Abstract
RNA interference screening identified XPO1 (exportin 1) among the 55 most vulnerable targets in multiple myeloma (MM). XPO1 encodes CRM1, a nuclear export protein. XPO1 expression increases with MM disease progression. Patients with MM have a higher expression of XPO1 compared with normal plasma cells (P<0.04) and to patients with monoclonal gammopathy of undetermined significance/smoldering MM (P< 0.0001). The highest XPO1 level was found in human MM cell lines (HMCLs). A selective inhibitor of nuclear export compound KPT-276 specifically and irreversibly inhibits the nuclear export function of XPO1. The viability of 12 HMCLs treated with KTP-276 was significantly reduced. KPT-276 also actively induced apoptosis in primary MM patient samples. In gene expression analyses, two genes of probable relevance were dysregulated by KPT-276: cell division cycle 25 homolog A (CDC25A) and bromodomain-containing protein 4 (BRD4), both of which are associated with c-MYC pathway. Western blotting and reverse transcription-PCR confirm that c-MYC, CDC25A and BRD4 are all downregulated after treatment with KPT-276. KPT-276 reduced monoclonal spikes in the Vk*MYC transgenic MM mouse model, and inhibited tumor growth in a xenograft MM mouse model. A phase I clinical trial of an analog of KPT-276 is ongoing in hematological malignancies including MM.
Keywords: Myeloma, CRM1, XPO1, KPT-276, CRM1 Inhibitors, exportin
INTRODUCTION
Multiple myeloma (MM) is a bone marrow-based multifocal plasma cell neoplasm causing severe organ damage including bone lesions, anemia and renal failure. It is encouraging that the development of novel drugs in recent years has led to major improvements in survival,1 especially for younger patients.2 Nevertheless, MM patients relapse after initial therapy and gain drug resistance. Therefore, there is an ongoing need for drugs that target novel pathways to move MM from a chronic to curable disease.
We have performed a genome-scale RNA interference (RNAi) lethality study in MM cells using 13 984 small interfering RNA to functionally identify critical molecular vulnerabilities without regard to preconceived mechanistic notions.3 Of the 55 most lethal genes identified, functional testing in HMCLs and non-MM cell lines indicated that proteasome subunits (A1, A3, A4, A6, C3, C4 and C5), MCL1, RRM1, USP8, TNK2, CKAP5, IK, KIF11, WBSCR22 and XPO1 are selectively vulnerable in myeloma.3 Proteasome inhibitors have been developed and are commonly used in the clinic,4 however, drugs that target the other potentially interesting therapeutic targets are not yet available.
Exportin 1 (XPO1) encodes CRM1 (chromosome maintenance region 1), a nuclear export protein that transports over 200 proteins with a canonical nuclear export sequence through the nuclear pore to the cytoplasm.5 CRM1 is the sole exporter of many tumor-suppressor proteins (TSPs),6 and functions as a proto-oncogene by transporting these tumor suppressors from the nucleus, where they are active, to the cytoplasm, where their activity is abrogated.7 By inhibiting CRM1 function, TSPs are retained in the nucleus and remain functional, thus potentially subverting loss of function.7 CRM1 expression increases in tumor versus normal cells in osteosarcoma,8 pancreatic9 and ovarian cancers,10 gliomas,11 mantle cell lymphoma12 and MM.13 CRM1 over expression is associated with poor prognosis and a decrease in overall survival. In this light, inhibitors have been synthesized to inhibit the CRM1 nuclear export pathway that is manipulated by cancer cells to promote proliferation and survival. The first CRM1 inhibitor, leptomycin B, showed potent CRM1 inhibition at nanomolar concentrations,6 however, it had no partial response and was toxic in a phase I clinical trial.14 Leptomycin B derivatives have been synthesized, and these compounds inhibit CRM1 at low concentration, without the toxicity observed with leptomycin B.6 CBS9106, a novel CRM1 inhibitor, decreased MM cell growth, induced cell cycle arrest at G1 and inhibited tumor growth in a xenograft model.15 Ratajone C, another novel compound, sensitized MM cells to topoisomerase II inhibitors such as doxorubicin and VP16 in vitro, however, it has yet to be tested in vivo.16 Although CRM1 inhibitors have had promising pre-clinical results in MM, these inhibitors are not currently in clinical trials.
A new class of small-molecule inhibitors, selective inhibitors of nuclear export (SINEs), has been designed to target CRM1. SINE compounds are specific, irreversible covalent inhibitors of CRM1. SINE compounds have shown anti-tumor activity in various malignancies,7,12,13,17–21 showing the importance of the CRM1 nuclear export function in promoting cancer cell survival. In hematological malignancies, SINE compounds increase apoptosis,12,13,17–19 decrease proliferation,7,12,18,19 cause G1 cell cycle arrest in vitro,12,17–19 inhibit tumor growth12,13,17,18 and increase survival in xenograft models.7,13,17–19 KPT-276, a SINE compound, is a small-molecule inhibitor of XPO1 and its gene product CRM1. KPT-276 has good bioavailability and pharmacokinetics. Because we have shown that XPO1 is a vulnerable target in MM, we tested the activity of KPT-276 against HMCL, patient samples and two mouse models of myeloma. Our results show that KPT-276 is an active anti-MM drug and reduces MM cell viability, causes cell cycle arrest, increases apoptosis in CD138+ cells from MM patients and inhibits disease progression in in vitro and in vivo models. Furthermore, pharmacodynamic analysis identifies regulators of c-MYC as potential downstream effect mediators.
MATERIALS AND METHODS
Cell lines and primary samples
Twelve human myeloma cell lines (KMS11, KMS12PE, KMS18, OPM1, OPM2, H929, JJN3, U266, RPMI-8226, SKMM2, OCI-MY5 and MM1.S) were maintained in RPMI supplemented with 5% fetal bovine serum, 1 mM glutamate and 1% penicillin/streptomycin. Primary cells from myeloma patients were obtained with approval from the Mayo Clinic Institutional Review Board and in accordance with the Declaration of Helsinki. Primary cells were also maintained in RPMI supplemented with 10% fetal bovine serum, 1 mM glutamate and 1% penicillin/streptomycin.
MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) assay
Cell lines were plated in 96-well microplates, at a final concentration of 2 × 105 cells/ml in 50μl culture medium. Dimethyl sulfoxide (DMSO) vehicle and KPT-276 was diluted in culture medium without antibiotics, and 50 μl of drug solution or vehicle was added to each well. MM cells were treated with concentrations ranging from 15.625 nM to 1000 nM. Cells were also treated with KPT-276 in combination with bortezomib, dexamethasone, melphalan (data not shown) or JQ1 to investigate synergy. MTT (Sigma-Aldrich, St Louis, MO, USA) was added to cells after 72 h of drug treatment, at 10 μl/well, and incubated at 37 °C. After 4 h, cells were lysed and absorbance was read at 490 nm using a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA, USA). IC50 and combination index values were calculated using CalcuSyn software (Biosoft, Cambridge, UK).
Cell cycle analysis
MM1.S cells were incubated with DMSO vehicle or various doses of KPT-276 for 24 h. Cells were washed with PBS and re-suspended in propidium iodide staining solution (1% sodium citrate, 50 mg/ml propidium iodide, 0.01% NP40 and 0.01 mg/ml RNaseA) for 15 min. Stained cells were measured on a BD Biosciences Fortessa cell analyzer (BD Biosciences, San Jose, CA, USA), and the final results were analyzed using FloJo software (Tree Star Inc., Ashland, OR, USA).
Apoptosis assay
MM bone marrow samples were obtained from MM patients with Mayo Clinic Institutional Review Board approval. Cells were treated with ACK lysis buffer to eliminate red blood cells. Unsorted whole bone marrow samples were cultured in RPMI supplemented with 10% fetal bovine serum and 1 mM glutamate. To obtain purified plasma cell samples, CD138 + populations were acquired with anti-CD138 antibodies on a StemCell Technologies Robocept (STEMCELL Technologies Inc., Vancouver, BC, Canada) cell sorter. CD138 +-sorted samples were also cultured in RPMI with 10% serum and 1 mM glutamate. Primary MM cells treated with DMSO vehicle or KPT-276 for 48 h, then were harvested and washed with PBS and re-suspended in Annexin V staining buffer. Cells were stained for 15 min with AnnexinV-Alexa Fluor 488 (Invitrogen) and CD138-PE (BD Biosciences) and analyzed on a Beckman Coulter CyAn flow cytometer. Flow cytometry data were further analyzed using FloJo software.
Immunoblotting
CD138+ cell pellets were obtained from the Mayo Clinic hematological malignancies biorepository. HMCL and CD138+ cells were lysed using NP40 lysis buffer (Cell Signaling Technologies, Danvers, MA, USA). Protein concentrations were determined using BCA Protein Assay Kit (Thermo Scientific Pierce, Rockford, IL, USA) following manufacturer’s instructions. Lysates were prepared by adding 3 × loading buffer and DTT, and run on 10% SDS-polyacrylamide gel electrophoresis. Proteins were transferred to Immobilon PVDF membrane (EMD Millipore, Billerica, MA, USA) and probed with primary antibodies against CRM1 (Sigma Aldrich), c-Myc (Santa Cruz Biotechnology, Dallas, TX, USA), β-actin (Cell Signaling Technology), Cdc25A (EMD Millipore) and bromodomain-containing protein 4 (BRD4; Santa Cruz Biotechnology). Blots were imaged using Western Lightening Plus-ECL chemiluminescence solution (PerkinElmer, Waltham, MA, USA), and images were captured with the Kodak X-OMAT 2000A Processor, using Kodak BioMax MR Film (Eastman Kodak Company, Rochester, NY, USA).
Reverse transcription-PCR
MM1.S and OCI-MY5 HMCLs were treated with an IC80 dose of KPT-276 and harvested at 6, 12, 24 and 48 h. RNA was extracted with the RNeasy Plus Mini Kit (Qiagen, Germantown, MD, USA) following the manufacturer’s protocol. cDNA was produced using the QuantiTect Reverse Transcription Kit (Qiagen). Reverse transcription-PCR was carried out using probes for BRD4 (Applied Biosystems, Grand Island, NY, USA), with β-actin as a control (Applied Biosystems) on the 7900HT Fast Real-Time PCR System (Applied Biosystems). Assay was done in triplicate, and comparative CT was used to analyze the results.
Gene expression profiling
Public data sets were used for analyzing the XPO1 expression level in normal plasma cells and different stages of myeloma progression (GEO series GSE6477) and for stratifying patients in TC classes (MMRF portal, http://www.broadinstitute.org/mmgp/home).
Two of the most sensitive (MM.1S and OCI-MY5) and resistant HMCLs (RPMI-8226 and KMS12PE) were treated with an IC80 dose of KPT-276 or vehicle for 6 h, in culture medium without antibiotics. Cells were harvested, and RNA was extracted with the RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s protocol. Gene expression profiling was done by the Mayo Clinic genomics core, using Hg_U133A_2 microarray array (Affymetrix, Santa Clara, CA, USA). Intensity values were extracted using the MAS5.0 algorithm with default parameters, and gene expression profiling was further analyzed with GeneSpring GX software (Agilent Technologies, Santa Clara, CA, USA). The raw data were filtered on flags (present, marginal or absent expression), including all probes that show present or marginal expression in at least one sample, vehicle or drug-treated. Then, we filtered on differentially expressed genes comparing the vehicle to the drug-treated cell lines (> 2-fold change). Any genes that changed in both the resistant and sensitive cell lines were excluded from further analyses, only focusing on changes observed in the sensitive but not in the resistant cell lines.
Xenograft mouse model
Athymic NCr-nu/nu mice were injected with MM1.S cells suspended in Matrigel (BD Biosciences) in the posterior flank. Mice were observed and drug treatment was initiated once tumors were palpable and able to be measured. KPT-276 was dissolved in 0.6% (w/v) Pluronic F-68 (Spectrum Chemical Manufacturing Corporation, New Brunswick, NJ, USA) and 0.6% (w/v) PVP K-29/32 vehicle solution. Drug was administered by oral gavage, 3 days/week for 12 days. Treatment lapsed because mice lost weight. After 10 days mice regained weight and treatment resumed, following the same dosing regimen. Tumors were measured with a Calimax caliper (Swiss Precision, Garden Grove, CA, USA) on days drug was administered. Tumor measurements were recorded and tumor volume calculated.
Vk*MYC mouse model
For in-vivo administration, KPT-276 was formulated by dissolving it in a 0.6% (w/v) Pluronic F-68 and 0.6% (w/v) PVP K-29/32 vehicle solution. Vk*MYC mice were dosed by oral gavage at 150 mg/kg, 3 days/week for 3 weeks. Tail vein bleeds were conducted at baseline and after 2, 3 and 4 weeks of drug treatment for serum collection. Serum protein electro-phoresis and densitometric analysis were performed to quantify reduction in the M-spike levels as previously indicated.22 The anti-myeloma activity of KPT-276 was plotted alongside the activity of other myeloma treatments, graphed as a percent reduction of M-spike levels at day 0.
RESULTS
RNAi and XPO1 specificity
We have previously published that XPO1/CRM1 was identified as one of the 55 most vulnerable targeting in MM, using high-throughput RNAi screening on 7622 druggable genes.3 XPO1 knockdown proved lethal in three representative HMCLs (KMS11, RPMI-8226 and JJN3), but had no effect on human embryonic kidney (293) cells, or lung cancer (A549) cells, showing that XPO1 expression is critical for MM cell survival.3
XPO1 expression
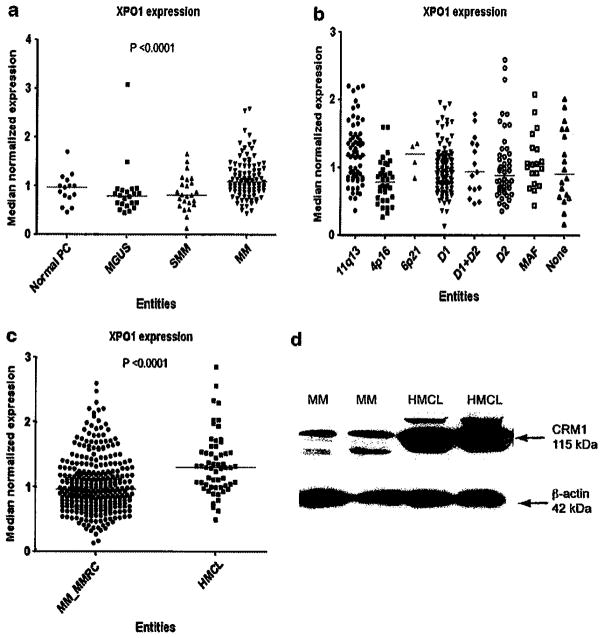
Normal plasma cells, monoclonal gammopathy of undetermined significance and smoldering MM share the same levels of XPO1 expression, whereas significantly higher levels can be found in active disease (Figure 1a). Next, we divided the MM cases in molecular subgroups using the TC classification.23 XPO1 expression showed the highest level in patients of subgroups 6p21 and 11q13, whereas the lowest expression level was found in 4q16 subgroup (Figure 1b). Therefore, TC class can provide a useful marker to determine if KPT-276 will improve disease outcome. It follows that a patient in the 6p21 subgroup will have a better outcome after treatment with KPT-276 than a patient in the 4q16 subgroup. We also observed a XPO1 gene and protein expression is higher in HMCL than in MM patients (Figures 1c and d). This confirms that an increase in XPO1 expression leads to a significant increase in CRM1 expression.
Figure 1.
XPO1 expression in myeloma data sets. XPO1 expression increases with disease progression, as shown in gene expression data sets from MM patients. XPO1 expression level is highest in myeloma as compared with normal plasma cells (PCs), monoclonal gammopathy of undetermined significance (MGUS) and smoldering myeloma (SMM) (a). XPO1 expression levels are also shown in relation to common mutations in MM. XPO1 expression is highest in 11q13 and 6p21 TC groups (b). XPO1 expression level is significantly higher in HMCL than in primary patient samples, as shown by gene (c) and protein level (d).
KPT-276 reduces viability of HMCLs and primary patient cells in vitro
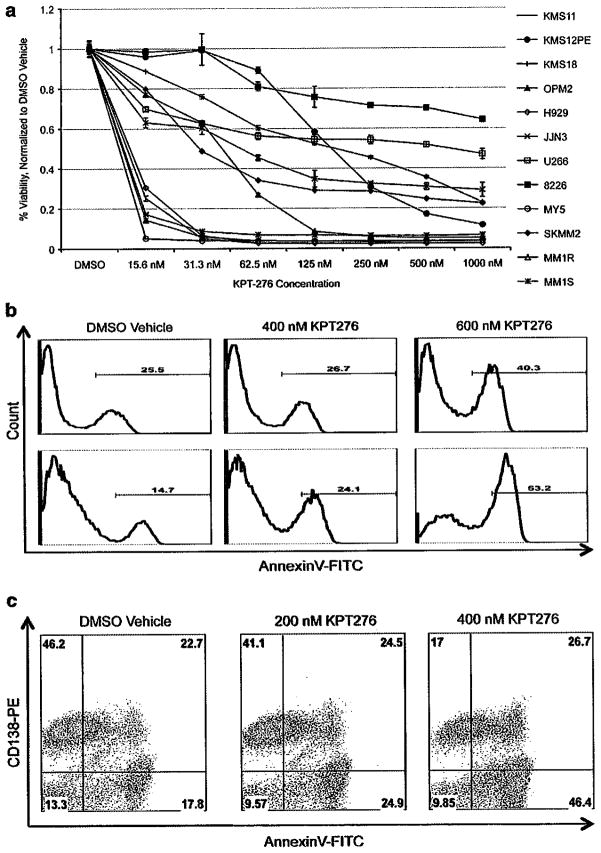
In order to test whether the XPO-1/CRM1 inhibitor KPT-276 had anti-MM activity, MTT assays were performed on a panel of 12 HMCLs. After 72 h of drug treatment, the viability of all HMCL decreased. Eleven of the twelve HMCLs treated achieved at least 50% reduction in viability at drug concentrations ≤ 1 uM, with a median IC50 value of approximately 160 nM (Figure 2a). To investigate whether or not KPT-276 had synergy with commonly used therapeutics, we tested KPT-276 in parallel with bortezomib, melphalan and dexamethasone. No synergy was seen for any of the drug combinations (data not shown).
Figure 2.
KPT-276 has anti-myeloma activity in vitro. Twelve HMCL were treated with vehicle or KPT-276 in nanomolar concentrations, and after 72 h cell viability was reduced in most cell lines by at least 50% (a). Bone marrow cells obtained from myeloma patients were also treated with KPT-276 for 48 h, and drug-treated cells stained positive for AnnexinV, as compared with vehicle control. CD138+ plasma cells stained positive for AnnexinV in both sorted CD138+ (b) and unsorted whole bone marrow (c). FITC, fluorescein isothiocyanate.
We also treated bone marrow cells obtained from MM patients. Results from two representative patients are shown in Figure 2b: the purified CD138+ plasma cells treated with KPT-276 had an increased apoptotic cell population, as compared with DMSO vehicle. To test the specificity of KPT-276 for CD138+ plasma cells, we also treated unsorted whole bone marrow samples with KPT-276. In a representative patient sample, the CD138+ plasma cell population was reduced from 46% in the DMSO-treated sample to 17% in the KPT-276-treated sample. We observed an increase in apoptotic cells, from 17.8% in the DMSO-treated sample to 46.4% in the KPT-276-treated sample. Plasma cells lose CD138+ status as they undergo apoptosis, which accounts for the increased population of AnnexinV-positive, CD138-negative cells in the KPT-276-treated sample. Therefore, increased apoptosis was observed for the CD138+ cell population, whereas the CD138-negative viable cell population did not have a significantly increased population of apoptotic cells, indicating that KPT-276 is specific for CD138+ tumor cells (Figure 2c).
Gene expression profiling reveals targets of KPT-276 Gene expression profiling was performed to investigate which genes are affected by KPT-276 inhibition of CRM1. Two HMCLs most sensitive to KPT-276 (IC50 ~15nM), OCI-MY5 and MM1.S, were treated with an IC80 dose of KPT-276 for 6 h. MM1.S and OCI-MY5 showed >2-fold expression change between conditions in 292 and 338 genes, respectively. Eleven genes were differentially expressed (>2-fold) in both HMCLs. Four genes, PRMT2, POGZ, SMARCC2 and C1orf53, were upregulated in both cell lines when treated with KPT-276 as compared with the DMSO vehicle. The remaining seven genes, FARSB, CDC25A, C4orf43, ZNF326, BXDC2, TPMT and RDH10 were downregulated in both cell lines when treated with KPT-276 as compared with the DMSO vehicle (Table 1). Although only reaching significance in one of the two cell lines, we were also struck by the appearance of BRD4 as downregulated in both treated cell lines.
Table 1.
Twelve genes up- or downregulated in two HMCL in response to KPT-276
| Gene Symbol | Fold Change MM1.S DMSO v. KPT-276 | Fold Change OCI-MY5 DMSO v. KPT-276 | Regulation DMSO v. KPT-276 |
|---|---|---|---|
| C1orf53 | 2.43 | 2.50 | Up |
| POGZ | 2.74 | 4.45 | Up |
| PRMT2 | 2.86 | 2.51 | Up |
| SMARCC2 | 2.14 | 2.22 | Up |
| BRD4 | 2.39 | 1.47 | Down |
| BXDC2 | 2.32 | 2.19 | Down |
| C4orf43 | 2.23 | 2.51 | Down |
| CDC25A | 2.19 | 2.01 | Down |
| FARSB | 2.07 | 2.54 | Down |
| RDH10 | 3.04 | 2.12 | Down |
| TPMT | 2.67 | 2.14 | Down |
| ZNF326 | 2.23 | 2.02 | Down |
Abbreviation: HMCL, human MM cell line.
KPT-276 treatment causes a reduction of c-Myc, CDC25A and BRD4 levels in vitro
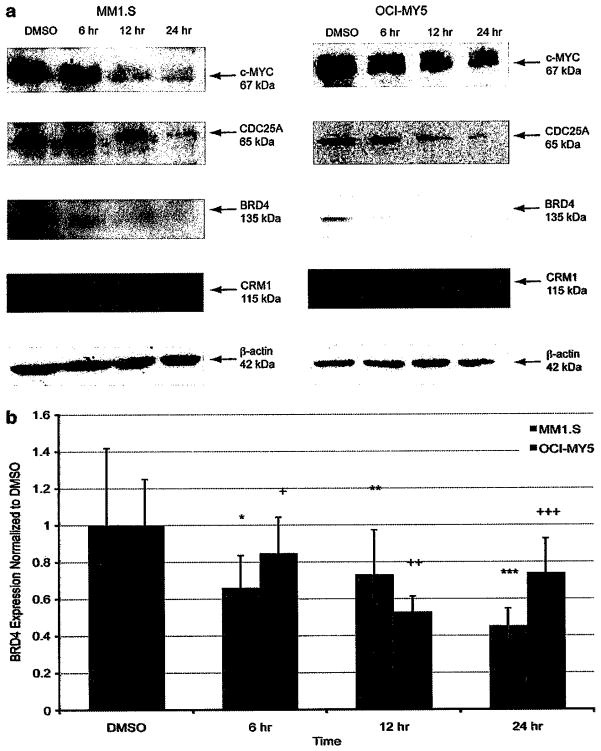
To validate the genes that were downregulated as shown by expression profiling, a time course experiment was performed to demonstrate that CDC25A, c-MYC and BRD4 levels are reduced upon KPT-276 treatment. c-Myc protein levels are decreased at 12 h post-KPT-276 treatment (Figure 3a). CDC25A protein levels also began to decrease approximately 12 h after initiation of drug treatment, which is expected as CDC25A transcription is twofold downregulated after 6 h of KPT-276 treatment (Figure 3a). BRD4 expression is also reduced following KPT-276 exposure, starting at 6 h post-administration (Figure 3a). BRD4 gene expression begins to decrease at 6 h post-treatment, and continues to decrease as shown by reverse transcription-PCR (Figure 3b). Although P-values are significant in MM1.S, BRD4 downregulation was validated in both MM1.S and OCI-MY5 by decreased protein expression.
Figure 3.
KPT-276 downregulates cell cycle genes. Cells were treated with KPT-276, then collected at time points spanning 24 h. For immunoblotting, lysates were run on SDS-polyacrylamide gel electrophoresis and probed with antibodies for CDC25A, c-Myc and BRD4. Western blots show CDC25A and c-MYC protein expression was reduced starting at 12 h post-treatment, and BRD4 expression was reduced starting at 6 h post-treatment (a). To analyze BRD4 expression in response to KPT-276, RNA was extracted for reverse transcription-PCR analysis. BRD4 expression levels decreased after 6 h of drug exposure in two sensitive cell lines (b), (P values: *0.274, **0.247, ***0.044, +0.192, ++0.088, +++0.186).
KPT-276 is synergistic with BRD4 inhibitor JQ1
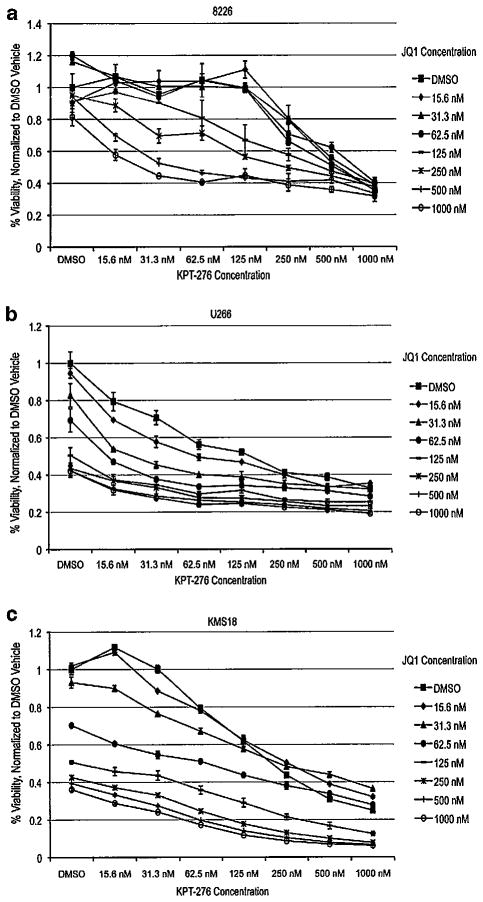
Because BRD4 was downregulated in response to KPT-276 in two cell lines, we investigated the effect of KPT-276 in combination with JQ1, a novel BRD4 small-molecule inhibitor. JQ1 has been shown to be toxic to HMCL as a single agent due to its inhibition of c-MYC (Delmore, et al.24). Three cell lines most resistant to KPT-276 and were treated with a combination of KPT-276 and JQ1, and a decrease in viability was observed at concentrations lower than treatment with KPT-276 alone (Figures 4a-c). Synergy was observed with JQ1 in all three cell lines, as indicated by combination index values (Table 2).
Figure 4.
KPT-276 is synergistic in combination with JQ1. Cells were treated with KPT-276 in combination with JQ1, both at nanomolar concentrations, for 72 h, then analyzed by MTT assay. HMCL viability was reduced at a greater rate in response to KPT-276 and JQ1 in combination, compared with treatment with either drug alone (a–c).
Table 2.
Combination index for KPT-276 and JQ1 in three KPT-276 resistant HMCL
| Cell line | KPT-276 IC50 (nM) | KPT-276 concentration (nM) | JQ1 concentration (nM) | CI |
|---|---|---|---|---|
| 8226 | 903 | 125 | 500 | 0.258 |
| U266 | 488 | 62.5 | 31.25 | 0.335 |
| KMS18 | 176 | 125 | 1000 | 0.063 |
Abbreviations: CI, combination index; HMCL, human MM cell line.
CI a measure of synergy derived from the multiple drug-effect equation of Chou-Talalay (Biosoft). A CI value between 0.3 and 0.7 indicates synergy, a CI value below 0.3 indicates strong synergy.
KPT-276 induces cell cycle arrest
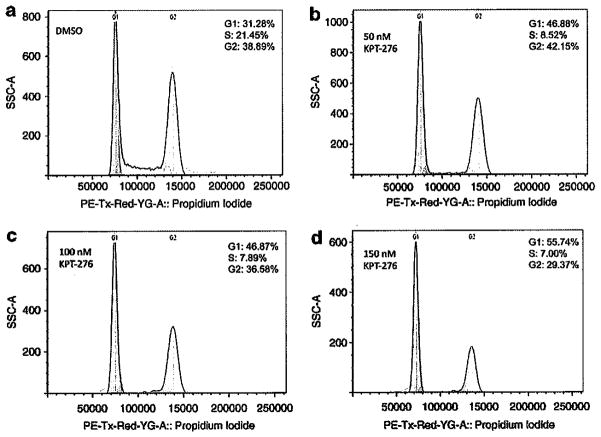
To further explore the influence of BRD4 and CDC25A, inhibition of which has been shown to cause G1/S phase cell cycle arrest, we next analyzed MM cells treated with KPT-276 for cell cycle arrest (Figure 5). Myeloma cells arrest in G1 in the drug-treated samples (Figures 5b-d) versus the DMSO-treated sample (Figure 5a). In cells treated with DMSO vehicle, 31% of cells were in G1, whereas cells treated with KPT-276 exhibited a higher population of cells (45–55%) in G1 phase. KPT-276-treated cells also had a reduced population in S phase (7–8.5%) compared with cells treated with DMSO, which had 21.5% of cells in S phase.
Figure 5.
KPT-276 induces cell cycle arrest in MM1.S cells. After treating MM1.S cells for 24 h with varying doses of KPT-276, cell cycle arrest was observed in all drug-treated samples. The population of cells arrested in G1 increased from 31% in vehicle-treated cells (a) to 56% in cells treated with the highest dose of KPT-276 (d). The population of cells in S-phase decreased in all drug-treated cells (7–8.5%) (b–d), as compared with cells treated with DMSO vehicle (21%) (a).
KPT-276 reduces disease progression in two mouse models of myeloma
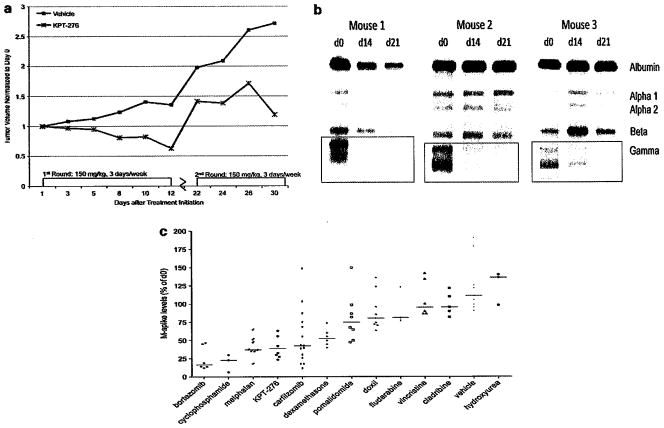
In a xenograft MM1.S MM model, the tumor volume significantly decreased after treatment with KPT-276. However, after 12 days, mice started losing weight and treatment had to be stopped. Treatment was suspended for 10 days, and the tumors re-grew. Despite the lapse in treatment, tumors were again responsive to re-initiation of therapy after the 10-day lapse (Figure 6a). Because of the possible association with c-MYC, the anti-MM activity of KPT-276 was demonstrated on the clinically predictive VK*MYC transgenic mouse model of MM.22 Compared with the serum M-spike levels at day 0, the M-spike levels of all treated mice decreased following KPT-276 treatment. Over 3 weeks of treatment, the M-spike was reduced by an average of 57% compared with day 0 levels, showing regression of the disease (Figure 6b). Treatment was stopped after 3 weeks because of significant weight loss. KPT-276 activity in the VK*MYC mouse was compared with the effect of other compounds in reducing M-spike levels. The anti-myeloma activity of KPT-276 in this mouse model after 2 weeks of treatment is comparable to melphalan and carfilzomib (Figure 6c).
Figure 6.
KPT-276 reduces immunoglobulin levels and tumor volume in mouse models of myeloma. KPT-276 decreased tumor volume in an MM1.S xenograft model. As soon as 12 days after treatment initiation, tumor volume decreased substantially as compared with tumors in vehicle-treated mice. Tumor growth was inhibited for up to 30 days (a). Three transgenic Vk*Myc mice were treated with KPT-276 for 3 weeks, and the M-spike, a disease marker, was reduced in all three mice (b). KPT-276 activity in Vk*MYC mice is comparable to melphalan and carfilzomib (c).
DISCUSSION
XPO1 has been identified as a top myeloma survival gene. Out of 6722 druggable genes, XPO1 was 1 of 55 genes essential to MM cell viability.3 XPO1 knockdown proved to be lethal in three HMCLs, but had no effect on human embryonic kidney (293) cells or lung cancer (A549) cells.3 Therefore, XPO1 is a vulnerable target in MM, and myeloma cell survival is dependent upon XPO1 expression. A recent publication has demonstrated that CRM1 knockdown by small hairpin RNA also decreases HMCL growth, survival and viability.13 We have shown that XPO1 expression correlates with disease progression; XPO1 expression in monoclonal gammopathy of undetermined significance and smoldering myeloma was comparable to expression in normal plasma cells, and increased when the disease progressed to MM. Until recently, no known drugs were available that specifically targeted XPO1.
A new class of SINE compounds has been developed to target XPO1, specifically its nuclear export function mediated by CRM1. When XPO1 function is inhibited, TSPs are retained and accumulate in the nucleus where they remain active.7 We show here that KPT-276, a novel SINE compound, is an active anti-MM compound in vitro. HMCL responded to KPT-276 treatment, with at least 50% reduction in viability at sub-micromolar concentrations. KPT-276 also drives MM patient CD138+ cells to apoptosis but normal CD138-negative cells are relatively unaffected, indicating that KPT-276 is relatively specific for MM cells. In our hands, no synergy was observed with KPT-276 in combination with dexamethasone, melphalan or bortezomib, in four HMCL tested. Despite the lack of synergy, our findings confirm the anti-MM activity of KPT-276, and also suggest that XPO1 is a valid target for small-molecule therapies.
In terms of pharmacodynamic markers, others have shown that KPT-276 increases total and nuclear p53 expression in HMCL.25 SINE treatment also induces a reduction of cell cycle protein expression, including cydin E, cyclin D1, CDK2, CDK4 and CDK6.26 KPT-185 and KPT-330, SINE analogs, decreased HMCL growth and survival, regardless of p53 status.13 These SINEs also induced apoptosis and nuclear accumulation of p53, IκB, p27 and p21.13 SINE compounds have also been shown to have specific anti-myeloma affects. For example, SINE compounds have recently been shown to decrease levels of c-Myc, plκB (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor), nuclear factor-κB, MCL-1 (myeloid cell leukemia sequence 1) and BCL-xL (BCL2-like 1), and also decrease secretion of IL-6 (interleukin 6), VEGF (vascular endothelial growth factor), MIP1β (chemokine (C–C motif) ligand 4) and IL-10 from bone marrow stromal cells.13 SINE compounds also reduced growth and survival of HMCL and primary cells in the presence of bone marrow stromal cells or osteoclasts.13 In addition to retaining TSP function and causing tumor cell apoptosis, SINE compounds also inhibited osteoclast formation and bone reabsorption via inhibition of nuclear factor-κB and activation of the signaling cascade in osteoclast precursors.13 The SINE compounds have shown successful inhibition of MM in vitro via inhibition of not only cell cycle proteins, but also genes known to be critical to MM oncogenesis. In our hands, we found KPT-276 to target additional genes that have important roles in MM.
Our data revealed that KPT-276 downregulates two c-Myc-related genes, CDC25A and BRD4. We have found that KPT-276 downregulates c-Myc itself, and recently published data corroborate this finding.13 MYC is a transcription factor that is often activated in MM, and activation of the MYC pathway drives tumor cell proliferation and survival.27 MYC activation is a common genetic event in myeloma patients, and MYC activation is associated with poor prognosis and shorter survival.28 Because MYC activation is implicated in MM progression and impacts patient survival, a drug that targets MYC and related genes is a potentially important therapy.
CDC25A, a MYC-related gene, is a tyrosine phosphatase that regulates G1/S phase progression by removing the inhibitory phosphate group on CDK2 at its catalytic subunit.29 In the rapid-response DNA repair pathway, CDC25A is ubiquitinated and degraded, and cells are arrested in G1 phase.29 CDC25A has three specific MYC-binding sites, and its transcription is dependent upon MYC.30 When c-Myc expression is increased, CDC25A expression is also increased four- to fivefold.30 MYC overexpression usually occurs as a consequence of chromosomal rearrangements in late-stage myeloma.31 Because c-MYC regulates the level of CDC25A, aberrant c-MYC overexpression leads to increased CDC25A expression. It appears that KPT-276 downregulates c-MYC, and subsequently CDC25A. A decrease in CDC25A transcription could restore the rapid-response checkpoint, with cells arrested in G1. CDC25A gene and protein levels were decreased upon KPT-276 treatment, and G1 cell cycle arrest was observed in HMCL.
BRD4, another gene downregulated in response to KPT-276, encodes a bromodomain that binds acetylated chromatin and regulates cell cycle, DNA replication and transcription.32 BRD4 expression is positively correlated with MM disease progression, and it is amplified often in MM patient samples.24 BRD4 is another cell cycle gene that has been linked to c-Myc activity in MM. BRD4 binds to acetyl-lysine pockets at the IgH enhancer region on the MYC locus, and promotes transcription and activation of MYC downstream targets.24 It has been recently shown that BRD4 downregulation by the small-molecule inhibitor JQ1 causes decreased MYC transcription, and a reduction of c-MYC protein levels,24 and we were able to replicate this data using KPT-276 that lowers BRD4 expression. Furthermore, BRD4 knockdown causes cells to arrest in G1 phase and enter apoptosis.33 The G1 arrest observed by knockdown of BRD4 was also observed in response to KPT-276 treatment in HMCL. Gene expression shows that a number of cell cycle genes, including CCND1, CCND2, ORCL2, MCM2, DHFR, TOP2A, PCNA-DNA, CHAF1A, HMGB1, AND1, RNABP1, POP1, NID1, NSH2 and PMS2, are all downregulated in response to BRD4 knockdown.33 Thus, by lowering BRD4 levels, KPT-276 may alter MM viability via MYC downregulation and/or lowering cyclin D levels. KPT-276 toxicity was increased when used in combination with JQ1, indicating that the effect of BRD4 inhibition by KPT-276 is further amplified by another BRD4 inhibitor. In conclusion, XPO1 inhibition via KPT-276 has promising anti-MM activity in vitro, as it reduces cell viability, induces apoptosis, causes G1 cell cycle arrest and downregulates genes implicated in MM oncogenesis.
KPT-276 inhibition of XPO1 in vivo also showed potent activity in two MM mouse models. In MM1.S xenograft tumors, tumor volume decreased by 40% within 12 days of treatment, whereas tumor volume increased by 36% in the vehicle-treated cohort. Because of substantial weight loss in the drug-treated mice, we had to suspend drug administration after 12 days. All mice recovered within 10 days and significant tumor growth was generally observed in this time. However, tumors rapidly responded to re-exposure to the drug, with a significant reduction in tumor volume in the drug-treated mice. Others have shown that KPT-276 causes cleavage of caspase 3 and increase in Annexin V-positive tumor cells, and inhibits tumor growth and increases survival in an MM1Sluc xenograft model.13 KPT SINEs also reduced bone lesions in vivo.13
KPT-276 was also active in the Vk*MYC mouse model of MM, which has a positive predictive value of 67% for the activity of single-agent compounds in clinical trials.22 The Vk*MYC mouse is genetically engineered to replicate the MYC locus rearrangements that activate MYC and cause monoclonal gammopathy of undetermined significance to progress to MM.22 These mice mimic human indolent disease, with anemia, renal and bone disease, and an M-spike that is detectable by serum protein electrophoresis.22 After 2 weeks, KPT-276 reduced the M-spike by an average of 52%, which is comparable to the anti-MM activity of melphalan and carfilzomib, two of the most potent FDA-approved drugs used in the clinic.
In summary, KPT-276 is an active anti-MM drug that causes decreased cell viability, increased apoptosis and cell cycle arrest in vitro in HMCL and patient samples, and is active in inhibiting myeloma tumor load in two mouse models. Cells treated with the compound show reduced levels of MYC, BRD4 and CDC25A, all cell cycle genes that are implicated in oncogenesis. A phase I clinical trial of a closely related SINE compound, KPT-330, is underway and promises to be of significant interest (Clinical Trials NCT01607892).
Acknowledgments
This work was supported by MMRF industry award grant. This study was supported in part by the Predolin Foundation. JS was supported by NIH. EB is a recipient of the Marriott Specialized Workforce Development Awards in Individualized Medicine, The Henry Predolin Foundation Career Development Award and the George Haub Family Career Development Award Fund in Cancer Research. JBE was supported by an MMRF fellowship Award. MK is supported by a research grant of the Deutsche Forschungsgemeinschaft (DFG). MC is recipient of a MMRF senior research award and PLB is supported by NIH CA136671 grant. KPT-276 was provided by Karyopharm Therapeutics Inc. (Natick, MA, USA). JQ1 was provided by the laboratory of Dr Jay Bradner (Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA).
Footnotes
CONFLICT OF INTEREST
AKS provides consulting services for Millennium and Celgene. MK and SS are founders of and hold equity in Karyopharm. DM is employed by Karyopharm. PLB provides consulting services for Onyx. The remaining authors declare no conflict of interest.
References
- 1.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–2520. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner H, Gondos A, Pulte D. Recent major improvement in long-term survival of younger patients with multiple myeloma. Blood. 2008;111:2521–2526. doi: 10.1182/blood-2007-08-104984. [DOI] [PubMed] [Google Scholar]
- 3.Tiedemann RE, Zhu YX, Schmidt J, Shi CX, Sereduk C, Yin H, et al. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012;72:757–768. doi: 10.1158/0008-5472.CAN-11-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellom ST, Jr, Shanker A. Development of proteasome inhibitors as therapeutic drugs. J Clin Cell Immunol. 2012;S5:5. doi: 10.4172/2155-9899.s5-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu D, Grishin NV, Chook YM. NESdb: a database of NES-containing CRM1 cargoes. Mol Biol Cell. 2012;23:3673–3676. doi: 10.1091/mbc.E12-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83:1021–1032. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapalombella R, Sun Q, Williams K, Tangeman L, Jha S, Zhong Y, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120:4621–4634. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, Dong Y, Lin F, Zhao H, Shen Z, Chen P, et al. The expression of CRM1 is associated with prognosis in human osteosarcoma. Oncol Rep. 2009;21:229–235. [PubMed] [Google Scholar]
- 9.Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH, Sun YJ. Prognostic value of CRM1 in pancreas cancer. Clin Inv Med. 2009;32:E315. [PubMed] [Google Scholar]
- 10.Noske A, Weichert W, Niesporek S, Roske A, Buckendahl AC, Koch I, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer. 2008;112:1733–1743. doi: 10.1002/cncr.23354. [DOI] [PubMed] [Google Scholar]
- 11.Shen A, Wang Y, Zhao Y, Zou L, Sun L, Cheng C. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery. 2009;65:153–159. doi: 10.1227/01.NEU.0000348550.47441.4B. discussion 159–160. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K, Wang M, Tamayo AT, Shacham S, Kauffman M, Lee J, et al. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exper Hematol. 2013;41:67–78. e64. doi: 10.1016/j.exphem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Tai YT, Landesman Y, Acharya C, Calle Y, Zhong MY, Cea M, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2013 doi: 10.1038/leu.2013.115. e-pub ahead of print 16 April 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74:648–649. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM, et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood. 2011;118:3922–3931. doi: 10.1182/blood-2011-01-333138. [DOI] [PubMed] [Google Scholar]
- 16.Turner JG, Marchion DC, Dawson JL, Emmons MF, Hazlehurst LA, Washausen P, et al. Human multiple myeloma cells are sensitized to topoisomerase II inhibitors by CRM1 inhibition. Cancer Res. 2009;69:6899–6905. doi: 10.1158/0008-5472.CAN-09-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT, et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol. 2013;161:117–127. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etchin J, Sun Q, Kentsis A, Farmer A, Zhang ZC, Sanda T, et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia. 2013;27:66–74. doi: 10.1038/leu.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120:1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue H, Kauffman M, Shacham S, Landesman Y, Yang J, Evans CP, et al. CRM1 Blockade by Selective Inhibitors of Nuclear Export (SINE) attenuates Kidney Cancer Growth. J Urol. 2012;189:2317–2326. doi: 10.1016/j.juro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azmi AS, Aboukameel A, Bao B, Sarkar FH, Philip PA, Kauffman M, et al. Selective inhibitors of nuclear export block pancreatic cancer cell proliferation and reduce tumor growth in mice. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.10.036. S0016-5085: 01552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chesi M, Matthews GM, Garbitt VM, Palmer SE, Shortt J, Lefebure M, et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood. 2012;120:376–385. doi: 10.1182/blood-2012-02-412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 24.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandarpa M, Kraftson SJ, Maxwell SP, McCauley D, Shacham S, Kauffman M, et al. CRM1 is highly expressed in myeloma plasma cells and its inhibition by KPT-SINE induces cytotoxicity by increasing p53 in the nucleus of multiple myeloma (MM) cells. ASH Annu Meet Abstr. 2011;118:1852. [Google Scholar]
- 26.Kong S-Y, Landesman Y, Jakubikova J, Sellitto MA, Cagnetta A, Cea M, et al. Blockade of Nuclear Export Protein CRM1 (chromosomal region maintenance 1, XPO1) by a Novel, Potent and Selective CRM1 Inhibitor KPT-185 Induces Significant Antitumor Activity Against Human Multiple Myeloma. ASH Annu Meet Abstr. 2011;118:2913. [Google Scholar]
- 27.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 28.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 30.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 31.Gabrea A, Leif Bergsagel P, Michael Kuehl W. Distinguishing primary and secondary translocations in multiple myeloma. DNA Repair. 2006;5:1225–1233. doi: 10.1016/j.dnarep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 33.Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]