SUMMARY
A clear relationship exists between visceral obesity and type 2 diabetes, whereas subcutaneous obesity is comparatively benign. Here we show that adipocyte-specific deletion of the coregulatory protein PRDM16 caused minimal effects on classical brown fat but markedly inhibited beige adipocyte function in subcutaneous fat following cold exposure or β3-agonist treatment. These animals developed obesity on a high fat diet, with severe insulin resistance and hepatic steatosis. They also showed altered fat distribution with markedly increased subcutaneous adiposity. Subcutaneous adipose tissue in mutant mice acquired many key properties of visceral fat, including decreased thermogenic and increased inflammatory gene expression and increased macrophage accumulation. Transplantation of subcutaneous fat into mice with diet-induced obesity showed a loss of metabolic benefit when tissues were derived from PRDM16 mutant animals. These findings indicate that PRDM16 and beige adipocytes are required for the “browning” of white fat and the healthful effects of subcutaneous adipose tissue.
INTRODUCTION
Obesity has become a global epidemic, contributing to increases in the prevalence of type 2 diabetes, hypertension, cardiovascular disease, and certain cancers. Generally, two broad categories of obesity are recognized: visceral (VISC) and subcutaneous (SubQ). The location where fat is deposited appears to have a great influence on the likelihood of an individual developing many of the sequelae of obesity (Gesta et al., 2007). Importantly, VISC adiposity is strongly associated with increased mortality, even in individuals with a normal body mass index (Pischon et al., 2008). SubQ adiposity, however, appears to be comparatively benign (Manolopoulos et al., 2010). The association between regional fat deposition and adverse health complications was first noted with pioneering clinical descriptions in the 1950s (Vague, 1956). It has also been recognized for centuries that men have a greater propensity for deposition of VISC fat, while premenopausal women have a greater tendency to accumulate fat in SubQ stores, though substantial variation exists in both sexes (Vague, 1947).
The relationship between site of adipose tissue accumulation and metabolic disease has been shown in several animal models. Transgenic mice overexpressing 11-β HSD-1 in adipose tissue develop VISC obesity along with insulin resistance, diabetes, and hyperlipidemia (Masuzaki et al., 2001). Conversely, transgenic mice overexpressing adiponectin or mitoNEET in adipose tissue develop remarkable SubQ obesity, but remain metabolically healthy (Kim et al., 2007; Kusminski et al., 2012). The functional importance of these adipose depots has been directly demonstrated in studies showing metabolic benefit by transplantation of SubQ fat or surgical removal of VISC fat (Gabriely et al., 2002; Tran et al., 2008).
These divergent metabolic effects of different adipose depots have raised interest in the unique properties of VISC and SubQ fat. VISC fat is notable for having a substantial degree of inflammation when obesity is present. Originally recognized as the secretion of TNFα and other inflammatory cytokines from fat tissue of obese animals (Hotamisligil et al., 1993), it is now known that there is a broad increase in a variety of immune cells in obese fat (Weisberg et al., 2003; Xu et al., 2003). On the other hand, SubQ fat is notable because it can easily “brown” when animals are stimulated with cold, β-adrenergic agonists, or other hormone-like stimuli (Vitali et al., 2012; Wu et al., 2013). This browning includes the induction of UCP1 mRNA and protein, and expression of a gene program that gives rise to uncoupled respiration and heat production. Some stimuli, such as the genetic ablation of RALDH1, cause considerable browning of the VISC depots in mice (Kiefer et al., 2012), but this phenomenon is much less common than the browning of SubQ depots.
It is now recognized that there are at least two distinct types of brown fat cells. Classical brown adipose tissue (BAT), epitomized by the interscapular depot in rodents, arises from a Myf-5+, muscle-like cell lineage. PRDM16, a transcriptional regulatory protein, appears to control this muscle-brown fat decision between days 9–12 of gestation in mice (Lepper and Fan, 2010; Seale et al., 2008). The postnatal role of PRDM16 in classical BAT has not been studied. In contrast, the UCP1+ cells that emerge in white fat depots under certain stimuli, termed beige or brite cells, do not come from a Myf5+ lineage. However, PRDM16 also appears to be involved in the development and function of beige cells (Ohno et al., 2012; Seale et al., 2011). Hence, beige and classical brown fat cells both express PRDM16 and UCP1 but they are quite distinct cell types. Beige fat cells have now been cloned and characterized (Wu et al., 2012). Importantly, the depots of “brown fat” visualized in humans share more molecular properties with rodent beige fat than with classical BAT (Lidell et al., 2013; Sharp et al., 2012; Wu et al., 2012). The kite-like structure in the interscapular region of human infants, however, seems to have characteristics of rodent classical BAT, and some classical brown fat cells may be retained in adult humans (Cypess et al., 2013; Jespersen et al., 2013; Lidell et al., 2013).
Analysis of the relative physiological importance of the classical BAT vs. the browning of white fat has been very difficult because many important browning agents, such as cold and β-adrenergic compounds, affect both types of brown fat cells. Similarly, while ablation of UCP1+ cells or mutation of Ucp1 illustrate the overall importance of brown fat in preventing obesity and diabetes in animals, the individual contributions of the two types of brown fat cells has been impossible to determine (Feldmann et al., 2009; Lowell et al., 1993). Agents that affect browning of white fat selectively, including transgenic expression of PRDM16, have caused metabolic benefit, suggesting that browning of white tissues could be important (Seale et al., 2011). On the other hand, it has been argued that there are insufficient beige cells to affect whole body physiology under ambient conditions (Nedergaard and Cannon, 2013).
We have now created an adipocyte-selective mutation in the Prdm16 gene that ablates the thermogenic program of beige fat cells, while leaving the classical BAT functionally intact. Mice lacking beige fat function develop obesity and insulin resistance when exposed to high fat diet and also develop hepatic steatosis. In addition, there is a striking change in the SubQ fat, which takes on many of the morphological and molecular characteristics of VISC fat. These data demonstrate that PRDM16 and beige fat cells are major regulators of systemic physiology. Moreover, PRDM16 determines some of the key functional differences between SubQ and VISC fat.
RESULTS
PRDM16 is Enriched in SubQ Adipose Tissues and Required for Thermogenic Gene Expression
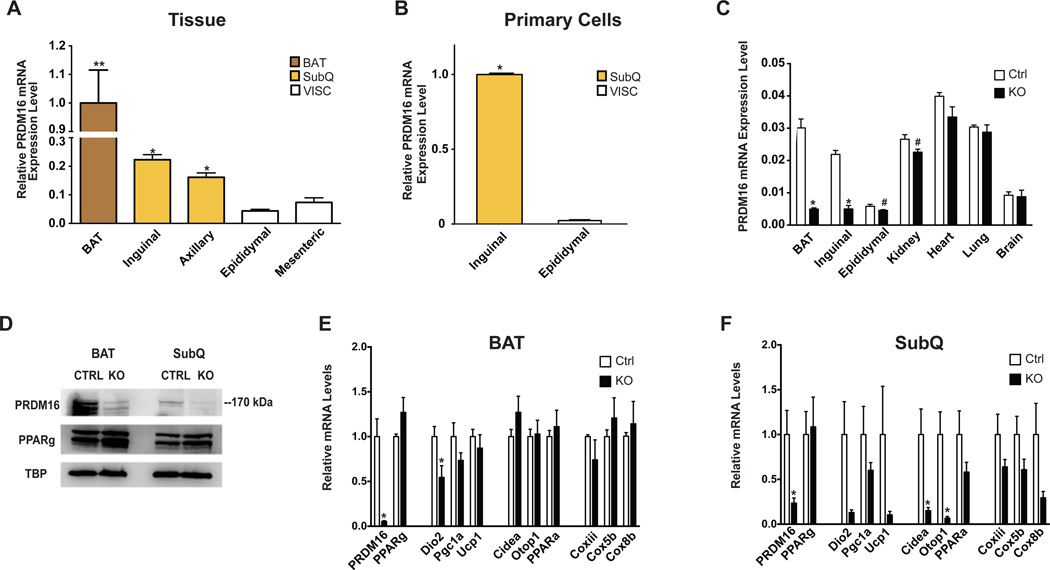
PRDM16 is involved in the development and function of classical brown and beige adipocytes. To determine how PRDM16 might be involved in the function of different adipose tissues that are considered “white”, we measured mRNA levels in multiple SubQ and VISC fat depots and compared these to the classical interscapular brown fat. Prdm16 was significantly elevated in two SubQ depots (inguinal and axillary) relative to the very low levels of expression in two VISC depots (epididymal and mesenteric). As expected, the highest levels of expression were seen in interscapular BAT (Figure 1A). This depot enrichment was maintained in primary adipocytes differentiated in vitro from the inguinal SubQ or epididymal VISC stromal vascular fraction (SVF) (Figure 1B). Expression of Ucp1, the prototypical thermogenic gene, was also enriched across SubQ depots, consistent with the presence of beige adipocytes in these pads (Figure S1A–B). These data suggested that PRDM16 could play a role in multiple SubQ depots.
Figure 1. PRDM16 Deletion in Adipocytes Results in Altered Subcutaneous Adipose Tissue Gene Expression.
(A) and (B) qPCR analysis of Prdm16 mRNA from multiple adipose tissues from 8–10 week old, male wild-type mice (n=9), normalized to mRNA expression in BAT (A) or from in vitro differentiated primary adipocytes, normalized to mRNA expression in inguinal cells (n=3) (B).
(C) qPCR analysis of Prdm16 mRNA from multiple tissues from 6–8 week old male Adipo-PRDM16 KO mice (n=4) and controls (n=4).
(D) PRDM16 protein in nuclear extracts from BAT or inguinal SubQ adipose tissue from Adipo-PRDM16 KO and controls. PPARγ and TBP protein are shown as controls.
(E) and (F) Normalized gene expression of thermogenic, brown adipose, and mitochondrial genes in BAT (E) and SubQ adipose tissue (F) from Adipo-PRDM16 KO and control mice at RT. Mice were males, 6–8 weeks old, n=5 per group.
Data are presented as mean ± SEM. *, p< 0.05. #, p=0.053 for kidney and 0.079 for epididymal fat. See also Figure S1.
We generated a conditional Prdm16 allele (Prdm16lox/lox) by engineering loxP sites flanking exon 9, which we have shown to be required for mediating the effects of PRDM16 on thermogenic gene expression (Seale et al., 2008) (Figure S1C). We initially crossed Prdm16lox/lox mice to Zp3-cre mice to generate a germline deletion. As expected, these animals had craniofacial defects and died within 24 hours of birth, confirming that this mutation created a null allele of Prdm16 (Bjork et al., 2010).
We next crossed Prdm16lox/lox and Adiponectin-cre mice (Eguchi et al., 2011) to generate mice with an adipocyte-specific knockout (KO) of Prdm16 (Adipo-PRDM16 KO). This promoter expresses cre recombinase after the early developmental time point when the classical brown fat/muscle fate decision has been established (Lepper and Fan, 2010). We performed metabolic characterization of Adiponectin-cre mice and confirmed that the Cre recombinase is not likely to contribute to the KO phenotype (Figure S1D). Analysis of mRNA expression from multiple tissues confirmed that the deletion was specific to white and brown adipose tissues (Figure 1C). Notably, PRDM16 mRNA levels in epididymal fat trended downward in KO mice, though expression in this depot was at the margin of detection. The trend towards decreased Prdm16 in the kidney of KO mice may reflect expression in adipose tissue surrounding this organ. Western blotting of nuclear extracts showed a marked reduction of PRDM16 protein in both classical BAT and inguinal SubQ adipose tissue from KO mice (Figure 1D). PRDM16 protein was too low to detect in the VISC depots.
PRDM16 deletion in the classical BAT resulted in a relatively normal pattern of gene expression (Figure 1E). The SubQ adipose tissue of Adipo-PRDM16 KO mice, however, showed reduced expression of a broad panel of thermogenic (Dio2, Pgc1a, Ucp1), brown adipose identity (Cidea, Otop1, PPARa), and mitochondrial electron transport genes (Coxiii, Cox5b, Cox8b) (Figure 1F). There was no alteration in the expression of Pparg, which is a general marker of adipose differentiation. Ucp1 mRNA was reduced by 90% in the inguinal SubQ depot. This did not reach statistical significance (p = 0.14) due to the large variance in Ucp1 mRNA levels in control mice. Adipo-PRDM16 KO mice also had decreased expression of a few of these genes in VISC adipose tissue, compared to controls, though the absolute expression level of these genes was very low (Figure S1E).
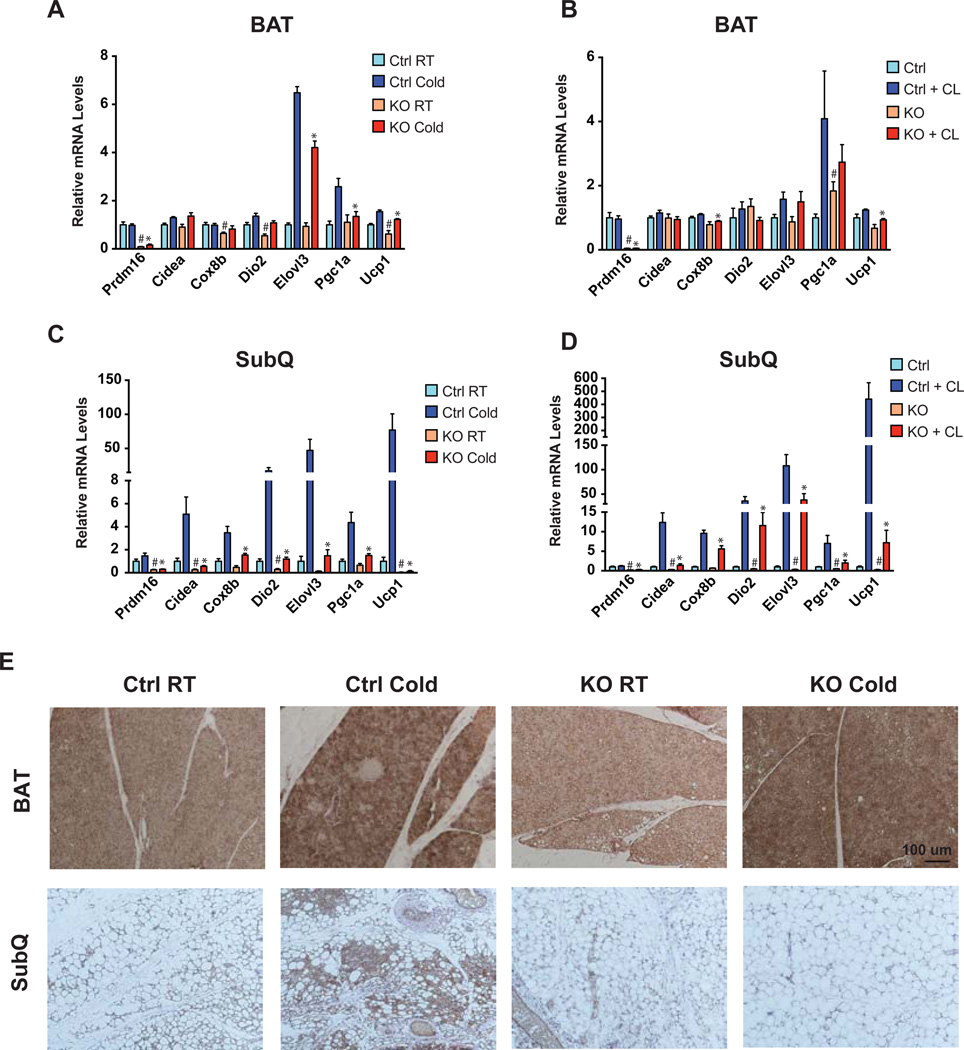
Since SubQ adipose tissues are particularly prone to inducing a thermogenic gene program (‘browning’), we examined the effects of two potent stimuli: cold exposure and treatment with a β–adrenergic agonist. In interscapular BAT, the thermogenic program was induced similarly in both control and Adipo-PRDM16 KO animals following cold exposure or CL 316,243 (hereafter referred to as CL) treatment. There were some small but statistically significant differences in the induction of several genes with cold and CL treatment (Figure 2A and 2B). In contrast, the very robust increase in the entire panel of thermogenic gene expression seen in control inguinal adipose tissues (cold: 74-fold induction of Ucp1, CL: >400-fold induction of Ucp1) was almost completely blocked in KO animals (Figure 2C and 2D). A similar pattern showing selective effects in SubQ adipose tissue was seen with prolonged cold exposure for 6 days (Figure S2A–B). Ucp1 showed blunted induction in VISC adipose tissue of the Adipo-PRDM16 KO mice following CL treatment, though the levels of thermogenic genes were overall very low in this depot (Figure S2C).
Figure 2. PRDM16 Regulates Beige Adipose Thermogenic Gene Expression.
(A–D) Normalized thermogenic gene expression in BAT (A and B) and SubQ adipose tissue (C and D) from Adipo-PRDM16 KO and control mice at RT and following 48 hours at 4°C (A and C) or following five daily injections of 1 mg/kg CL (B and D). Cold exposure was done with male 6–8 week old mice, n=5–6 per group. CL treatment was done with female 6–8 week old mice, n=5–6 per group. Data are presented as mean ± SEM. *, p< 0.05 KO vs. control cold exposed or CL treated. #, p< 0.05 KO vs. control room temperature or untreated.
(E) Representative images from UCP1 immunohistochemistry on sections of interscapular BAT or inguinal SubQ adipose tissue from Adipo-PRDM16 KO and control mice at RT or following 48 hours at 4°C. Images are shown at 10x magnification. Scale bar =100 microns.
See also Figure S2.
The morphology and UCP1 content of these adipose depots was also examined in control and Adipo-PRDM16 KO mice. No differences in UCP1 were seen in the BAT between mice of either genotype, with or without cold exposure (Figure 2E). The SubQ adipose tissue from Adipo-PRDM16 KO mice, however, contained larger adipocytes and fewer small, multilocular UCP1+ adipocytes (Figure 2E). Moreover, cold exposure resulted in the emergence of numerous small, multilocular UCP1+ adipocytes in controls, while KO mice showed no change in appearance relative to RT (Figure 2E). Of note, Adipo-PRDM16 KO mice showed no difference in core temperature following 6 hours (Figure S2D) or 96 hours at 4°C (36.04°C ± 0.19 in KOs vs. 35.78°C ± 0.26 in controls, p=0.44). This further indicates that these mice have little apparent defect in the function of BAT since ablation of classical BAT or UCP1 is associated with severe cold intolerance (Enerback et al., 1997; Lowell et al., 1993).
We next asked whether these changes in the thermogenic program of SubQ adipose tissue were cell autonomous. SubQ adipocytes from KO mice differentiated in vitro also showed a significant reduction in the expression of most thermogenic (81% reduction in Ucp1), brown fat identity (97% reduction in Cidea), and mitochondrial genes (80% reduction in Cox8b), strongly suggesting a cell autonomous role for PRDM16 in browning (Figure S2E). Cells from Adipo-PRDM16 KO mice also showed a blunted response to stimuli that induce browning, such as isoproterenol and FGF21 (Figure S2F–G) (Fisher et al., 2012; Wu et al., 2013).
PRDM16 Regulates Adipose Tissue and Whole Body Energy Expenditure
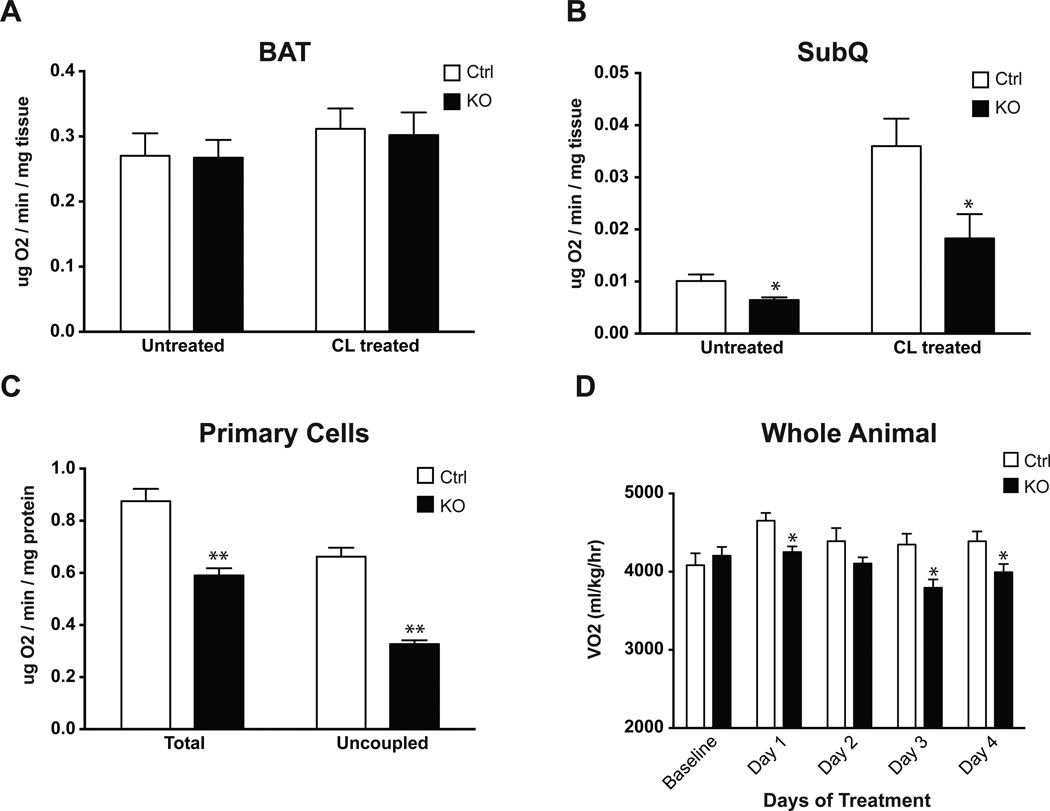
Given the preferential effect of this PRDM16 deletion on thermogenic gene expression in beige adipocytes, we assessed the physiological effect of this ablation, using O2 consumption as a readout. SubQ and BAT pads were removed from mice and O2 consumption was measured. These assays were performed both in untreated animals and after 5 days of CL treatment. Importantly, O2 consumption in interscapular BAT was unchanged between KOs and controls in both basal and stimulated states (Figure 3A). In contrast, O2 consumption in SubQ adipose tissue from Adipo-PRDM16 KO mice was significantly reduced, both at baseline (36% reduced) and following stimulation with CL (49% reduced) (Figure 3B). O2 consumption in VISC adipose tissue was below the limit of detection in most animals. Assays in primary SubQ adipocytes showed a significant reduction in both total (33% reduced) and uncoupled (51% reduced) respiration in KO cells compared to controls (Figure 3C). Taken together, these results indicate that adipose-selective deletion of PRDM16 results in a large defect in a broad program of thermogenesis in SubQ adipose tissue, while classical BAT is relatively unaffected.
Figure 3. Altered Subcutaneous Adipose Tissue and Whole Body O2 Consumption in Adipo-PRDM16 KO Mice.
(A and B) O2 consumption in brown (A) and SubQ adipose tissue (B) from control and Adipo-PRDM16 KO mice. Tissues from untreated animals and animals treated with 5 daily injections of 1 mg/kg CL were analyzed. CL treatment was done with female 6–8 week old mice, n=5–6 per group.
(C) Total and uncoupled respiration in primary SubQ adipocytes from control and Adipo-PRDM16 KO mice. N=6 per group.
(D) O2 consumption in 8–10 week old Adipo-PRDM16 KO and control male mice, n=8 per group. Data were recorded at baseline and for 6 hours following daily injection of 0.1 mg/kg CL.
Data are presented as mean ± SEM. *, p< 0.05. **, p<0.001. See also Figure S3.
This selective ablation of beige cell thermogenic function offered a unique opportunity to study the role of these cells and the browning of white fat on whole body physiology. When Adipo-PRDM16 KO mice were fed a chow diet, no difference was observed in food intake or activity between the mutant and control groups (Figure S3A–B). Adipo-PRDM16 KO mice also showed no difference in O2 consumption, but did have a significantly increased respiratory exchange ratio (RER), suggesting a decrease in utilization of fatty acid oxidation as an energy substrate (Figure S3C–D). We also studied these animals after injection with CL. Control animals showed a significant increase in O2 consumption (6.5–13.9%) following each injection. KO mice showed no increase in O2 consumption, suggesting that ablation of beige adipocyte function can affect whole body energy expenditure (Figure 3D).
Adipo-PRDM16 KO Mice Develop Obesity and Insulin Resistance
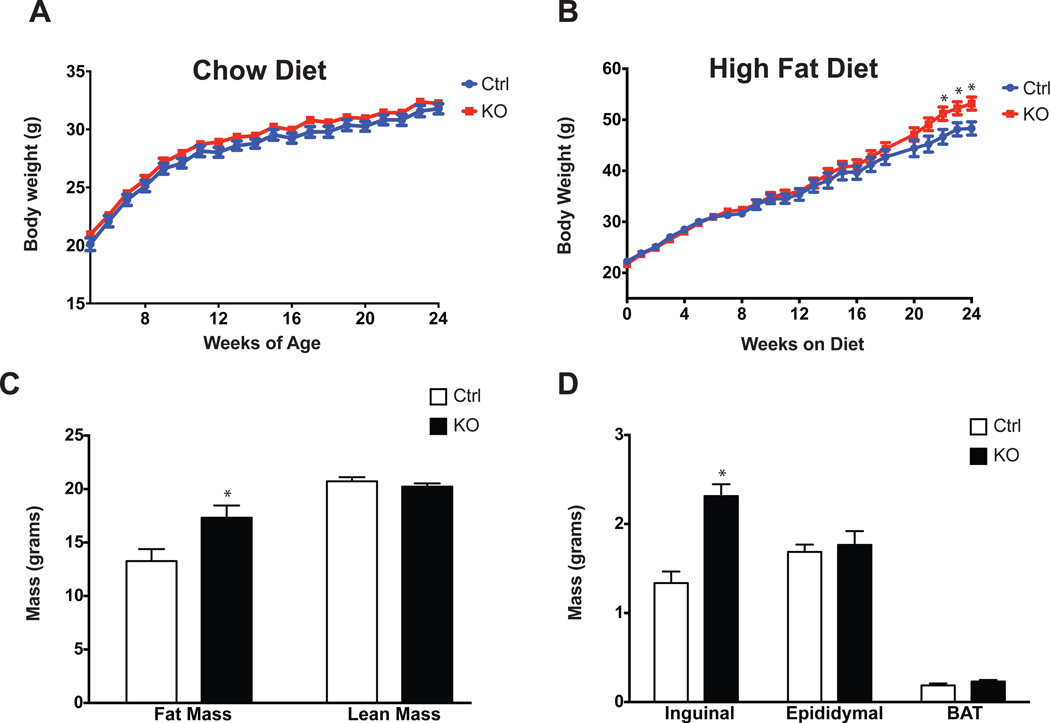
Animals were then subjected to a physiological challenge in the form of a high fat, high carbohydrate diet. Adipo-PRDM16 KO mice showed no difference in body weight from control animals on a chow diet, but demonstrated increased weight gain on a high fat diet (HFD) (Figure 4A and 4B). At thermoneutrality, KOs showed no difference in body weight compared to controls, over 7 weeks on either a chow or HFD (Figure S4A–B). Of note, Ucp1 KO mice show significant weight gain over this time period at thermoneutrality, presumably due to defects in classical brown fat function (Feldmann et al., 2009).
Figure 4. Obesity and Altered Fat Distribution in Adipo-PRDM16 KO Mice.
(A and B) Body weights of Adipo-PRDM16 KO and control mice on standard chow (A) or HFD (B). HFD was started at 4 weeks of age. Experiments were done with male mice, n=12–15 per group.
(C) Body composition of Adipo-PRDM16 KO and control mice following 16 weeks on HFD. N=12 for controls and N=19 for KOs.
(D) Weights of individual fat pads from Adipo-PRDM16 KO and control mice following 18 weeks on HFD. N=11 per group.
Data are presented as mean ± SEM. *, p< 0.05. See also Figure S4.
Body composition analysis was done after 16 weeks on HFD, when the weight curves had just started to diverge. Mutant animals had significantly increased fat mass, with no change in lean body mass (Figure 4C). These mice also showed an interesting change in the distribution of fat mass: KO animals had SubQ fat mass (inguinal depot) nearly twice that of controls, while the VISC (epididymal) and BAT masses were unchanged (Figure 4D). Metabolic analysis of mice after 2 weeks of HFD showed no significant differences in food intake, activity, O2 consumption, or RER compared to control animals (Figure S4C–4F). O2 consumption was also unchanged after longer periods on HFD, likely reflecting the fact that indirect calorimetry is insufficiently sensitive to detect small differences in energy expenditure, which could result in modest obesity over a prolonged period (Butler and Kozak, 2010).
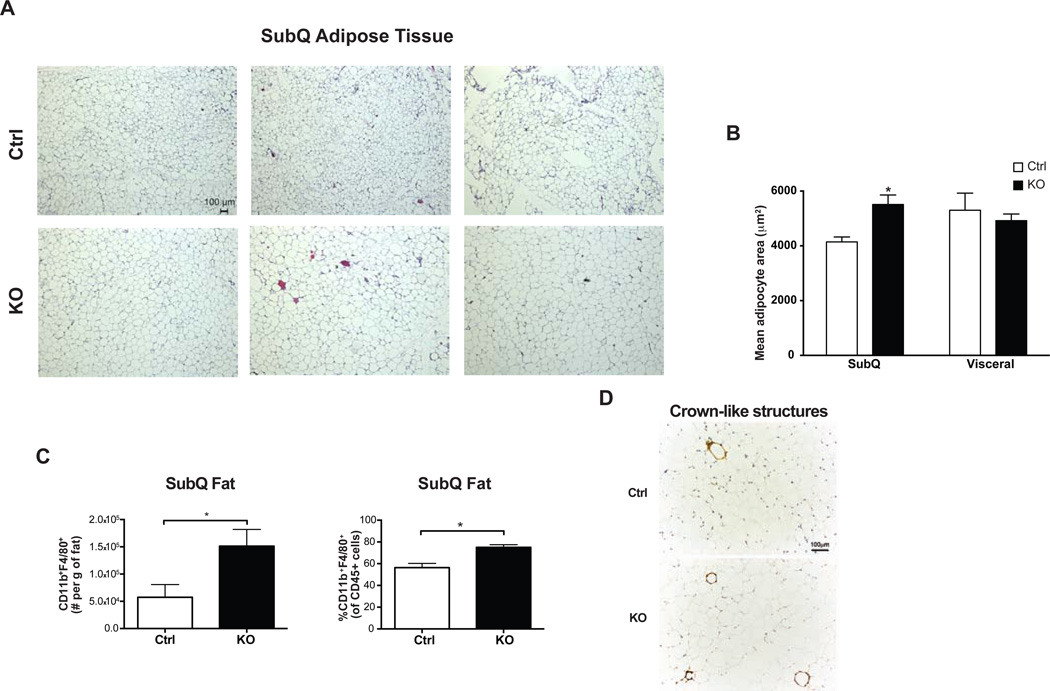
SubQ adipocytes in high-fat fed Adipo-PRDM16 KO mice were markedly larger than those in control mice (Figure 5A). Quantitative assessment confirmed a 33% increase in mean adipocyte area in the SubQ depot of mutant animals to a size equivalent to adipocytes in the VISC depot (Figure 5B). KO mice had no difference in mRNA expression levels of general markers of adipose differentiation, though they did have a trend towards increased Glut4 mRNA levels compared to controls (Figure S5A). The morphology and size of VISC epididymal adipocytes was unchanged between control and KO animals (Figure S5B). We used flow cytometry to quantitate the major myeloid and lymphoid subsets in the adipose tissue of high-fat fed mice. When compared to control animals, SubQ adipose tissue from KOs had a significant increase in the fraction and number of CD11b+F4/80+ macrophages (Figure 5C), along with increased crown-like structures (Figure 5D). These alterations were restricted to SubQ adipose tissue, as there were no differences in the representation of CD11b+F4/80+ macrophages in the VISC adipose tissue or spleen from KO animals (Figure S5C). No other immune cell subsets, including T regulatory cells, which have been shown to play a role in metabolic disease (Cipolletta et al., 2012), were altered in adipose tissue or lymphoid organs from KO mice (Figure S5D).
Figure 5. Increased Cell Size and Macrophage Accumulation in Subcutaneous Adipose Tissue from Adipo-PRDM16 KO Mice.
(A and B) Cell size in inguinal SubQ adipose tissue from male Adipo-PRDM16 KO and control mice following 18 weeks on HFD. Representative images from hematoxylin and eosin (H&E) stained sections. Images are shown at 5x magnification. Scale bar =100um. (B). Mean adipocyte area from inguinal SubQ and epididymal VISC adipose tissue. N=3 per group.
(C) Flow cytometry quantitation of CD11b+F4/80+ macrophages in SubQ adipose tissue from Adipo-PRDM16 KO and control mice following 6 weeks on HFD.
(D) Morphological analysis for crown-like structures using Mac-2 staining from Adipo-PRDM16 KO and control mice following 6 weeks on HFD.
For (C) and (D), experiments was done with male mice 8–10 weeks old at start of HFD, n=5–6 per group.
Data are presented as mean ± SEM. *, p< 0.05. See also Figure S5.
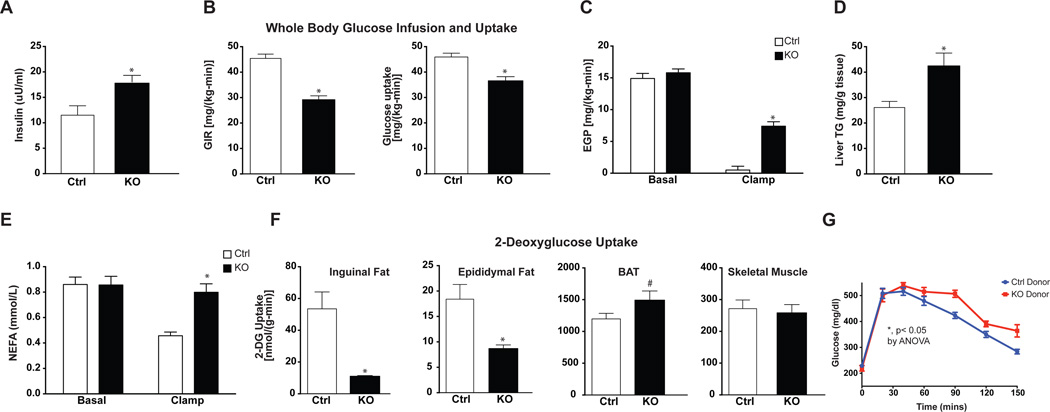
We next assessed glucose homeostasis in Adipo-PRDM16 KO mice. We performed hyperinsulinemic-euglycemic clamps, a sensitive method for assessing whole body and tissue-specific insulin sensitivity. These studies were performed at a time well before the weight curves of KO animals diverged from controls (6 weeks on high-fat diet). There was no difference in fasting plasma glucose between groups (Figure S6A). However, fasting plasma insulin levels were significantly increased (55% increased, p< 0.05) in Adipo-PRDM16 KO mice, suggestive of insulin resistance (Figure 6A). During the clamp, plasma glucose levels were matched between groups at approximately 120 mg/dl (Figure S6A). Adipo-PRDM16 KO mice showed significant whole-body insulin resistance as evidenced by a markedly reduced glucose infusion rate (64% of controls) and decreased whole body glucose uptake (Figure S6B and Figure 6B). Mutant mice also demonstrated significant hepatic insulin resistance with decreased insulin-stimulated suppression of endogenous glucose production (53% in KOs vs. 95% in controls) (Figure 6C). This primary alteration in adipose tissue was also associated with hepatic steatosis in KO animals (Figure 6D). Moreover, KO animals showed a failure in insulin-mediated suppression of lipolysis (6% in KOs vs. 45% in controls) (Figure 6E). Although control and Adipo-PRDM16 KO mice were dosed with the same amount of insulin, clamped insulin levels were modestly increased in the KOs (p=0.08, Figure S6C), suggesting impaired insulin clearance consistent with insulin resistance. Gene expression analysis from a separate cohort of mice following 6 weeks HFD, showed marked reductions of many thermogenic genes in KO SubQ adipose tissue compared to controls. Some of these genes showed a modest, but significant, reduction in classical BAT (Figure S6D).
Figure 6. Insulin Resistance, Hepatic Steatosis, and Altered Glucose Uptake in Adipo-PRDM16 KO Mice.
(A–E) Hyperinsulinemic-euglycemic clamp studies on Adipo-PRDM16 KO and control mice after 6 weeks on HFD.
(A) Fasting insulin levels.
(B) Glucose infusion rate and whole body glucose uptake.
(C) Basal and insulin-stimulated endogenous glucose production.
(D) Liver triglyceride content.
(E) Basal and insulin-stimulated levels of non-esterified fatty acids.
(F) 2-deoxyglucose uptake in tissues from Adipo-PRDM16 KO and control mice.
Data are presented as mean ± SEM. *, p< 0.05, # p= 0.09. Clamp studies were done with male mice, 8 weeks old at the start of HFD. N=7–8 per group.
(G) Intraperitoneal glucose tolerance test (1.5 mg/kg) in male, high fat-fed recipients of Adipo-PRDM16 KO or control SubQ adipose tissue. The GTT was done 6 weeks following transplantation. N=6 per group. *, p <0.05 by ANOVA.
Data are presented as mean ± SEM.
See also Figure S6.
A bolus of 14C-2-deoxyglucose was administered to determine tissue-specific rates of glucose transport. Marked reductions in glucose uptake were observed in the SubQ (79% decreased) and VISC fat (53% decreased) (Figure 6F). Interestingly, glucose uptake in the classical BAT was actually higher in Adipo-PRDM16 KO mice. Skeletal muscle glucose uptake was no different between control and KO mice. These data indicate that deletion of PRDM16 in adipocytes results in selectively dysfunctional beige adipose function with strong effects on whole body and tissue-specific insulin sensitivity.
Adiponectin levels showed no difference between control and KO mice (Figure S6E). Moreover, PRDM16 null adipose cells do not appear to have cell autonomous defects in insulin signaling, as levels of total and phosphorylated Akt were unchanged between control and KO primary SubQ adipocytes (Figure S6F).
Loss of Metabolic Benefit With Transplantation of Subcutaneous Fat from Adipo-PRDM16 KO Mice
This spectrum of phenotypes in Adipo-PRDM16 KO mice suggested that PRDM16 might be required for the healthful actions of SubQ adipose tissue. To address this, we transplanted SubQ fat pads (Gunawardana and Piston, 2012) from weanling pups of the two PRDM16 genotypes into high fat fed wild-type recipients and animals were then maintained on HFD. All recipients gained weight equally following the transplants. A glucose tolerance test was done at 6 weeks, and a highly significant difference was observed between the groups, with worse glucose tolerance in recipients of KO tissue (Figure 6G). There was no significant difference in insulin tolerance between these groups. This result has been replicated in a separate cohort of transplant recipients. The inclusion of a donor group from PRDM16 overexpressing, transgenic mice showed that glucose tolerance appears to track with PRDM16 levels in donor tissue (Figure S6G).
SubQ Adipose Tissue of Adipo-PRDM16 KO Mice Acquires a Partial VISC Adipose Phenotype
The combination of adipose and hepatic insulin resistance and the presence of larger adipocytes in KO SubQ adipose tissue with increased macrophage accumulation suggested a phenotype more typical of VISC obesity. These observations indicated that Adipo-PRDM16 KO SubQ adipose tissue might have developed some of the deleterious characteristics of VISC adipose tissue. To address this in an unbiased fashion we used whole genome microarrays to define depot-specific gene expression, and then examined whether this molecular profile appeared ‘visceralized’ in the SubQ adipose tissue of Adipo-PRDM16 KO animals.
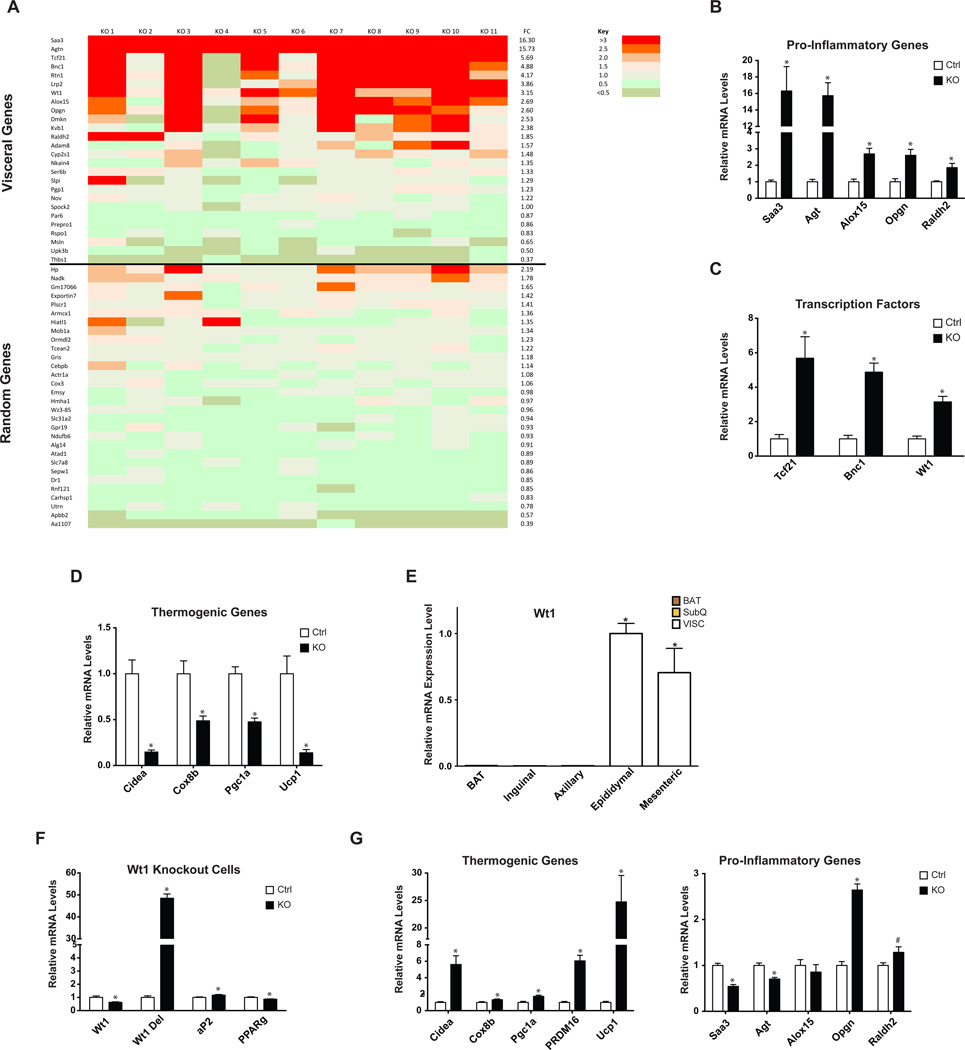
Depot-enriched gene expression was studied by comparing RNA expression in adipose tissue from two SubQ depots (inguinal and axillary) to that from two VISC depots (epididymal and mesenteric). We defined a VISC gene set as containing genes with >3-fold increased expression and a p-value <0.05 between the VISC and SubQ depots. 60 genes met these criteria. We then determined which of these genes had expression that was enriched >2-fold in fractionated adipocytes from VISC vs. SubQ adipose tissue, to avoid studying differences due to other cell types. 26/60 genes met this requirement. We then studied the expression of these 26 visceral-selective genes in SubQ adipose tissue from control or Adipo-PRDM16 KO mice fed a HFD. 11/26 were >2-fold increased with p-value <0.05 in KOs vs. controls. We then selected 30 genes at random that were expressed in SubQ adipocytes and used these as a comparator. Only 1/30 was >2-fold increased with a p-value <0.05 in KO vs. control adipose tissue.
These data are shown as a heat map with the color indicating the fold-change for each gene of this VISC fat gene expression set in SubQ adipose tissue from 11 KO mice relative to that of the control group (Figure 7A). The acquisition of a VISC-selective expression profile is highly significant (11/26 vs. 1/30 genes, p =0.0004 by chi-squared). In addition this VISC profile contains a number of pro-inflammatory genes of known functional importance: Serum amyloid A3 (Saa3), Angiotensinogen (Agt), 12/15 lipoxygenase (Alox15), Osteoprotegerin (Opgn), and Retinaldehyde dehydrogenase 2 (Raldh2) (Figure 7B). The VISC-selective expression profile also incudes three transcription factors (Tcf21, Bnc1, and Wt1) (Figure 7C). Two of these pro-inflammatory genes (Agt and Raldh2) were further increased in VISC adipose tissue from KO mice, compared to controls, though the expression of the three VISC-enriched transcription factors was unchanged (Figure S7A–B). Of note, more than half of these pro-inflammatory genes and transcription factors were significantly upregulated in PRDM16 KO SubQ fat cells differentiated in vitro (Figure S7C–D). This strongly suggests that the altered molecular profile in PRDM16 null adipose tissue is not simply a consequence of the metabolic dysregulation seen in these animals.
Figure 7. Induction of a Visceral Adipose Tissue Gene Expression Profile in Adipo-PRDM16 KO Subcutaneous Adipose Tissue.
(A) Heat map showing relative expression of VISC selective and randomly selected genes in SubQ adipose tissue from Adipo-PRDM16 KO mice after 18 weeks on HFD. Each row depicts an individual gene. Each column depicts the expression in a KO sample relative to the average expression in control mice. Average fold change is indicated in the column to the right. Fold change for each individual sample is color-coded according to the key.
(B) Normalized expression of pro-inflammatory genes and (C) transcription factors.
(D) Normalized expression of thermogenic genes.
For (A)–(D), gene expression was measured in male mice following 18 weeks HFD. N=11 per group.
(E) qPCR analysis of Wt1 mRNA from multiple adipose tissues from 8–10 week old, male wild-type mice, normalized to mRNA expression in epididymal white adipose tissue. N=9 per group.
(F–G) Normalized expression of Wt1 (F) and thermogenic and pro-inflammatory genes
(G) in primary VISC Wt1lox/lox;cre+ adipocytes. N=6 per group.
Data are presented as mean ± SEM. *, p< 0.05, #, p=0.058. See also Figure S7.
High-fat feeding also resulted in significantly decreased expression of typical thermogenic genes in SubQ adipose tissue of Adipo-PRDM16 KOs (Figure 7D), suggesting a coordinated balance between the molecular phenotype of VISC and SubQ adipose tissue. We considered whether PRDM16 might normally induce thermogenesis and repress inflammation, while a VISC-selective transcription factor might induce inflammation and repress thermogenesis. The three transcription factors identified in our VISC-enriched profile (shown above) were immediate candidates for conferring these depot-specific properties. We focused on Wt1 because it showed cell autonomous induction in the absence of PRDM16 (Figure S7D) and is strikingly enriched in VISC adipose tissues (Figure 7E).
We obtained Wt1lox/lox mice (Gao et al., 2006) and crossed them to Adiponectin-cre mice. We isolated adipose tissue SVF from Wt1lox/lox;cre+ (KO) and Wt1lox/lox;cre- (control) VISC fat and differentiated them in vitro. We detected a significant reduction in the native Wt1 transcript and a corresponding increase in a deleted transcript in KO cells compared to controls (Figure 7F). Both control and KO cells differentiated equally, with no morphological differences. There were small but significant differences in markers of adipocyte differentiation. Importantly, deletion of Wt1 resulted in a striking induction of thermogenic genes (5.6-fold for Cidea, 6.1-fold for Prdm16, and 24.7-fold for Ucp1) and repression of several pro-inflammatory genes (46% for Saa3 and 30% for Agt) compared to controls (Figure 7G). These data strongly suggest that reciprocal regulation of PRDM16 and Wt1 is at least partly responsible for determining whether adipose expresses a SubQ or VISC gene program. Interestingly, in a differentiation time course in primary adipocytes from Adipo-PRDM16 KO mice, successive reduction in Prdm16 mRNA levels is closely associated with progressive increases in Wt1 mRNA levels (Figure S7E).
DISCUSSION
We have described here a new mouse model with specific ablation of beige adipocyte function. Adipo-PRDM16 KO mice have significant reductions in thermogenic gene expression and O2 consumption of white adipose tissue, both in the basal state and following stimulation with cold and the β3-adrenergic agonist CL. On the other hand, both thermogenic gene expression and O2 consumption in classical BAT appears unaffected by PRDM16 deletion. Importantly, the direct measure of tissue-specific glucose uptake during hyperinsulinemic-euglyemic clamps indicated that there is essentially no functional deficiency in classical BAT. In fact, glucose uptake was elevated in KO BAT, suggesting this tissue might be compensating for the absence of functional beige adipose cells. This may represent a mirror image of the recent report of hyperactive beige adipose tissue in the setting of dysfunctional classical brown fat (Schulz et al., 2013). While deletion of PGC1α in adipocytes also leads to impaired thermogenic function in SubQ adipose tissue and metabolic defects (Kleiner et al., 2012; Pardo et al., 2011), the magnitude of the defect appears far greater in the Adipo-PRDM16 KO mice. Pgc1a expression is reduced in our KOs, consistent with PRDM16 being upstream of PGC1a (Seale et al., 2007).
The physiological relevance of beige fat has been questioned, since expression of Ucp1 mRNA and protein is at least an order of magnitude lower in these cells than in classical BAT (Nedergaard and Cannon, 2013). While mice with more beige fat cells have shown a protection from metabolic diseases (Seale et al., 2011; Vegiopoulos et al., 2010), the functional role of this cell type when present in normal amounts has not been clear. As shown here, animals deficient in competent beige adipocytes develop late-onset obesity on HFD. This is associated with increased fat mass, including a striking increase in SubQ adipose stores. While we were unable to detect differences in whole body O2 consumption in the basal state, KO animals showed significantly reduced energy expenditure following treatment with a β3-adrenergic agonist.
Hyperinsulinemic-euglycemic clamp studies on beige cell-deficient animals before the development of obesity showed significant insulin resistance, specifically affecting white adipose tissues and liver. Along with this, mutant animals developed hyperinsulinemia and hepatic steatosis. This model of insulin resistance is unusual in that glucose uptake in skeletal muscle seems unaffected. Beige adipocytes may have as yet undescribed actions on other tissues, with recent studies suggesting they regulate bone metabolism (Rahman et al., 2013).
This mouse model of beige fat deficiency differs in some interesting ways from mouse models with specific loss of classical BAT function. Mice with defective BAT develop obesity and insulin resistance (Feldmann et al., 2009; Hamann et al., 1996; Tseng et al., 2008). However, such animals tend to have defects in maintaining core temperature. When our KO mice were placed at 4°C, they showed no alteration in body temperature.
In the absence of PRDM16, mice developed marked enlargement of the SubQ adipose tissue. Interestingly, domestic pigs, which have abundant SubQ adipose tissue, lack functional UCP1 (Trayhurn et al., 1989). Similarly, Adipo-PRDM16 KO mice have virtually no UCP1 in their SubQ adipocytes. Histological examination of the SubQ fat from these mice demonstrated the presence of larger, unilocular cells, which become equivalent in size to VISC fat cells. KO mice also showed a virtual absence of small, multilocular UCP1+ adipocytes. PRDM16 deficient animals not only lose the thermogenic program characteristic of beige adipocytes, but additionally, the SubQ adipose tissue acquires some of the deleterious characteristics of VISC fat, including the accumulation of macrophages. It is not yet clear whether acquisition of this VISC phenotype is causally related to loss of the thermogenic function of beige adipocytes or reflects a separate function of PRDM16 in adipocytes. The physiological significance of this change in depot phenotype, however, is made clear by our fat transplantation studies. Animals that received PRDM16-deficient SubQ pads had significantly worse glucose tolerance than recipients of control fat pads, recapitulating experimental animal models with increased VISC fat (Masuzaki et al., 2001).
Since reduced thermogenesis in SubQ adipose tissue of Adipo-PRDM16 KO mice is coupled to increased inflammatory gene expression and macrophage accumulation, it is not possible to disentangle the relative contribution of each of these alterations to the overall phenotype. Our data suggests that PRDM16 specifically affects the function of beige fat cells in the SubQ depots and that this is the proximal defect, occurring prior to acquisition of VISC properties upon high-fat feeding. Additionally, a recent paper showed that PRDM16 is only detected in multilocular and paucilocular cells within the omental fat of humans with pheochromocytoma (Frontini et al., 2013), consistent with its enrichment and selective expression in beige adipocytes. However, we cannot exclude possible effects from a very small amount of PRDM16 in true white adipocytes.
A comprehensive molecular analysis showed significant expression of a VISC gene signature in the SubQ fat of PRDM16 KO mice. This signature was developed based on objective criteria, using multiple depots of each type, and employing fat cell purification to avoid confounding by other cell types. These data clearly suggest that deletion of PRDM16 and ablation of beige adipocytes not only affects thermogenesis, but also results in substantial ‘visceralization’ of the SubQ adipose tissue. This analysis establishes with statistical significance (p<0.0004) that deletion of PRDM16 results in acquisition of at least some aspects of the molecular phenotype of VISC adipose tissue. Of note, several of these VISC markers expressed in KO SubQ pads are of interest in metabolic diseases: Agt (Kalupahana et al., 2012), SAA3 (Kwon et al., 2012), Alox15 (Nunemaker et al., 2008; Sears et al., 2009), Opgn (Skopkova et al., 2007), and Raldh2 (Frey and Vogel, 2011).
Three transcription factors (WT1, TCF21, BNC1) were among the visceral signature genes induced in Adipo-PRDM16 KO SubQ adipose tissue. WT1 and TCF21 have known roles in urogenital development, and KO models of these genes have similar phenotypes (Kreidberg et al., 1993; Quaggin et al., 1999), suggesting that WT1 and TCF21 may function in the same pathway. WT1 is also a known tumor suppressor, which is mutated in Wilms’ tumor (Haber et al., 1990). A role for WT1 in adipose biology has not previously been reported. Deletion of Wt1 in primary VISC adipocytes resulted in downregulation of genes in the VISC signature and upregulation of thermogenic genes characteristic of SubQ/beige adipocytes. These data suggest an epistatic relationship between a thermogenic program regulated by PRDM16 and a VISC-specific program regulated by WT1. This appears analogous to the regulation of thermogenesis vs. lipid storage by PRDM16 and TLE3 (Villanueva et al., 2013).
Thus, it is now clear that beige fat cells make an important contribution to whole body physiology of mice. In the absence of functional beige fat, mice are prone to obesity, insulin resistance, and hepatic steatosis when challenged with HFD. In light of the presence of beige fat cells in normal adult humans (Lidell et al., 2013; Sharp et al., 2012; Wu et al., 2012), these cells are an attractive target for the treatment of obesity and type 2 diabetes. This influence of beige fat on liver metabolism is particularly interesting since there has been much more attention recently to the frequency and importance of hepatic steatosis as an important comorbidity in the Metabolic Syndrome (Angulo, 2002). Identification of pharmacological activators of PRDM16 targeted to beige adipocytes could hold promise as a new class of therapeutics.
EXPERIMENTAL PROCEDURES
Animals
Animal experiments were performed according to procedures approved by the Dana-Farber Cancer Institute and Yale University School of Medicine IACUC. The generation of Adipo-PRDM16 KO mice and the other mouse strains used is described in the extended experimental procedures.
Molecular Studies
qPCR and Western blotting were done according to standard methods. Antibodies used were PRDM16 (sheep anti-PRDM16, R&D), PPARγ (Cell Signaling), TBP (Santa Cruz), AKT and P-AKT (Cell Signaling), and Actin (Cell Signaling). Microarray hybridization and scanning were performed by the Dana-Farber Cancer Institute microarray core facility using Affymetrix Mouse Genome 430A 2.0 Gene Chip arrays. The GEO accession number for microarray data reported in this paper is GSE53307.
Histological Analysis
Tissues were fixed in 4% paraformaldehyde. Paraffin embedding and sectioning were done by the Dana-Faber/Harvard Cancer Center Research Pathology core facility. Adipocyte size was calculated as described (Vitali et al., 2012). The mean area of 200 random adipocytes (100 per section) from each animal was calculated using the Nikon LUCIA image program (version 4.61; Laboratory Imaging).
Immunohistochemistry
Immunohistochemistry was performed as previously described (Giordano et al., 2013) and as in the extended experimental procedures
Respiration
Tissue respiration was performed using a Clark electrode (Strathkelvin Instruments). Freshly isolated tissues were isolated from untreated mice or after 5 daily injections of 1.0 mg/kg CL (Sigma). Tissues were minced and placed in respiration buffer. For each adipose depot, readings were taken with three separate pieces of tissue of equivalent size. O2 consumption was normalized to tissue weight. For assays on cultured cells, fully differentiated adipocytes were trypsinized and a single cell suspension was placed in respiration buffer. Total respiration was measured and then oliogmycin was added to measure uncoupled respiration.
Flow Cytometry
Epididymal VISC and inguinal SubQ adipose tissue and spleen were excised and digested for 20 min with collagenase type II (Sigma). Cell suspensions were filtered through a 40-micron sieve, and the SVF was collected after centrifugation at 450g for 10 min. For T-cell analysis, cells were stained with anti-CD45 (clone 30-F11), -CD3 (145-2C11), -CD4 (GK1.5), -CD8 (5H10) and -CD25 (PC61) (BioLegend); and were fixed, permeabilized and intracellularly stained for Foxp3 (FJK-16s) and GATA3 (TWAJ) (eBiosciences). For myeloid cell analysis, cells were stained with anti-CD45, -CD3 (145-2C11), -CD11b (M1/70), -CD11c (N418), F4/80 (CI:A3-1), and anti-Ly6c (HK1.4) (BioLegend). B cells were stained with anti-CD45, and anti-CD19 (6D5) (BioLegend). Cells were analyzed using an LSRII instrument (BD Bioscience) and FlowJo software.
Metabolic Phenotyping
Metabolic phenotyping and hyperinsulinemic-euglycemic clamps were performed as described (Jurczak et al., 2012; Ye et al., 2012) and as in the extended experimental procedures. All experiments were done with male mice, with the exception of the CL gene expression and tissue respiration studies. Energy expenditure was analyzed using a Comprehensive Lab Animal Monitoring System (Columbus Instruments). Fat and lean mass was measured by MRI. Cold exposure and thermoneutrality experiments were done at either 4°C or 30°C. Total adiponectin levels were measured by ELISA (Millipore).
Fat Transplant Studies
Inguinal SubQ adipose tissue was removed from 2–3 week old Adipo-PRDM16 KO, littermate control mice, or aP2-PRDM16 transgenic mice (donors) and transplanted into the SubQ space of C57Bl6 mice (recipients) that had been on HFD for 6 weeks. Transplants were performed as described (Gunawardana and Piston, 2012). Glucose tolerance tests were done 6 weeks post-transplant. Animals were fasted for 4 hours and then received intraperitoneal glucose (1.5 mg/kg). Insulin tolerance tests were done 8 weeks post-transplant. Animals were fasted for 4 hours and then received intraperitoneal Humulin R insulin (1.0 U/kg, Lilly).
Cell Culture
For primary adipocytes, SVF from inguinal or epididymal fat from 5–6 week old mice was prepared and differentiated for 6–8 days as described (Kajimura et al., 2009). Where indicated, cells were treated with isoproterenol 10uM for 6hr (Sigma) or FGF21 100 ng/ml overnight (R&D).
Statistics
The student’s t test was used for single comparisons. Two-way ANOVA with repeated measures was used for the GTT studies. Chi-squared was used to compare the VISC-selective gene set to a randomly selected set of genes. Unless specified, * indicates p< 0.05.
Supplementary Material
Adipocyte-specific deletion of PRDM16 inhibits beige adipocyte function
Mutant animals develop dietary obesity, insulin resistance, and hepatic steatosis
Mutant subcutaneous fat acquires cellular and molecular properties of visceral fat
PRDM16 may determine key differences between subcutaneous and visceral fat
ACKNOWLEDGEMENTS
We thank Dr. Evan Rosen (Beth Israel Deaconess Medical Center) for providing Adiponectin-Cre mice, Dr. Vicki Huff (MD Anderson) for providing Wt1lox/lox mice, and the Nikon Imaging Center at Harvard Medical School for use of microscopy equipment. We thank Dr. Vamsi Mootha (Massachusetts General Hospital) for guidance on data analysis and Drs. Rana Gupta, Pere Puigserver, Evan Rosen, and members of the Spiegelman lab for helpful discussions. P.C. was supported by the American Heart Association grant 11FTF7510004. D.P.K. was supported by a National Science Foundation Graduate Research Fellowship. This work was supported by NIH grants DK040936, DK045735, DK059635 (G.I.S.); NIH grant DK092541 (D.M.); and by NIH grant DK031405 and the JPB foundation (B.M.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Angulo P. Nonalcoholic fatty liver disease. N. Engl. J. Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum. Mol. Genet. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AA, Kozak LP. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes. 2010;59:323–329. doi: 10.2337/db09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos-Flier E, et al. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey SK, Vogel S. Vitamin A metabolism and adipose tissue biology. Nutrients. 2011;3:27–39. doi: 10.3390/nu3010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontini A, Vitali A, Perugini J, Murano I, Romiti C, Ricquier D, Guerrieri M, Cinti S. White-to-brown transdifferentiation of omental adipocytes in patients affected by pheochromocytoma. Biochim. Biophys. Acta. 2013;1831:950–959. doi: 10.1016/j.bbalip.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Gabriely I, Ma XH, Yang XM, Atzmon G, Rajala MW, Berg AH, Scherer P, Rossetti L, Barzilai N. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: an adipokine-mediated process? Diabetes. 2002;51:2951–2958. doi: 10.2337/diabetes.51.10.2951. [DOI] [PubMed] [Google Scholar]
- Gao F, Maiti S, Alam N, Zhang Z, Deng JM, Behringer RR, Lecureuil C, Guillou F, Huff V. The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proc. Natl. Acad. Sci. USA. 2006;103:11987–11992. doi: 10.1073/pnas.0600994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Giordano A, Murano I, Mondini E, Perugini J, Smorlesi A, Severi I, Barazzoni R, Scherer PE, Cinti S. Obese adipocytes show ultrastructural features of stressed cells and die of pyroptosis. J. Lipid Res. 2013;54:2423–2436. doi: 10.1194/jlr.M038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardana SC, Piston DW. Reversal of type 1 diabetes in mice by brown adipose tissue transplant. Diabetes. 2012;61:674–682. doi: 10.2337/db11-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Buckler AJ, Glaser T, Call KM, Pelletier J, Sohn RL, Douglass EC, Housman DE. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms' tumor. Cell. 1990;61:1257–1269. doi: 10.1016/0092-8674(90)90690-g. [DOI] [PubMed] [Google Scholar]
- Hamann A, Flier JS, Lowell BB. Decreased brown fat markedly enhances susceptibility to diet-induced obesity, diabetes, and hyperlipidemia. Endocrinol. 1996;137:21–29. doi: 10.1210/endo.137.1.8536614. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homoe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Jurczak MJ, Lee AH, Jornayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, Kahn M, Samuel VT, Glimcher LH, Shulman GI. Dissociation of inositol-requiring enzyme (IRE1alpha)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J. Biol. Chem. 2012;287:2558–2567. doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalupahana NS, Massiera F, Quignard-Boulange A, Ailhaud G, Voy BH, Wasserman DH, Moustaid-Moussa N. Overproduction of angiotensinogen from adipose tissue induces adipose inflammation, glucose intolerance, and insulin resistance. Obesity. 2012;20:48–56. doi: 10.1038/oby.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer FW, Vernochet C, O'Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, Stulnig TM, Cohen DE, Kahn CR, et al. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat. Med. 2012;18:918–925. doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner S, Mepani RJ, Laznik D, Ye L, Jurczak MJ, Jornayvaz FR, Estall JL, Chatterjee Bhowmick D, Shulman GI, Spiegelman BM. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proc. Natl. Acad. Sci. USA. 2012;109:9635–9640. doi: 10.1073/pnas.1207287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Kusminski CM, Holland WL, Sun K, Park J, Spurgin SB, Lin Y, Askew GR, Simcox JA, McClain DA, Li C, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med. 2012;18:1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon EY, Shin SK, Cho YY, Jung UJ, Kim E, Park T, Park JH, Yun JW, McGregor RA, Park YB, et al. Time-course microarrays reveal early activation of the immune transcriptome and adipokine dysregulation leads to fibrosis in visceral adipose depots during diet-induced obesity. BMC Genom. 2012;13:450. doi: 10.1186/1471-2164-13-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Leinhard OD, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. Evidence for two types of brown adipose tissue in humans. Nat. Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Manolopoulos KN, Karpe F, Frayn KN. Gluteofemoral body fat as a determinant of metabolic health. Int. J. Obes. 2010;34:949–959. doi: 10.1038/ijo.2009.286. [DOI] [PubMed] [Google Scholar]
- Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, Flier JS. A transgenic model of visceral obesity and the metabolic syndrome. Science. 2001;294:2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. UCP1 mRNA does not produce heat. Biochim. Biophys. Acta. 2013;1831:943–949. doi: 10.1016/j.bbalip.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Nunemaker CS, Chen M, Pei H, Kimble SD, Keller SR, Carter JD, Yang Z, Smith KM, Wu R, Bevard MH, et al. 12-Lipoxygenase-knockout mice are resistant to inflammatory effects of obesity induced by Western diet. Amer. J. Physiol. Endocrinol. Metab. 2008;295:E1065–E1075. doi: 10.1152/ajpendo.90371.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo R, Enguix N, Lasheras J, Feliu JE, Kralli A, Villena JA. Rosiglitazone-induced mitochondrial biogenesis in white adipose tissue is independent of peroxisome proliferator-activated receptor gamma coactivator-1alpha. PloS One. 2011;6:e26989. doi: 10.1371/journal.pone.0026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K, van der Schouw YT, Spencer E, Moons KG, Tjonneland A, et al. General and abdominal adiposity and risk of death in Europe. N. Engl. J. Med. 2008;359:2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- Rahman S, Lu Y, Czernik PJ, Rosen CJ, Enerback S, Lecka-Czernik B. Inducible Brown Adipose Tissue, or Beige Fat, Is Anabolic for the Skeleton. Endocrinol. 2013;154:2687–2701. doi: 10.1210/en.2012-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, Cypess AM, Mishina Y, Gussoni E, Tseng YH. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears DD, Miles PD, Chapman J, Ofrecio JM, Almazan F, Thapar D, Miller YI. 12/15-lipoxygenase is required for the early onset of high fat diet-induced adipose tissue inflammation and insulin resistance in mice. PloS One. 2009;4:e7250. doi: 10.1371/journal.pone.0007250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PloS One. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopkova M, Penesova A, Sell H, Radikova Z, Vlcek M, Imrich R, Koska J, Ukropec J, Eckel J, Klimes I, et al. Protein array reveals differentially expressed proteins in subcutaneous adipose tissue in obesity. Obesity. 2007;15:2396–2406. doi: 10.1038/oby.2007.285. [DOI] [PubMed] [Google Scholar]
- Tran TT, Yamamoto Y, Gesta S, Kahn CR. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trayhurn P, Temple NJ, Van Aerde J. Evidence from immunoblotting studies on uncoupling protein that brown adipose tissue is not present in the domestic pig. Can. J. Physiol. Pharmacol. 1989;67:1480–1485. doi: 10.1139/y89-239. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vague J. La differenciation sexuelle: facteur determinant des formes: de l’obesite. La Presse Med. 1947;55:339–340. [PubMed] [Google Scholar]
- Vague J. The degree of masculine differentiation of obesities: a factor determining predisposition to diabetes, atherosclerosis, gout, and uric calculous disease. Am. J. Clin. Nutr. 1956;4:20–34. doi: 10.1093/ajcn/4.1.20. [DOI] [PubMed] [Google Scholar]
- Vegiopoulos A, Muller-Decker K, Strzoda D, Schmitt I, Chichelnitskiy E, Ostertag A, Berriel Diaz M, Rozman J, Hrabe de Angelis M, Nusing RM, et al. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science. 2010;328:1158–1161. doi: 10.1126/science.1186034. [DOI] [PubMed] [Google Scholar]
- Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, Hu Y, Peng X, Xu F, Saez E, et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali A, Murano I, Zingaretti MC, Frontini A, Ricquier D, Cinti S. The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. J. Lipid Res. 2012;53:619–629. doi: 10.1194/jlr.M018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Kleiner S, Wu J, Sah R, Gupta RK, Banks AS, Cohen P, Khandekar MJ, Bostrom P, Mepani RJ, et al. TRPV4 is a regulator of adipose oxidative metabolism, inflammation, and energy homeostasis. Cell. 2012;151:96–110. doi: 10.1016/j.cell.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.