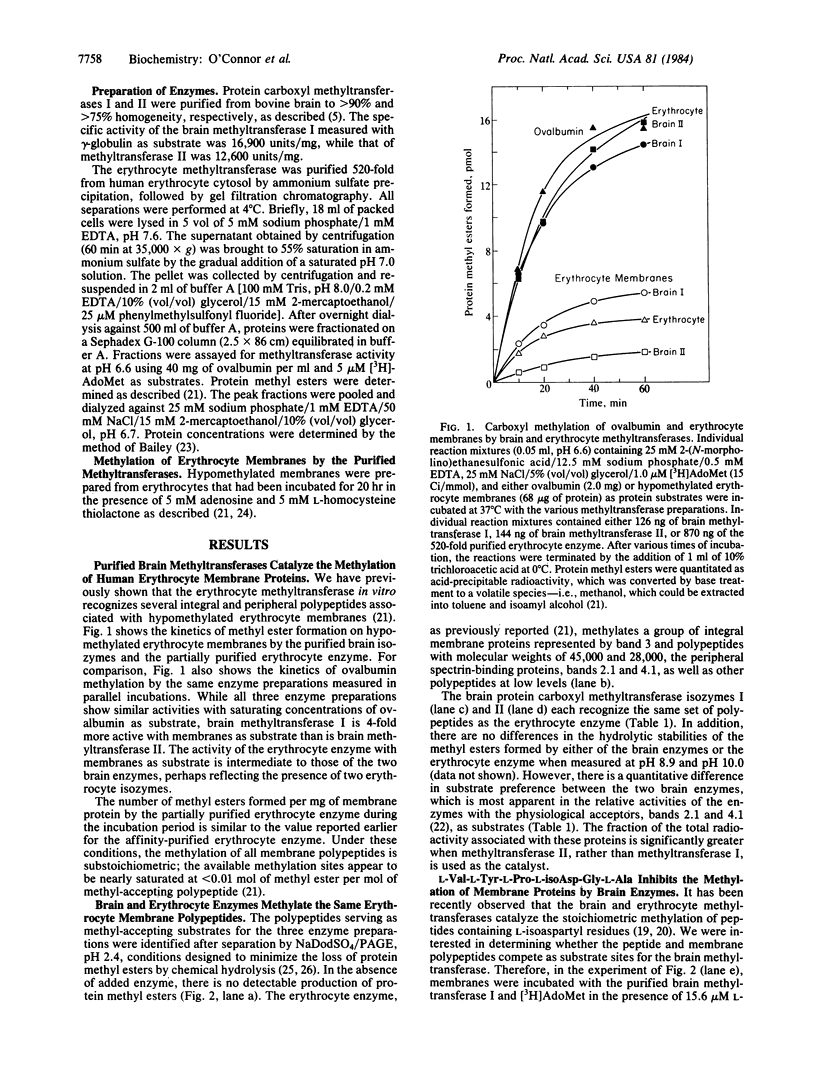
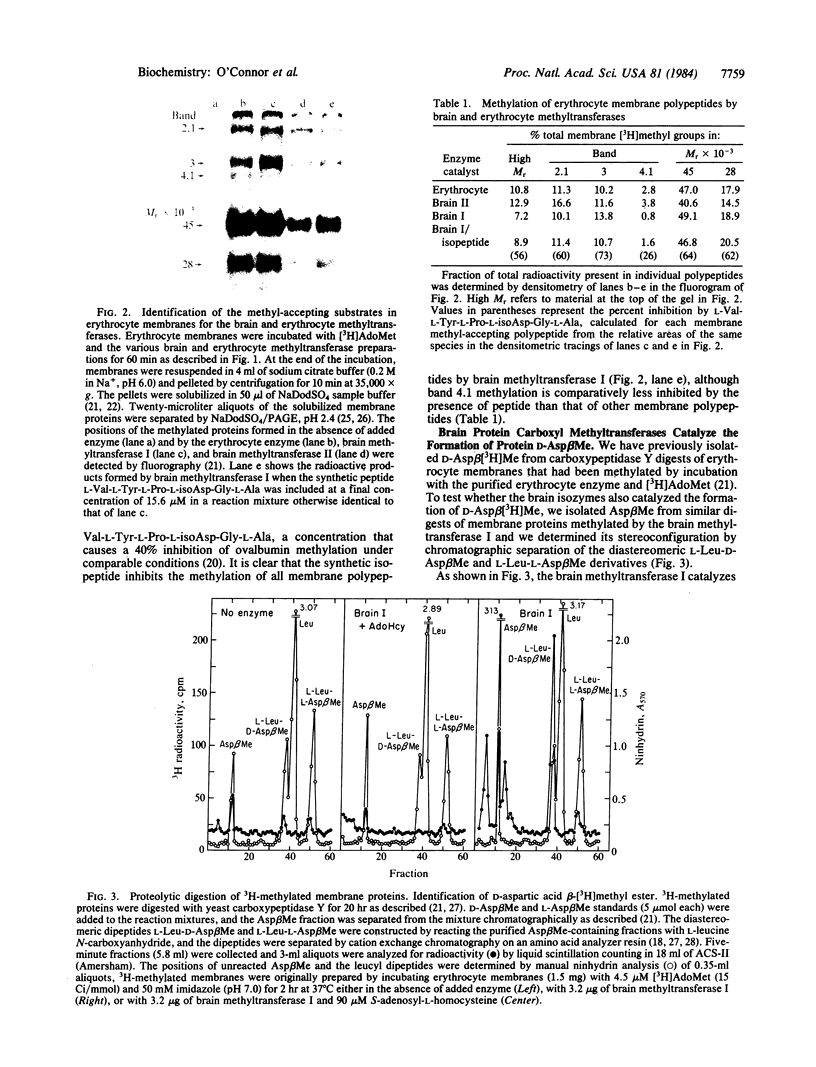
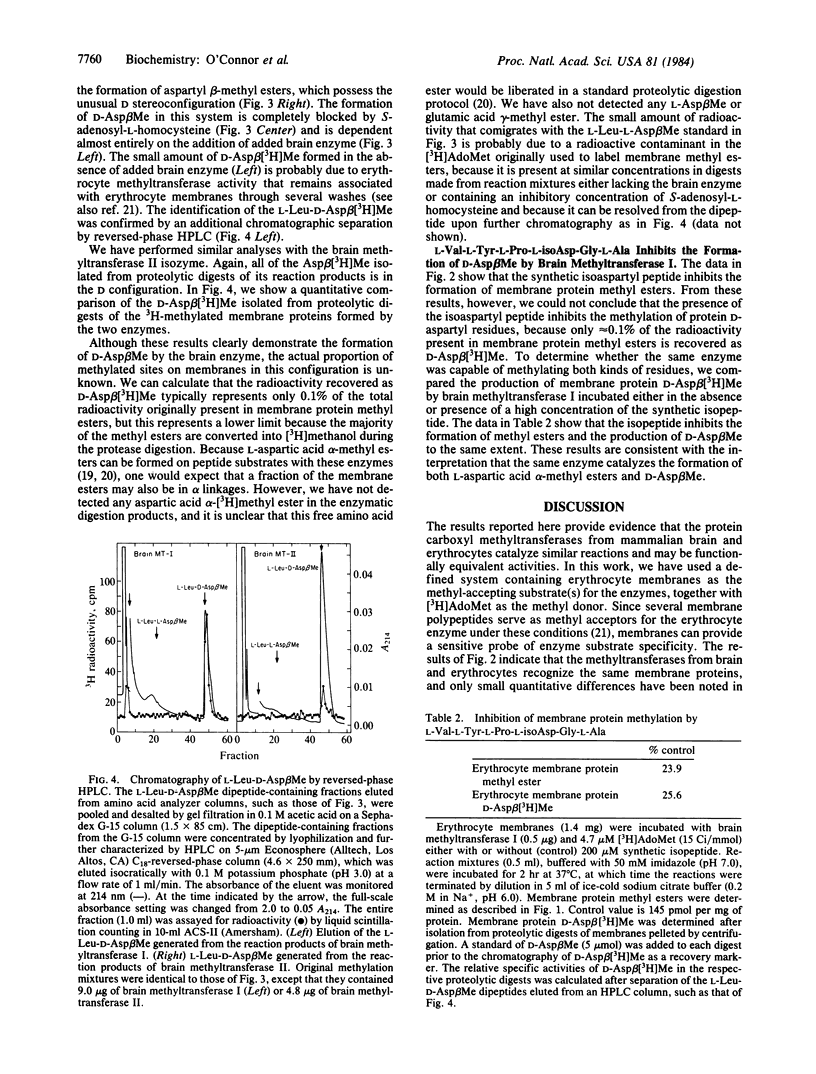
Abstract
Two purified isozymes of protein carboxyl methyltransferase from bovine brain catalyze the substoichiometric transfer of methyl groups in vitro from S-adenosyl-L-[methyl-3H]methionine to several erythrocyte membrane proteins, which include bands 2.1, 3, and 4.1, as well as several integral membrane polypeptides. D-Aspartic acid beta-[3H]methyl ester has been isolated from proteolytic digests of these methylated proteins, suggesting that protein D-aspartyl residues can serve as methyl-acceptor sites for the two brain enzymes. This formation of D-aspartic acid beta-[3H]methyl ester is competitively inhibited by the peptide L-Val-L-Tyr-L-Pro-L-isoAsp-Gly-L-Ala, which contains an L-aspartyl residue in an unusual beta-peptide linkage. Since this peptide is a stoichiometric substrate for the brain methyltransferases, it appears that one enzymatic activity can catalyze methyl ester formation at both D-aspartyl and L-isoaspartyl sites. In these respects, the activity of both brain isozymes closely resembles those previously described for the erythrocyte enzyme. The results are discussed in terms of a model in which derivatized aspartyl residues in proteins, arising by either racemization or isomerization, are recognized by the methyltransferase; the enzyme may function in either the metabolism or correction of the altered structures. The presence of a similar enzyme in both translationally active (brain) and inactive (erythrocyte) tissues suggests that the reactions are of general importance to cellular integrity.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aswad D. W., Deight E. A. Endogenous substrates for protein carboxyl methyltransferase in cytosolic fractions of bovine brain. J Neurochem. 1983 Dec;41(6):1702–1709. doi: 10.1111/j.1471-4159.1983.tb00883.x. [DOI] [PubMed] [Google Scholar]
- Aswad D. W., Deight E. A. Purification and characterization of two distinct isozymes of protein carboxymethylase from bovine brain. J Neurochem. 1983 Jun;40(6):1718–1726. doi: 10.1111/j.1471-4159.1983.tb08147.x. [DOI] [PubMed] [Google Scholar]
- Aswad D. W. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10714–10721. [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Inhibition of protein carboxyl methylation by S-adenosyl-L-homocysteine in intact erythrocytes. Physiological consequences. J Biol Chem. 1984 Jun 10;259(11):7115–7122. [PubMed] [Google Scholar]
- Barber J. R., Clarke S. Membrane protein carboxyl methylation increases with human erythrocyte age. Evidence for an increase in the number of methylatable sites. J Biol Chem. 1983 Jan 25;258(2):1189–1196. [PubMed] [Google Scholar]
- Billingsley M. L., Roth R. H. Dopamine agonists stimulate protein carboxylmethylation in striatal synaptosomes. J Pharmacol Exp Ther. 1982 Dec;223(3):681–688. [PubMed] [Google Scholar]
- Billingsley M. L., Velletri P. A., Roth R. H., DeLorenzo R. J. Carboxylmethylation of calmodulin inhibits calmodulin-dependent phosphorylation in rat brain membranes and cytosol. J Biol Chem. 1983 May 10;258(9):5352–5357. [PubMed] [Google Scholar]
- Billingsley M., Kuhn D., Velletri P. A., Kincaid R., Lovenberg W. Carboxylmethylation of phosphodiesterase attenuates its activation by ca2+-calmodulin. J Biol Chem. 1984 May 25;259(10):6630–6635. [PubMed] [Google Scholar]
- Clarke S., McFadden P. N., O'Connor C. M., Lou L. L. Isolation of D-aspartic acid beta-methyl ester from erythrocyte carboxyl methylated proteins. Methods Enzymol. 1984;106:330–344. doi: 10.1016/0076-6879(84)06033-x. [DOI] [PubMed] [Google Scholar]
- Clarke S., Sparrow K., Panasenko S., Koshland D. E., Jr In vitro methylation of bacterial chemotaxis proteins: characterization of protein methyltransferase activity in crude extracts of Salmonella typhimurium. J Supramol Struct. 1980;13(3):315–328. doi: 10.1002/jss.400130305. [DOI] [PubMed] [Google Scholar]
- Diliberto E. J., Jr, Axelrod J. Regional and subcellular distribution of protein carboxymethylase in brain and other tissues. J Neurochem. 1976 Jun;26(6):1159–1165. doi: 10.1111/j.1471-4159.1976.tb07001.x. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Avruch J. Four gel systems for electrophoretic fractionation of membrane proteins using ionic detergents. J Supramol Struct. 1972;1(1):66–75. doi: 10.1002/jss.400010110. [DOI] [PubMed] [Google Scholar]
- Flynn D. D., Kloog Y., Potter L. T., Axelrod J. Enzymatic methylation of the membrane-bound nicotinic acetylcholine receptor. J Biol Chem. 1982 Aug 25;257(16):9513–9517. [PubMed] [Google Scholar]
- Gagnon C., Kelly S., Manganiello V., Vaughan M., Odya C., Strittmatter W., Hoffman A., Hirata F. Modification of calmodulin function by enzymatic carboxylic methylation. Nature. 1981 Jun 11;291(5815):515–516. doi: 10.1038/291515a0. [DOI] [PubMed] [Google Scholar]
- Kim S., Choi J., Jun G. J. Purification of protein methylase II from human erythrocytes. J Biochem Biophys Methods. 1983 Aug;8(1):9–14. doi: 10.1016/0165-022x(83)90016-7. [DOI] [PubMed] [Google Scholar]
- Kim S., Nochumson S., Chin W., Paik W. K. A rapid method for the purification of S-adenosylmethionine: protein-carboxyl O-methyltransferase by affinity chromatography. Anal Biochem. 1978 Feb;84(2):415–422. doi: 10.1016/0003-2697(78)90059-3. [DOI] [PubMed] [Google Scholar]
- Kloog Y., Saavedra J. M. Protein carboxylmethylation in intact rat posterior pituitary lobes in vitro. J Biol Chem. 1983 Jun 10;258(11):7129–7133. [PubMed] [Google Scholar]
- Manning J. M., Moore S. Determination of D- and L-amino acids by ion exchange chromatography as L-D and L-L dipeptides. J Biol Chem. 1968 Nov 10;243(21):5591–5597. [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Methylation at D-aspartyl residues in erythrocytes: possible step in the repair of aged membrane proteins. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2460–2464. doi: 10.1073/pnas.79.8.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. D., Jr, Clarke S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. Detection of a new site of methylation at isomerized L-aspartyl residues. J Biol Chem. 1984 Sep 10;259(17):10722–10732. [PubMed] [Google Scholar]
- O'Connor C. M., Clarke S. Methylation of erythrocyte membrane proteins at extracellular and intracellular D-aspartyl sites in vitro. Saturation of intracellular sites in vivo. J Biol Chem. 1983 Jul 10;258(13):8485–8492. [PubMed] [Google Scholar]
- Polastro E. T., Deconinck M. M., Devogel M. R., Mailier E. L., Looza Y. B., Schnek A. G., Léonis J. Purification and some molecular properties of protein methylase II from equine erythrocytes. Biochem Biophys Res Commun. 1978 Apr 14;81(3):920–927. doi: 10.1016/0006-291x(78)91439-0. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C., Clarke S. Methylation of membrane proteins in human erythrocytes. Identification and characterization of polypeptides methylated in lysed cells. J Biol Chem. 1981 Mar 25;256(6):3067–3076. [PubMed] [Google Scholar]
- Terwilliger T. C., Koshland D. E., Jr Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J Biol Chem. 1984 Jun 25;259(12):7719–7725. [PubMed] [Google Scholar]
- Unger C., Jahn R., Söling H. D. Is protein carboxymethylation involved in stimulus-secretion coupling? FEBS Lett. 1981 Jan 26;123(2):211–214. doi: 10.1016/0014-5793(81)80289-x. [DOI] [PubMed] [Google Scholar]



