miyagawa, redesigned the transgene construct to take advantage of insulator (CTRL) and extended LAP2 (LATP2) elements of the latency promoter to achieve for the first time transgene expression in the absence of ICP0 and all other IE gene activity (Fig. 5). This vector design was capable of long-term gene expression without cytotoxicity or interference with cell division and represented the final breakthrough to make HSV a widely useful gene delivery technology. In 2014, this new vector will be exploited to carry large transgenes that are beyond the capacity of other commonly used vector systems.
FIG. 5.
Third-generation replication-defective HSV vector design. (A) Vector deleted for the joint and IE genes with the remaining LAT locus containing an EGFP reporter gene cassette under control of the recombinant CMV enhancer–chicken β-actin promoter, splice donor, and intron–human β-globin splice acceptor (CAG) promoter. A mCherry reporter gene cassette under control of the ubiquitin C (UbC) promoter is expressed during vector production in complementing cells but silent in noncomplementing cells. (B) EGFP is active long-term in human dermal fibroblasts (28 days); some loss of green cells results from cell division. (C) Mouse astrocytes in culture (25 days); astrocytes are identified using a specific antibody against glial fibrillary acidic protein (GFAP), and in (D) hippocampal neurons after direct intracranial injection using a stereotactic frame (4 days).
Amplicon vectors
A second strategy for creating replication-defective HSV vectors was based on the finding that repeat high-dose passage of HSV resulted in the accumulation of defective interfering viruses that contained an origin of replication and packaging signals but had lost the complete set of lytic functions needed to support productive virus replication (Spaete and Frenkel, 1985). This discovery led to the creation of amplicon vectors by Xandra Breakefield, Cornel Fraefel, and Alberto Epstein that resembled defective interfering viruses and could be propagated by amplicon transfection followed by replication-defective “helper” virus infection (Fig. 3A). The helper is complemented by an essential gene introduced into the cell line as described above. Amplicon production was improved by selective removal of packaging signals from the helper genome to avoid amplicon stock contamination with packaged helper virus, a system requiring cotransfection of amplicon and helper (Fraefel et al., 2000; Zaupa et al., 2003). Amplicons are advantaged by the ability to accommodate very large sequences, exceeding 100 kb. However, difficulties in high-titer vector production and stability have dissuaded me from using these vectors in clinical trials, but they remain powerful tools for preclinical studies.
Replication-competent vectors
Our interest in treating cancer began with the generation of replication-defective vectors that contained single or multiple transgenes to activate anticancer drugs, improve distribution of cancer-killing metabolites, and induce natural killing mechanisms (Moriuchi et al., 1998) (Palella et al., 1989; Marconi et al., 2000). While these vectors proved very effective in animal models, we were concerned that they would not reach cells distant from the site of vector inoculation. A resurgence of interest in using replication-competent HSV and many other virus types for treating cancer by selective lytic replication in tumors (oncolysis) has encouraged us to develop these vectors for treating gliomas and other tumor types. Virus replication was envisioned to enhance intratumoral vector distribution and therefore effectiveness.
In the last 10 years, conditionally replicating HSV vectors were designed based on the removal of accessory genes that contribute to neuropathogenesis. The most well studied of these genes is the late gene designated ICP34.5 (Chambers et al., 1995; Mineta et al., 1995). Its product, neurovirulence factor ICP34.5 (a.k.a. γ34.5), prevents activation of the protein kinase R pathway allowing virus replication in the face of interferon responses (Simonato et al., 2013). Many tumors have defective innate immune activity allowing for tumor-specific growth of γ34.5 deletion mutants. Despite effective treatment of animal models of human glioblastoma using such mutants, successful treatment of human brain tumors has been limited (Markert et al., 2009). Nevertheless, the arming of these vectors with genes that induce antitumor immunity is a promising strategy, and indeed one such vector has proven effective in treating melanoma (Hobbs et al., 2001). A γ34.5 deletion vector expressing granulocyte macrophage colony-stimulating factor was developed by Rob Coffin at BioVex, and Amgen has used this vector (T-VEC) to treat melanoma with promising phase 3 clinical trial results (Bartlett et al., 2013). T-VEC may become the first FDA-approved oncolytic vector for cancer therapy, paving the way for other oncolytic vectors.
We have taken the view that we may be able to improve the oncolytic activity of HSV vectors by targeting virulent wild-type viruses to tumor cells without compromising any viral function particularly those that resist innate immunity. Using this approach, Justus Cohen and Hiroaki Uchida have created vectors that are fully retargeted for preferential infection of tumor cells (Uchida et al., 2013). To further tighten the tumor selectivity of these vectors, we took advantage of naturally occurring cellular microRNAs that are highly expressed in normal tissue but completely absent in tumor cells. For example, miR-124 is one of the most highly expressed genes in brain but is not expressed in glioblastoma, making it ideal for this type of vector design. Paola Grandi recently guided the introduction of multiple copies of the miR-124 recognition site into the 3′ UTR of ICP4 to prevent virus replication in normal neurons, thereby increasing the safety of EGFR-retargeted vectors for the treatment of glioblastoma (Mazzacurati et al., submitted manuscript in revision). Such strategies should be broadly useful for cancer therapies and these vectors can carry multiple transgenes that enhance intratumoral vector spread and induce antitumor immunity.
HSV Vectors and Gene Therapy
In 1989, I moved to the University of Pittsburgh School of Medicine as chair of the Department of Molecular Genetics and Biochemistry. It was a great time since there was ample NIH support for gene therapy. I had the good fortune to work with a talented group of burgeoning gene therapists in Pittsburgh that included Paul Robbins, Chris Evans, Xiao Xiao, Jude Samulski, Eric Hoffman, Leaf Huang, Ray Frizzell, and Barry London, and together we established one of the early molecular medicine centers. Studies began on strategies to treat cystic fibrosis, muscular dystrophy, arthritis, liver cirrhosis, and heart disease. David Fink became my close collaborator for many years in the development of HSV vectors for treatment of nervous system disease (Glorioso and Fink, 2009). Since HSV establishes latency in peripheral nerve ganglia, David and I agreed to focus our research efforts on developing gene therapies for peripheral neuropathies (Goins et al., 1999; Chattopadhyay et al., 2004) and chronic pain (Goss et al., 2001, 2002). In the mid-1990s, Steve Wilson (University of South Carolina) joined my lab while on sabbatical and introduced the idea of treating pain using HSV vector-mediated expression of enkephalin. Preproenkephalin gene delivery results in protein processing to produce met and leu opiate peptides that impede pain-related neurotransmitter release through binding of the mu and delta receptors found on both nociceptors and second-order neurons in the spinal cord. Thus, we reasoned that HSV expression of preproenkephalin in the primary afferent could substantially dampen pain-signaling at the site where nociceptor activation was occurring (Fig. 6)—in other words, treating pain at its source. This strategy proved effective in blocking both neuropathic and inflammatory pain in multiple animal models of chronic pain (Wilson et al., 1999; Goss et al., 2001, 2002). Transient expression of enkephalin prevented pain, and longer-term treatment could be achieved by repeat dosing. Importantly, enkephalin gene therapy did not lead to tolerance and could be observed even when tolerance to morphine was induced by high-dose administration (Hao et al., 2003). These features were essential since early phase human clinical trials for treatment of cancer-related pain would encounter these two issues. Data from the phase I trial looked very promising (Fink et al., 2011) and the results of the phase II trial are in review. These trials were carried out by Diamyd AB, a public company located in Stockholm, Sweden, in which I was a shareholder and consultant.
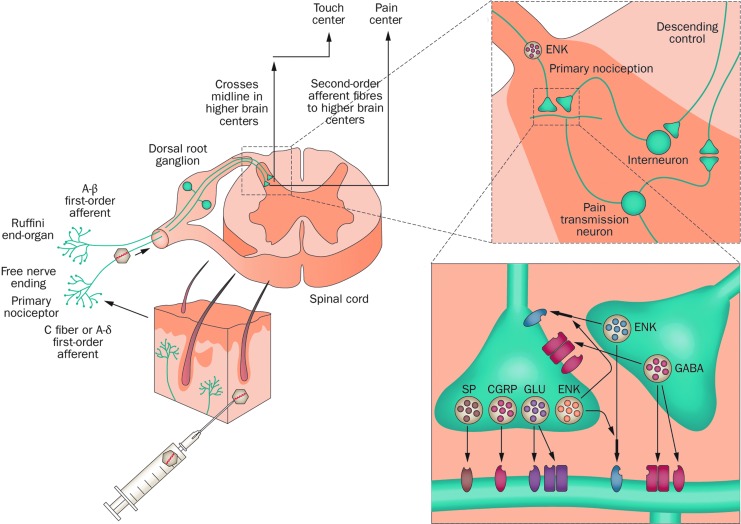
FIG. 6.
Pain gene therapy using an HSV preproenkephalin-expressing vector. Normal pain signaling pathway control involves signals from peripheral tissues or organs that stimulate the primary first-order afferents that innervate those sites. This leads to the release of various neurotransmitters, including opioid peptides such as ENK, that alter pain signaling to the brain by ascending second-order neurons within the dorsal horn of the spinal cord. The ENK-expressing HSV vector is injected into skin followed by virus uptake into primary afferents; retrograde axonal transport results in HSV vector-mediated production and release of ENK (depicted in orange) from DRG terminals within the dorsal horn. Binding to the specific opioid receptors present on ascending pain transmission neurons and interneurons leads to a block in pain signal transmission. CGRP, calcitonin gene-related peptide; DRG, dorsal root ganglion; ENK, enkephalin; GABA, glutamic acid decarboxylase; GLU, glutamate; SP, substance P. (Figure reproduced with permission from Simonato et al., 2013.)
Despite the utility of enkephalin and other therapies (Liu et al., 2004; Hao et al., 2005, 2007, 2009), the consequences of long-term pain-relieving transgene expression using a form of the latency promoter remained uncertain and untested in patients. I came around to the view that long-term, regulated therapy would be essential for pain gene therapy to become a practical solution for chronic pain treatment. In 2005, one of my faculty members, Mike Cascio, described to me his work on the physical structure of the glycine receptor (GlyR), a common chloride channel. We learned that the GlyR was absent in sensory nerves and that the alpha subunit alone could be distributed along axons and was sufficient for channel activation by glycine. Activation resulted in hyperpolarization of the neuron with subsequent silencing, raising the question whether delivery of the GlyRα gene to sensory nerves with subsequent glycine administration to the skin would block pain. The exciting answer was yes for both neuropathic and inflammatory pain, that is, regardless of the cause of pain (Goss et al., 2011). Thus, we had created a “nerve-silencing” tool that could be regulated by a drug, providing a novel drug-regulated approach to gene therapy. We also found that glycine could be administered systemically to treat bladder pain (Goss et al., 2011). These studies have now been extended to the use of a mutant form of the GlyR that is exclusively activated by ivermectin (Lynagh and Lynch, 2010), an approved drug for treatment of helminth infections in humans (Omura, 2008), and it appears feasible that ivermectin-inducible GlyR activation will create a powerful and highly specific means of controlled, long-term pain treatment.
The Future of HSV Vectors
Gene therapy has made remarkable strides over the last 30 years. Successful therapies have been forthcoming for treating cancer, metabolic disorders, and conditions affecting the nervous system, retina, and the immune system. A number of new vector systems have been developed with broad potential. The commercialization of gene therapy is at its beginning, but the expectation is that genetic intervention will take its place alongside other forms of standard medical practice. I am convinced that HSV vectors will play very important roles in the future of this field. In addition, I am excited about the possibility of exploiting our most advanced HSV vectors for cellular reprogramming and transdifferentiation of normal and disease tissues in vitro and in vivo. Single HSV vector expression of the complete set of Yamanaka reprogramming genes has been achieved (Miyagawa et al., unpublished) and illustrates the power of our new vector system. Such application of HSV gene transfer will expedite strategies for tissue regeneration. The cell division-dependent, hit-and-run capability of our vectors is expected to be ideal for this purpose. We have also been able to create expression libraries using HSV vectors (Wolfe et al., 2010) that can be used, for example, to identify new reprogramming and transdifferentiation genes. We have recently used an HSV-based cellular cDNA library to discover activities that regulate ion channels and control pain responses in nociceptors (Srinivasan et al., 2007; Reinhart et al., unpublished). Finally, I believe that gene delivery to the brain represents the most important frontier for HSV-mediated gene therapy and provides a unique opportunity to study complex processes such as learning and memory and to treat complex genetic and acquired diseases, including brain degeneration, epilepsy, and cancer.
Acknowledgments
I wish to thank my many students and fellows over the years whose diligence and intelligence have facilitated this research, and my colleagues at the University of Michigan and Pittsburgh whose insights and encouragement have inspired us. Also I wish to thank Justus Cohen, Paola Grandi, Bill Goins, and Yoshi Miyagawa for careful reading of the manuscript and helpful suggestions. Also I am grateful to Gianluca Verlengia and Michele Simonato for the photomicrographs of astrocyte cell culture and brain infections with the HSV:EGFP vector. Over the years this work has been sponsored by the NIH and private foundations, including the Juvenile Diabetes Research Foundation, Muscular Dystrophy Association, McGowan Institute for Regenerative Medicine, Commonwealth of Pennsylvania Department of Health, Cystic Fibrosis Foundation, Cure Huntington Disease Initiative Foundation, and Alliance for Cancer Gene Therapy.
References
- Bartlett D.L., Liu Z., Sathaiah M., et al. (2013). Oncolytic viruses as therapeutic cancer vaccines. Mol. Cancer 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R., Gillespie G.Y., Soroceanu L., et al. (1995). Comparison of genetically engineered herpes simplex viruses for the treatment of brain tumors in a scid mouse model of human malignant glioma. Proc. Natl. Acad. Sci. USA 92, 1411–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M., Goss J., Wolfe D., et al. (2004). Protective effect of herpes simplex virus-mediated neurotrophin gene transfer in cisplatin neuropathy. Brain 127, 929–939 [DOI] [PubMed] [Google Scholar]
- Fink D.J., Wechuck J., Mata M., et al. (2011). Gene therapy for pain: results of a phase I clinical trial. Ann. Neurol. 70, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraefel C., Jacoby D.R., and Breakefield X.O. (2000). Herpes simplex virus type 1-based amplicon vector systems. Adv. Virus Res. 55, 425–451 [DOI] [PubMed] [Google Scholar]
- Glorioso J.C., and Fink D.J. (2009). Herpes vector-mediated gene transfer in the treatment of chronic pain. Mol. Ther. 17, 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso J.C., Mata M., and Fink D.J. (2003). Gene therapy for chronic pain. Curr. Opin. Mol. Ther. 5, 483–488 [PubMed] [Google Scholar]
- Goins W.F., Sternberg L.R., Croen K.D., et al. (1994). A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J. Virol. 68, 2239–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goins W.F., Lee K.A., Cavalcoli J.D., et al. (1999). Herpes simplex virus type 1 vector-mediated expression of nerve growth factor protects dorsal root ganglion neurons from peroxide toxicity. J. Virol. 73, 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin A.L., Sandri-Goldin R.M., Levine M., and Glorioso J.C. (1981). Cloning of herpes simplex virus type 1 sequences representing the whole genome. J. Virol. 38, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss J.R., Mata M., Goins W.F., et al. (2001). Antinociceptive effect of a genomic herpes simplex virus-based vector expressing human proenkephalin in rat dorsal root ganglion. Gene Ther. 8, 551–556 [DOI] [PubMed] [Google Scholar]
- Goss J.R., Harley C.F., Mata M., et al. (2002). Herpes vector-mediated expression of proenkephalin reduces bone cancer pain. Ann. Neurol. 52, 662–665 [DOI] [PubMed] [Google Scholar]
- Goss J.R., Cascio M., Goins W.F., et al. (2011). HSV delivery of a ligand-regulated endogenous ion channel gene to sensory neurons results in pain control following channel activation. Mol. Ther. 19, 500–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S., Mata M., Goins W., et al. (2003). Transgene-mediated enkephalin release enhances the effect of morphine and evades tolerance to produce a sustained antiallodynic effect in neuropathic pain. Pain 102, 135–142 [DOI] [PubMed] [Google Scholar]
- Hao S., Mata M., Wolfe D., et al. (2005). Gene transfer of glutamic acid decarboxylase reduces neuropathic pain. Ann. Neurol. 57, 914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S., Mata M., Glorioso J.C., and Fink D.J. (2007). Gene transfer to interfere with TNFalpha signaling in neuropathic pain. Gene Ther. 14, 1010–1016 [DOI] [PubMed] [Google Scholar]
- Hao S., Wolfe D., Glorioso J.C., et al. (2009). Effects of transgene-mediated endomorphin-2 in inflammatory pain. Eur. J. Pain 13, 380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs W.E., Brough D.E., Kovesdi I., and DeLuca N.A. (2001). Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75, 3391–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wolfe D., Hao S., et al. (2004). Peripherally delivered glutamic acid decarboxylase gene therapy for spinal cord injury pain. Mol. Ther. 10, 57–66 [DOI] [PubMed] [Google Scholar]
- Lynagh T., and Lynch J.W. (2010). An improved ivermectin-activated chloride channel receptor for inhibiting electrical activity in defined neuronal populations. J. Biol. Chem. 285, 14890–14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi P., Krisky D., Oligino T., et al. (1996). Replication-defective herpes simplex virus vectors for gene transfer in vivo. Proc. Natl. Acad. Sci. USA 93, 11319–11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi P., Tamura M., Moriuchi S., et al. (2000). Connexin 43-enhanced suicide gene therapy using herpesviral vectors. Mol. Ther. 1, 71–81 [DOI] [PubMed] [Google Scholar]
- Markert J.M., Liechty P.G., Wang W., et al. (2009). Phase Ib trial of mutant herpes simplex virus G207 inoculated pre-and post-tumor resection for recurrent GBM. Mol. Ther. 17, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D.J. (1987). The genome of herpes simplex virus: structure, replication and evolution. J. Cell Sci. Suppl. 7, 67–94 [DOI] [PubMed] [Google Scholar]
- McGeoch D.J., Dalrymple M.A., Davison A.J., et al. (1988). The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69 (Pt 7), 1531–1574 [DOI] [PubMed] [Google Scholar]
- Mineta T., Rabkin S.D., Yazaki T., et al. (1995). Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat. Med. 1, 938–943 [DOI] [PubMed] [Google Scholar]
- Moriuchi S., Oligino T., Krisky D., et al. (1998). Enhanced tumor cell killing in the presence of ganciclovir by herpes simplex virus type 1 vector-directed coexpression of human tumor necrosis factor-alpha and herpes simplex virus thymidine kinase. Cancer Res. 58, 5731–5737 [PubMed] [Google Scholar]
- Omura S. (2008). Ivermectin: 25 years and still going strong. Int. J. Antimicrob. Agents 31, 91–98 [DOI] [PubMed] [Google Scholar]
- Palella T.D., Hidaka Y., Silverman L.J., et al. (1989). Expression of human HPRT mRNA in brains of mice infected with a recombinant herpes simplex virus-1 vector. Gene 80, 137–144 [DOI] [PubMed] [Google Scholar]
- Perry L.J., and McGeoch D.J. (1988). The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69 (Pt 11), 2831–2846 [DOI] [PubMed] [Google Scholar]
- Samaniego L.A., Wu N., and DeLuca N.A. (1997). The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71, 4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard A.A., and DeLuca N.A. (1991). A second-site revertant of a defective herpes simplex virus ICP4 protein with restored regulatory activities and impaired DNA-binding properties. J. Virol. 65, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonato M., Bennett J., Boulis N.M., et al. (2013). Progress in gene therapy for neurological disorders. Nat. Rev. Neurol. 9, 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R.R., and Frenkel N. (1985). The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc. Natl. Acad. Sci. USA 82, 694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Huang S., Chaudhry S., et al. (2007). An HSV vector system for selection of ligand-gated ion channel modulators. Nat. Methods 4, 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J.G. (1987). Defining herpes simplex genes involved in neurovirulence and neuroinvasiveness. Curr. Eye Res. 6, 63–67 [DOI] [PubMed] [Google Scholar]
- Uchida H., Marzulli M., Nakano K., et al. (2013). Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol. Ther. 21, 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.P., Yeomans D.C., Bender M.A., et al. (1999). Antihyperalgesic effects of infection with a preproenkephalin-encoding herpes virus. Proc. Natl. Acad. Sci. USA 96, 3211–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe D., Craft A.M., Cohen J.B., and Glorioso J.C. (2010). A herpes simplex virus vector system for expression of complex cellular cDNA libraries. J. Virol. 84, 7360–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.A., Nelson N.J., McGeoch D.J., and Challberg M.D. (1988). Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Virol. 62, 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F., and Schaffer P.A. (1995). An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69, 6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaupa C., Revol-Guyot V., and Epstein A.L. (2003). Improved packaging system for generation of high-level noncytotoxic HSV-1 amplicon vectors using Cre-loxP site-specific recombination to delete the packaging signals of defective helper genomes. Hum. Gene Ther. 14, 1049–1063 [DOI] [PubMed] [Google Scholar]