Abstract
Background
Glioblastoma multiforme (GBM) contains a population of cells that exhibit stem cell phenotypes. These cancer stem cells (CSCs) may be a source of therapeutic resistance, although support for this important concept is limited.
Methods
We determined whether early-passage GBM CSCs respond differently than patient-matched, genotypically similar non-CSCs to clinically relevant single or serial doses of temozolomide (TMZ), radiation therapy (XRT), or alternating TMZ treatment and XRT, which is the standard of care for GBM patients.
Results
Despite the phenotypic differences, including the presence of stem cell markers and formation of intracranial tumors, the CSCs and matched non-CSCs were equally resistant to TMZ in a majority of patients, using 2 independent assays. TMZ response was consistent with methylated O6-DNA methylguanine-methyltransferase (MGMT) and MGMT protein levels in both culture types. In contrast, CSCs were unexpectedly more responsive to XRT compared with matched non-CSCs from 2 patients despite having relatively equal resistance to TMZ. However, for the majority of culture pairs from individual patients, responses in CSCs were indistinguishable from non-CSC cultures.
Conclusions
In our patient-matched primary cultures, response to TMZ was tightly linked to the individual tumor's MGMT status and independent of their phenotypic differences. TMZ and XRT together revealed no additive benefit compared with monotherapy for either culture type, in contrast to the notion that the CSC population is more resistant to XRT. If the tumor cell response in vitro mirrors therapeutic response in larger patient cohorts, these rapid assays in primary cultures could allow empirical selection of efficacious therapeutic agents on a patient-specific basis.
Keywords: cancer stem cell, glioblastoma multiforme, radiation response
Glioblastoma multiforme (GBM) displays molecular heterogeneity among patients and within individual tumors. Inter- and intratumoral heterogeneity is a major confounding factor for achieving durable therapeutic response. Intratumoral heterogeneity at the cellular level includes cell subpopulations referred to as cancer stem cells (CSCs), which have properties similar to neural progenitors, such as the ability to differentiate into multiple CNS cell lineages.1,2 CSC-enriched cultures derived from primary GBM can be propagated in vitro as neurospheres in suspension2,3 or as adherent monolayers,4 as well as in vivo as xenografts.3,4 Interestingly, the ability of dissociated primary tumors to establish viable CSC suspension cultures has been associated with worse overall survival for patients from whom the cultures were derived,5,6 suggesting that the tumor CSC component is a significant contributor to tumor malignancy. Enhancer of zeste homolog 2 and signal transducer and activator of transcription 3, which both show elevated expression in GBM, preferentially interact in CSCs, and this interaction appears to help maintain a state of stemness.7 CSCs and non-CSCs cultured from the same tumor also exhibit differences in their histone profiles, though how the epigenetic differences relate to differences in culture phenotypes such as drug response is unknown.8 Such paired non-CSC and CSC cultures allow controlled comparisons of genotypically similar but phenotypically distinct cells for molecular, biologic, and therapeutic response characteristics. Here we use these cultures to directly address the hypothesis that CSCs are more resistant than non-CSCs to therapy in a genetically controlled setting.
The standard of care for GBM patients is resection, followed by chemotherapy and radiation therapy (XRT). The most commonly used chemotherapeutic agent is temozolomide (TMZ), an orally delivered DNA alkylator that crosses the blood–brain barrier and undergoes spontaneous conversion to the active form 3-methyl-(triazen-1-yl)imidazole-4-carboxamide (ie, MTIC).9 The overall survival of GBM patients who receive TMZ correlates with the methylation status of O6-DNA methylguanine-methyltransferase (MGMT), a DNA repair protein that preferentially removes the TMZ-induced methyl group adduct at O6-guanine.10 In addition to MGMT, GBM may be inherently resistant to TMZ or may develop increased resistance during the course of TMZ therapy. A testable hypothesis to account for GBM TMZ resistance is that it is conferred by tumor CSC subpopulations and that CSCs undergo preferential expansion during or after treatment.11 This resistance could be the result of both intrinsic factors such as increased drug efflux and extrinsic factors such as hypoxic microenvironments.12 A similar mechanism accounted for GBM resistance to radiation therapy.13 However, there is disagreement regarding the relative importance of the GBM CSC component to therapeutic resistance, as indicated by reports suggesting that CD133+ CSC populations may be more sensitive to TMZ14 or XRT15 than tumor-matched CD133–14 or serum-derived but unmatched tumor cultures that are depleted of CSCs.15 Genetic differences between cultures were not controlled for in each case, nor did these studies examine response to combined XRT and TMZ treatment. To directly address the influence of CSCs on GBM response to standard-of-care therapy in a genetically controlled manner, we established CSC-enriched and non-CSC culture pairs from 10 patients with primary GBM. To augment these analyses, we established and tested CSC cultures from an additional 15 primary GBM patients.
Materials and Methods
Primary Tumors and Cell Culture
The Brain Tumor Research Center Tissue Bank at the University of California, San Francisco (UCSF) provided the human tumor samples. The samples were collected during surgery from consenting patients assigned nonidentifying numbers (SF#) according to the protocol approved by the UCSF Committee on Human Research. Primary GBM was dissociated and grown as GBM neural stem cells as previously described4 or as a monolayer in Dulbecco's modified Eagle's medium (DMEM):F12 (1:1) with 10% fetal bovine serum (Invitrogen). Human GBM cell lines U87, U251, and LN229 were cultured in DMEM:F12 (1:1) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. Early-passage cultures (P2–P10) were used for all experiments described, with time to initial testing ∼2 weeks after tissue was collected. Cultures were established from 25 patients, and assays were performed on all 25 CSC cultures and non-CSCs from 10 patients.
Fingerprint Analysis
The target locus containing the repeat region was amplified by PCR with one of the primers in the pair labeled with a fluorescent tag marker. An in-house DNA fingerprinting marker panel, 13 loci with high heterogeneity of alleles, was used: D1S3721, D3S1259, D5S1453, D6S2436, D7S1802, D8S1108, D9S2157, D13S325, D14S1434, D15S652, D17S974, D18S1364, Amel. The amplified products were pooled together and analyzed via capillary electrophoresis on a 3730xl DNA Analyzer (Applied Biosystems). The resulting data files were analyzed on GeneMapper (Applied Biosystems) with size-appropriate allele calls made to produce genotype calls.
TMZ Treatment
TMZ treatment was described previously16 with the following modifications. Unsynchronized cells were treated with TMZ for 3 h. After treatment, cells were incubated in fresh media for 4 days and then harvested for cell cycle analysis as previously described.16 For multiple treatments with TMZ, unsynchronized cells were treated with 50 μM TMZ for 3 h, after which cells were incubated with fresh media. This was repeated for 2 more days, and the cells were then harvested at several different time points.
Irradiation of Cells
Irradiation was performed at room temperature in a Mark I-68 cesium-137 irradiator (J. L. Shepherd & Associates) at a dose rate of 2.26 Gy/min.
Immunocytochemistry
Cells were grown on 8-well chamber slides, fixed with 4% paraformaldehyde, and then washed with phosphate buffered saline. The following primary antibodies were used: nestin (R&D Systems), sex determining region Y–box 2 (Sox2; R&D Systems), glial fibrillary acidic protein (Millipore), and neuron-specific class III beta-tubulin (TuJ1; Covance).
Cell Surface Expression Analysis
Cells were treated with either Accutase (CSCs) or trypsin (non-CSCs). After inactivation, cells were incubated first with Fc Block (Miltenyi) for 10 min at 4°C, and then with an antibody against CD133 (Miltenyi) for 30 min. Cells were washed 3× with fluorescence activated cell sorting (FACS) buffer and then analyzed on a FACSCalibur flow cytometer (BD Biosciences).
Cell Cycle Analysis
Cell cycle analysis was performed as previously described.16 Briefly, primary cultures (1 × 106) were treated with dimethyl sulfoxide or TMZ for 3 h and washed, and new medium was added. At the appropriate time, cells were fixed with methanol. Prior to cell cycle analysis, fixed cells were stained with propidium iodide (100 μg/mL). Cells were collected on a BD FACSCalibur, and cell cycle analysis was performed using FlowJo software.
Methylation-specific PCR
Methylation-specific PCR of MGMT was performed as previously described,17 except that only one round of PCR was performed, using the internal set of primers. PCR products were electrophoresed on 4%–20% Tris/borate/EDTA polyacrylamide gels and stained with SYBR Safe (Invitrogen) for 30 min for visualization.
Protein Analysis
Cells were lysed in 1× radioimmunoprecipitation assay buffer. Primary antibodies used were MGMT (Cell Signaling) and glyceraldehyde 3-phosphate dehydrogenase (Cell Signaling), and secondary antibody was horseradish peroxidase–conjugated goat anti-rabbit IgG (Cell Signaling). Blots were developed with ECL Plus (GE Healthcare).
Cell Viability Assay
Primary cultures were plated in a 96-well plate, allowed to attach for 24 h, and then treated with dimethyl sulfoxide or TMZ for 3 h. Cells were then washed, medium was replaced, and cells either were allowed to recover for 24 h or received 2 Gy of ionizing radiation. This regimen was repeated 2 additional times. To assay cell viability, plates were removed at 24, 48, or 72 h after the final treatment and incubated in Cell TiterGlo reagent (Promega). Plates were then read on a GloMax plate reader, and the average of 12 wells was computed for each dose and each time point.
Colony Formation Assay
Primary cultures were plated in a 96-well plate and allowed to attach for 24 h. Cells then received 2 Gy of ionizing radiation for 3 consecutive days. Cells were allowed to grow for 14–21 days and were stained with crystal violet, and then colonies with at least 25 cells were counted to determine the surviving fraction.
Results
Characterization of CSC and Patient-matched Non-CSC Cultures
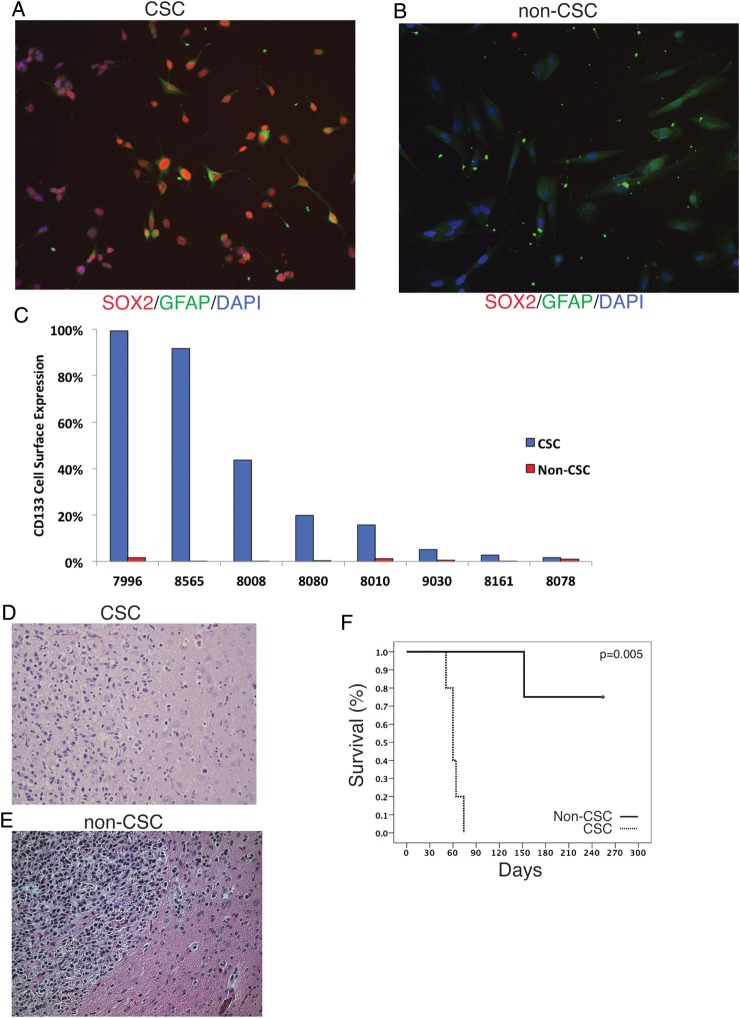
To determine whether primary GBM CSCs were more resistant to standard-of-care therapy than stem cell–depleted cultures from the same patient, we established early-passage CSC-enriched and CSC-depleted (non-CSC) cultures from primary GBM tissue of 10 patients. For 15 additional patients, we established CSC cultures only. Genotyping microsatellite markers in the primary tumor tissue, the matching CSCs and non-CSCs confirmed the common origin and genotypic similarity for each sample trio (Supplementary Table S1). The neural stem cell markers Sox2 and nestin were expressed in >90% of cells in CSC cultures, while all but one of the matched non-CSC cultures were negative for these markers (Fig. 1A and B and Table 1). Furthermore, culturing CSCs in conditions that promote differentiation into either neurons (without growth factor) or astrocytes (with 5% normal goat serum) resulted in cells that stained positive for TuJ1 or glial fibrillary acidic protein, respectively (Supplementary Fig. S1A–F), thereby confirming the multidifferentiation potential of cells within the CSC-enriched cultures. We also examined expression of CD133, a cell surface marker associated with cancer stemness. CSC cultures displayed varying levels of CD133 expression by FACS analysis (2%–98.9% positive), consistent with previous results,18 while non-CSC cultures did not express detectable CD133 on the cell surface (Fig. 1C).
Fig. 1.
Validation of phenotypic differences between patient-matched CSC and non-CSC cultures of primary GBM. (A) Adherent CSC cultures express the stem cell marker Sox2, while (B) the patient-matched non-CSC culture does not express stem markers and has a more differentiated morphology. (C) Expression of CD133 for CSC and patient-matched non-CSC cultures for 8 patients. (D) Intracranial injection of 300 000 CSCs resulted in tumors that exhibit an invasive pattern of growth characteristic of primary GBM, while (E) injection of 300 000 patient-matched non-CSCs resulted in only 1 tumor, and it had a circumscribed border and lacked evidence of invasion into the surrounding parenchyma. (F) Kaplan–Meier analysis of the 2 cohorts of mice implanted orthotopically with the CSC or non-CSC cultures (D, E) demonstrates greater tumor-forming ability in the CSC (4/4) relative to the patient-matched non-CSC culture (1/4). GFAP, glial fibrillary acidic protein; DAPI, 4′,6′-diamidino-2-phenylindole.
Table 1.
Summary of stem cell markers and response to therapy in paired CSC and non-CSC cultures
| Cell Line |
Sox2 | Nestin | CD133 | |
|---|---|---|---|---|
| 7996 | CSC | |||
| 7996 | Non-CSC | |||
| 8161 | CSC | |||
| 8161 | Non-CSC | |||
| 8279 | CSC | |||
| 8279 | Non-CSC | |||
| 8565 | CSC | |||
| 8565 | Non-CSC | |||
The ability of these cultures to generate tumors in vivo was also examined. Five of 8 CSC lines tested formed intracranial tumors in immune-compromised mice. CSC cultures injected intracranially gave rise to tumors that were diffusely invasive and had features typical of high-grade gliomas, including pleomorphic nuclei, high mitotic index, and migration along white matter tracts (Fig. 1D and F, Supplementary Fig. S1G), though they lacked evidence of angiogenesis. For one CSC culture that gave rise to intracranial tumors, we also tested the patient-matched non-CSC culture for the ability to form tumors. Three out of 4 mice did not develop tumors from the non-CSCs, and in the single instance that a tumor was generated, it had much longer latency and was circumscribed and less invasive than the tumors from patient-matched CSC cultures (Fig. 1E and F, Supplementary Fig. S1H). These results show that our CSC cultures were phenotypically distinct from the patient-matched, non-CSC cultures, consistent with prior reports.3,4
Response of Patient-matched CSC and Non-CSC Cultures to TMZ
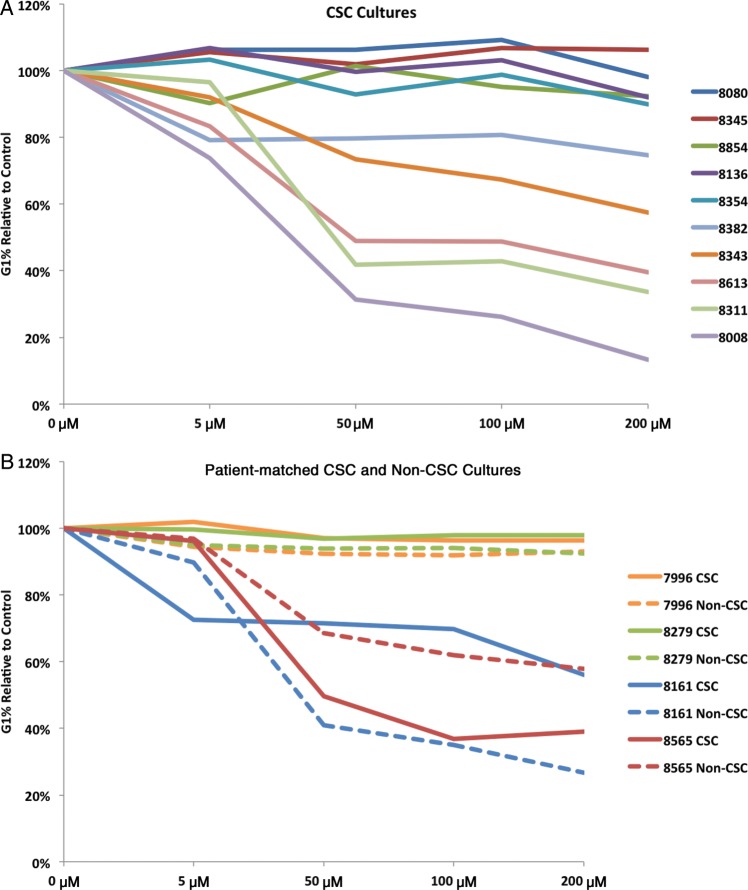
The standard of care for GBM patients is surgical resection followed by the administration of TMZ and XRT to reduce residual disease. TMZ alkylates DNA at the N7 and O6 positions of guanine residues, resulting in DNA damage, which if left unrepaired leads to G to A mutations and/or growth arrest or apoptosis.9,16,19,20 We first determined whether the primary cultures would respond to TMZ doses that were achievable in plasma (50 μM) or cerebrospinal fluid (5 μM),21 and then whether responses in CSCs were distinct from those in matched non-CSC cultures. As controls for TMZ response, we treated the established, serum-grown glioma cell lines U87, LN229, and U251 (Supplementary Fig. S2A) with increasing doses of TMZ, which resulted in a G2/M arrest and a concurrent decrease in G1 in all 3 cultures, as previously shown.16 In contrast, our 25 primary CSC cultures exhibited a range of responses to TMZ, similar to serum-grown non-CSC cultures treated with bis-cloroethylnitrosourea22 (Fig. 2A and Supplementary Fig. S2B). Unexpectedly, when we examined the response of non-CSC cultures, they were very similar to that of their matched CSC counterparts (Fig. 2B and Supplementary Fig. S2C). This was true for almost all of the patient-derived CSCs along the continuum of the response level. These results show that despite their reproducible phenotypic differences in vitro and in vivo, corresponding CSC and non-CSC cultures have a similar TMZ responsiveness in vitro.
Fig. 2.
Response to TMZ treatment is patient specific and independent of the CSC phenotype. (A) Cell cycle analysis of 10 patient-derived CSC cultures shows a spectrum of response to physiologically achievable (5–50 µM) and elevated (100–200 µM) doses of TMZ as determined by a decrease in cells in G1. (B) TMZ response in CSC is similar to matched non-CSC cultures and is patient specific.
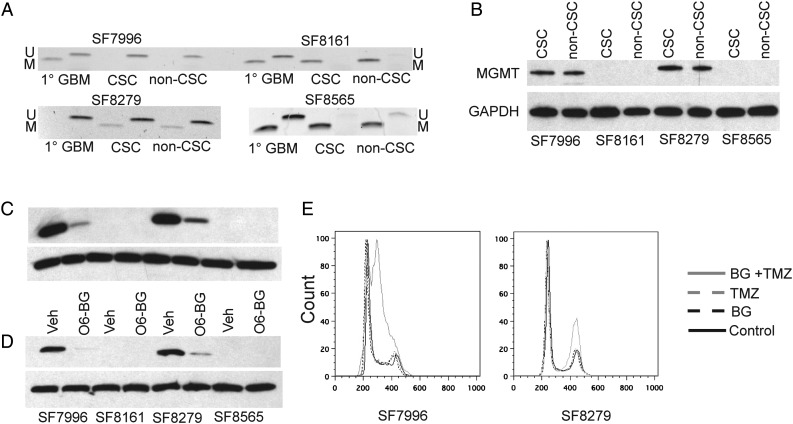
MGMT is a DNA repair protein that removes TMZ-induced methyl group adducts from O6-guanine.9,23 DNA methylation within the promoter and first exon/enhancer24 of MGMT is associated with closed chromatin, loss of SP1 transcription factor binding, and silencing of MGMT gene expression in many GBM cell lines25,26 and in some but not all studies correlates with reduced MGMT expression in tumor tissue.17 Hypermethylation of MGMT is associated with longer overall survival in GBM patients given XRT or the combination of TMZ treatment and XRT.10 We examined MGMT methylation status in primary GBM cell cultures to determine whether any association was evident between MGMT methylation and cell culture response to TMZ and whether the methylation status differed between corresponding CSC and non-CSC pairs. Using methylation-specific PCR, we found that the TMZ-responsive glioma cell lines U87, LN229, and U251 all displayed MGMT hypermethylation (Supplementary Fig. S3A), as previously described.27,28 Within each CSC and non-CSC cell culture pair, we observed similar TMZ response, MGMT methylation status, and MGMT expression (Fig. 3A, Supplementary Fig. S3B). Specifically, MGMT protein was detected in cultures with unmethylated MGMT but was not detected in cultures with MGMT hypermethylation (Fig. 3B), and cultures with hypermethylated MGMT showed the most substantial response to TMZ treatment. Pharmacologic inhibition of MGMT using the MGMT-specific inhibitor O6-benzylguanine29 resulted in a decrease of MGMT protein in both CSC (Fig. 3C) and matched non-CSC cultures (Fig. 3D) and, when used in combination with TMZ, resulted in cell cycle arrest compared with use of TMZ alone (Fig. 3E). Thus, our results demonstrate that MGMT methylation and protein expression are important determinants of response to TMZ in our GBM cultures, as shown for GBM patients and unpaired CSC or serum-grown cultures, while additionally discovering that these associations are not affected by the enrichment or depletion of the GBM CSCs.30–34
Fig. 3.
MGMT is an important determinant of response to TMZ in primary cultures of CSC and non-CSC. (A) Methylation-specific PCR of the MGMT locus for 4 primary GBM tumors and their patient-matched cultures; M, methylated; U, unmethylated. (B) Western blot analysis of MGMT in the same cultures as (A); GAPDH, glyceraldehyde 3-phosphate dehydrogenase. The presence of MGMT protein is associated with unmethylated MGMT and correlates with lack of response to TMZ. (C, D) Incubation of the primary cultures with O6-benzylguanine (BG) significantly reduces the level of MGMT protein in (C) MGMT-expressing CSC and (D) non-CSC cultures; Veh, vehicle. (E) Inhibition of MGMT protein with O6-benzylguanine in TMZ-resistant primary cultures results in cell cycle arrest after TMZ exposure.
Response of Cultures to Combined TMZ and XRT
Glioblastoma patients receive both TMZ and XRT, yet rarely has this combination treatment been studied. When this standard of care has been studied, long-term, serum-grown glioma cell lines have been used.35–37 To examine how primary cultured cells respond to the standard of care, and the extent to which they differ from established serum-grown cultures, we first examined the response of the 3 serum-grown, established glioma cell lines to a single dose of TMZ, XRT, or the combination of TMZ + XRT (Supplementary Fig. S4A). Cell cycle analysis at both 3 and 5 days posttreatment and cell viability analysis at 5 days posttreatment demonstrated that the glioma lines exhibited variable response to TMZ but were relatively nonresponsive to XRT (Supplementary Fig. S4B–E). We next expanded the analysis to 4 of our patient-matched culture pairs and 3 additional tumors with CSC cultures only. Surprisingly, there was no significant difference in response for TMZ, XRT, or the combination between most of the patient-matched cultures (Supplementary Fig. S4F–I). However, for 1 patient (#7996), the CSC culture had a modestly increased response to XRT compared with the matched non-CSC cultures in both assays. Furthermore, the combination of TMZ and XRT did not have an increased effect for the patient-matched cultures or for 3 unpaired CSC cultures (Supplementary Fig. S4J–M).
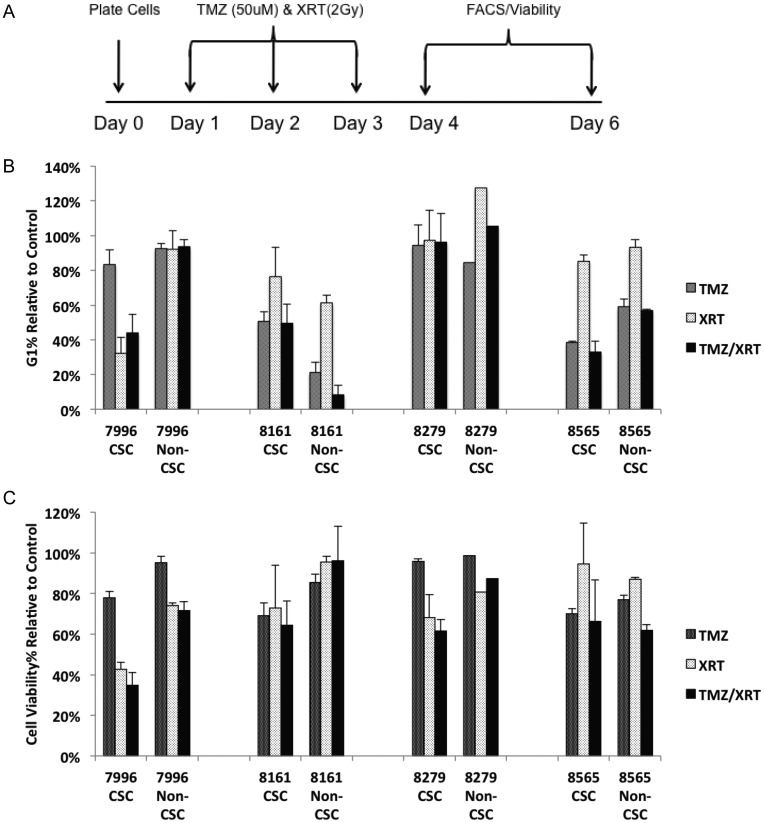
GBM patients typically receive TMZ and fractionated daily XRT for 6 weeks. To begin to recapitulate the in vivo therapeutic schedule, CSC and non-CSC matched cultures from 10 patients, including the 4 cultures used for single-dose TMZ or XRT, were treated daily with XRT or combined TMZ + XRT for 3 days (Fig. 4A, Supplementary Fig. S5). Most notably, the magnitude of response was larger for the multiday treatment compared with the single-dose treatment. However, the majority of patient-matched cultures again did not exhibit significant differences in response between CSC and non-CSC cultures. The non-CSC culture from 1 patient (#8161) was more responsive to TMZ than the CSC culture (similar to Fig. 2B) when assayed by cell cycle, but the 2 cultures had equivalent responses when assayed for cell viability. Finally, CSC cultures from 3 patients exhibited greater response to TMZ (#8008, #8010) or XRT (#7996, #8010) than their matched non-CSC cultures.
Fig. 4.
Response to XRT is more pronounced in a subset of CSC cultures relative to patient-matched non-CSC cultures. (A) The TMZ and XRT in vitro treatment regimen. (B, C) Response of a panel of patient-matched CSC and non-CSC cultures to TMZ, XRT, and TMZ + XRT as assayed by cell cycle analysis at (B) day 4 and cell viability analysis at (C) day 6 posttreatment.
We then examined whether the daily administration of combined TMZ + XRT resulted in an increased effect on response. Again, 3 individual CSCs in particular (#7996, #8008, and #8010) demonstrated a significantly greater response to the combination of TMZ + XRT compared with their matched non-CSCs (Fig. 4B and C), consistent with the single-dose experiments but inconsistent with the general concept that CSC-enriched, CD133-expressing, and tumorigenic cultures are more resistant to therapy than non-CSC and CD133– cells derived from the same tumor. For the remaining cultures there was very little additional effect of combined TMZ + XRT on either CSC or non-CSC cultures relative to treatment with TMZ or XRT alone. We also performed colony formation assays following XRT using the same treatment schedule, and the responses (data not shown) were consistent with cell cycle and cell viability assays. The data indicate that these 10 individual patient cultures were sensitive to either TMZ or XRT, and there is no additional antitumor effect resulting from combined treatment relative to the efficacious monotherapy. In GBM patients, combined TMZ + XRT provides a minimal increase (2.5 mo) in overall survival compared with XRT alone in unselected GBM patients, but a more significant increase (8.1 mo) in MGMT methylated subsets.10
Response to Therapy Is Independent of CD133 Status
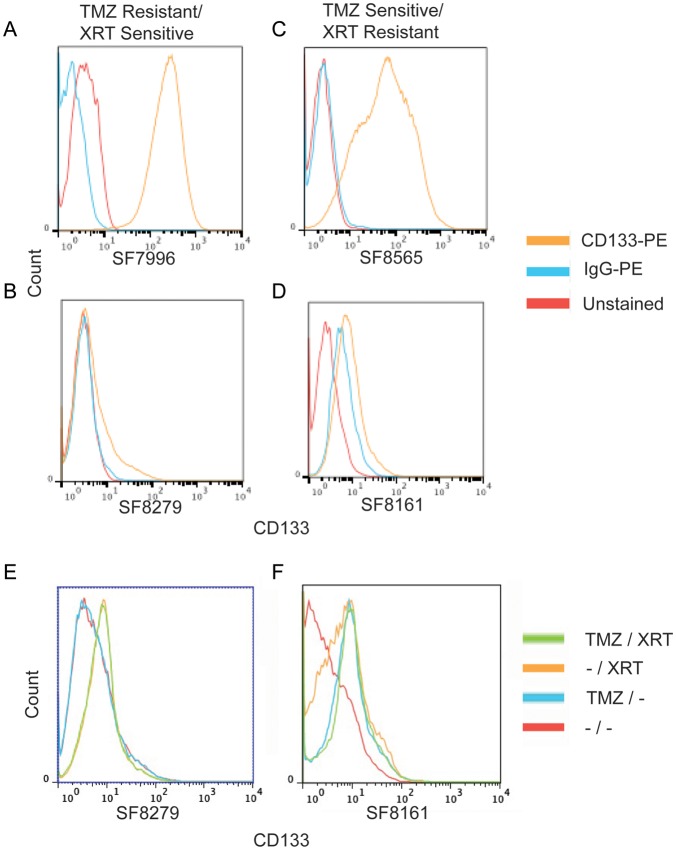
Previous studies have suggested that tumor cultures enriched for CD133-expressing CSCs are less sensitive to XRT13 and TMZ.38 In contrast, another study found that CD133+ sorted primary GBM stem cells were more sensitive to XRT compared with genetically unrelated cancer cell lines.15 In our individual CSC-enriched cultures, cell surface expression of CD133 ranged from 98.9% to 2% (Fig. 1C) of the cells in the culture, and the percent of cells expressing CD133 was stable over multiple passages for individual cultures (data not shown). However, no consistent association between CD133 expression and TMZ or XRT response was evident among the cultures we examined (Fig. 5A–D). CSC cultures that were more responsive to XRT than to TMZ had either high (Fig. 5A) or low (Fig. 5C) levels of CD133, as was the case for cultures that were more responsive to TMZ than to XRT, and showed high or low CD133 expression, respectively (Fig. 5B and D).
Fig. 5.
CD133 cell surface expression does not predict response to TMZ or XRT but increases after treatment. (A–D) CSC cultures were assayed for cell surface expression of CD133. PE, phycoerythrin. Examples of cultures responsive to XRT but not to TMZ are shown, with (A) high levels and (B) low levels of CD133 expression. Similarly, we also found cultures responsive to TMZ but not to XRT with (C) high levels and (D) low levels of CD133 expression. (E, F) CSC cultures with a constitutive low level of cell surface expression of CD133 exhibit modestly increased levels of CD133 expression after treatment with TMZ, XRT, or both.
Despite the lack of association between extent of CD133 expression and response to therapy, it is nonetheless possible that the CD133+ component within an individual culture may be more therapy resistant than the CD133– cell component. We therefore determined whether treatment with TMZ, XRT, or the combination affects the percent of cells expressing CD133. For cultures with high basal levels of CD133, there was no detectable effect of TMZ, XRT, or both on the percent of cells expressing CD133. However, the 2 cultures with little to no expression of CD133 exhibited a small increase in CD133 expression after either TMZ treatment or XRT (Fig. 5E and F). Given the short, 4- to 6-day time frame from TMZ treatment to assaying CD133 cell surface expression and the ∼2-day doubling time of these primary cells, it is very unlikely that the increase of CD133 is the result of a selective outgrowth of preexisting TMZ- or XRT-resistant CD133+ cells. Rather, it is attributable to a modestly increased expression of CD133 in cells with little or no expression. Thus, our results indicate that response to therapy is independent of the level of preexisting CD133 cell surface expression but that either TMZ treatment or XRT can increase the percentage of cells with detectable cell surface expression of CD133.
Discussion
In this study we examined patient-matched CSC and non-CSC cultures of early-passage primary GBM for their response to standard-of-care treatment. Contrary to previous reports, we found that GBM CSC cultures were not necessarily more resistant, and in some cases were more sensitive, to either TMZ or XRT than their non-CSC counterparts. Furthermore, high expression of CD133 was not correlated with tumor cell response to treatment. In cultures from several patients, only one therapeutic modality had a measurable effect. However, our in vitro studies do not address the regional intratumoral heterogeneity commonly found in GBM.39,40 It is possible that CSCs derived from one region of GBM might respond differently to TMZ and/or XRT than CSCs from another region. Thus, incorporating cultures initiated from different regions of one patient's tumor into future studies could provide a more in-depth understanding of an individual's response to therapy. Combined with a larger cohort, these studies could provide empirical evidence for precision medical approaches to therapeutic decisions, perhaps in combination with genomic analysis of the samples.
As one of the few prognostic molecular biomarkers for GBM patients, methylation of MGMT is associated with longer overall survival in patients receiving TMZ and XRT.10,41 However, exceptions to this relationship have been noted, such that individual patients with methylated MGMT may have short overall survival despite receiving standard-of-care treatment, and vice versa.10 Such observations as well as results from other avenues of investigation suggest that methylation of MGMT is not the sole determinant for predicting overall survival when GBM patients are treated with TMZ and that further analysis of individual tumor-specific characteristics may be useful. We sought to determine whether different culture conditions that yield phenotypically distinct cell populations influence the response of cells to TMZ. Our results provide new, additional support for the common speculation that MGMT methylation in patient samples is mechanistically linked to overall survival via suppressed MGMT expression and consequent tumor cell response to TMZ. Cells grown from different regions of a GBM tumor could have different response profiles, though a prior study concluded that there is little or no intratumoral heterogeneity of MGMT methylation in GBM.42 It will also be of interest to address the effects of sustained TMZ treatment and whether TMZ-responsive primary cultures acquire resistance to TMZ, as occurs in many patients over time.
Whereas MGMT methylation status did not correlate with culture type, it is possible that other epigenetic modifications segregate with CSCs. H3K4me1, H3K4me3, H3K36me3, H3K27me3, and H3K27ac patterns differed consistently between multiple pairs of CSC and patient-matched non-CSC cultures.8 Expanding these findings to a larger cohort of patient-derived cultures and including global DNA methylation analysis could unveil new epigenetic regulation of the CSC phenotype.
Previous work with GBM neurospheres in nonadherent cultures found that CD133+ fractions were more resistant to XRT than CD133– fractions.13 Here, we have used a complementary approach involving culture conditions that either enrich or deplete CSCs in primary adherent cultures established from patient tumors. We found that several CSC-enriched cultures exhibited increased rather than decreased response to XRT compared with matched CSC-depleted cultures. This was unexpected given that CD133+ cells sorted from GBM cancer cell lines are more resistant to XRT than the CD133– non–stem cell subpopulations from the same cell lines13 and given that our CSC populations were CD133+, while the non-CSCs were negative for CD133 on the cell surface. Furthermore, in a genetically engineered mouse model of GBM, treatment with TMZ appears to target the rapidly dividing cells but not the relatively quiescent, neural stem cell–like cells.11 In the context of previous studies, our results therefore suggest that adherent cultures of GBM CSCs are not more resistant to TMZ but in some cases are more responsive to XRT than matched non-CSCs, although substantial variation in response to XRT was evident across samples from different patients. This underscores the concept that inherent molecular differences among tumors are important in determining the extent of XRT response.
Exactly how the in vitro response to the combination of TMZ + XRT compares with the antitumor response in the same patients is quite difficult to address directly. A recent study found that xenografts from CSCs were more radiation resistant than the same CSCs grown in vitro due to their decreased susceptibility to the induction of DNA double strand breaks and enhanced ability to repair breaks.43 Another study compared CD133+ and CD133– cells in intracranial xenografts and found that when given XRT, the CD133+ cells had fewer γH2AX and 53BP1 foci, measures of radiation response, compared with the CD133– cells.44 Neither of these studies tested the standard-of-care regimen, however. Thus, it is likely that the cellular environment in vivo influences the relative response of CSCs versus non-CSCs. While our study did not address how CSC- and non-CSC–derived xenografts respond to XRT, our results do support equivalent XRT responsiveness in CSC-enriched versus CSC-depleted GBM cultures with presumably very similar genetic backgrounds. It will be very interesting to determine whether the response of CSC- versus non-CSC–derived xenografts to TMZ, XRT, or the combination differs from the response observed in vitro.
Patient primary cultures have been used for many years in an attempt to empirically predict patient response to multiple classes of therapeutics, which represents an attractive approach for drug screening for the purpose of “personalizing” patient treatment.4,22,45,46 Primary cultures could also be beneficial for the examination of the response spectrum of many combinations of therapeutic agents. Our initial therapeutic testing of primary cultures is completed in the typical time interval between surgery and the onset of adjuvant therapy. It remains important to determine the most appropriate tumor culture condition in which to perform these studies.
GBM CSCs reproduce gene expression profiles and tumor pathology of corresponding primary tumors more faithfully than do established glioma lines or serum-derived primary lines.3,4 Results presented here support the use of CSCs to preserve GBM cell tumorigenicity and invasive growth when implanted in the brains of athymic mice. The preservation of tumor genetic, molecular, and phenotypic characteristics could be instrumental in developing model systems from which drug screening results would more successfully translate to a clinical setting.
Supplementary Material
Funding
This work was supported by an institutional training grant from the National Institutes of Health (T32 CA108462-07) to S.D.F. and grants from Accelerated Brain Cancer Cure, the Goldhirsh Foundation, and the Grove Foundation to J.F.C. J.F.C. is supported by the Karen Osney Brownstein Endowed Chair in Molecular Neuro-Oncology.
Supplementary Material
Acknowledgments
We thank Dr Mitchell Berger, Dr Andrew Parsa, Dr Manish Aghi, and Dr Michael McDermott for providing surgical specimens, Dr Michael Barnes and Dr Andrew Bollen for neuropathological analysis of xenograft sections, Cynthia Cowdrey and Alvin Au from the Brain Tumor Research Core for help in acquiring primary tissue, Jon Woo and the Genomics Core Facility in the Institute for Human Genetics for DNA fingerprint analysis, and members of the Costello lab for helpful discussions.
Conflict of interest statement. None declared.
References
- 1.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 2.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 3.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 4.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Laks DR, Masterman-Smith M, Visnyei K, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27(4):980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panosyan EH, Laks DR, Masterman-Smith M, et al. Clinical outcome in pediatric glial and embryonal brain tumors correlates with in vitro multi-passageable neurosphere formation. Pediatr Blood Cancer. 2010;55(4):644–651. doi: 10.1002/pbc.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rheinbay E, Suva ML, Gillespie SM, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Reports. 2013;3(5):1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6(7):2585–2597. [PubMed] [Google Scholar]
- 10.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beier D, Schulz JB, Beier CP. Chemoresistance of glioblastoma cancer stem cells—much more complex than expected. Mol Cancer. 2011;10:128. doi: 10.1186/1476-4598-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 14.Beier D, Rohrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68(14):5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 15.McCord AM, Jamal M, Williams ES, Camphausen K, Tofilon PJ. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin Cancer Res. 2009;15(16):5145–5153. doi: 10.1158/1078-0432.CCR-09-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61(5):1957–1963. [PubMed] [Google Scholar]
- 17.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59(4):793–797. [PubMed] [Google Scholar]
- 18.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 20.Roos WP, Batista LF, Naumann SC, et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26(2):186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 21.Ostermann S, Csajka C, Buclin T, et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin Cancer Res. 2004;10(11):3728–3736. doi: 10.1158/1078-0432.CCR-03-0807. [DOI] [PubMed] [Google Scholar]
- 22.Rosenblum ML, Gerosa M, Dougherty DV, et al. Age-related chemosensitivity of stem cells from human malignant brain tumours. Lancet. 1982;1(8277):885–887. doi: 10.1016/s0140-6736(82)92154-7. [DOI] [PubMed] [Google Scholar]
- 23.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50(19):6119–6129. [PubMed] [Google Scholar]
- 24.Harris LC, Remack JS, Brent TP. Identification of a 59 bp enhancer located at the first exon/intron boundary of the human O6-methylguanine DNA methyltransferase gene. Nucleic Acids Res. 1994;22(22):4614–4619. doi: 10.1093/nar/22.22.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14(10):6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269(25):17228–17237. [PubMed] [Google Scholar]
- 27.Mueller W, Nutt CL, Ehrich M, et al. Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene. 2007;26(4):583–593. doi: 10.1038/sj.onc.1209805. [DOI] [PubMed] [Google Scholar]
- 28.van Nifterik KA, van den Berg J, van der Meide WF, et al. Absence of the MGMT protein as well as methylation of the MGMT promoter predict the sensitivity for temozolomide. Br J Cancer. 2010;103(1):29–35. doi: 10.1038/sj.bjc.6605712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc Natl Acad Sci U S A. 1990;87(14):5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20(9):2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 31.Pepponi R, Marra G, Fuggetta MP, et al. The effect of O6-alkylguanine-DNA alkyltransferase and mismatch repair activities on the sensitivity of human melanoma cells to temozolomide, 1,3-bis(2-chloroethyl)1-nitrosourea, and cisplatin. J Pharmacol Exp Ther. 2003;304(2):661–668. doi: 10.1124/jpet.102.043950. [DOI] [PubMed] [Google Scholar]
- 32.Barvaux VA, Ranson M, Brown R, McElhinney RS, McMurry TB, Margison GP. Dual repair modulation reverses temozolomide resistance in vitro. Mol Cancer Ther. 2004;3(2):123–127. [PubMed] [Google Scholar]
- 33.Hermisson M, Klumpp A, Wick W, et al. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96(3):766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]
- 34.Spiegl-Kreinecker S, Pirker C, Filipits M, et al. O6-methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro Oncol. 2010;12(1):28–36. doi: 10.1093/neuonc/nop003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Nifterik KA, Van den Berg J, Slotman BJ, Lafleur MV, Sminia P, Stalpers LJ. Valproic acid sensitizes human glioma cells for temozolomide and gamma-radiation. J Neurooncol. 2012;107(1):61–67. doi: 10.1007/s11060-011-0725-z. [DOI] [PubMed] [Google Scholar]
- 36.Barazzuol L, Jena R, Burnet NG, et al. Evaluation of poly (ADP-ribose) polymerase inhibitor ABT-888 combined with radiotherapy and temozolomide in glioblastoma. Radiat Oncol. 2013;8:65. doi: 10.1186/1748-717X-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrabi S, Combs SE, Brons S, Haberer T, Debus J, Weber KJ. Temozolomide in combination with carbon ion or photon irradiation in glioblastoma multiforme cell lines—does scheduling matter? Int J Radiat Biol. 2013;89:692–697. doi: 10.3109/09553002.2013.791406. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton MG, Roldan G, Magliocco A, McIntyre JB, Parney I, Easaw JC. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102(2):255–260. doi: 10.1007/s11060-010-0307-5. [DOI] [PubMed] [Google Scholar]
- 43.Jamal M, Rath BH, Williams ES, Camphausen K, Tofilon PJ. Microenvironmental regulation of glioblastoma radioresponse. Clin Cancer Res. 2010;16(24):6049–6059. doi: 10.1158/1078-0432.CCR-10-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamal M, Rath BH, Tsang PS, Camphausen K, Tofilon PJ. The brain microenvironment preferentially enhances the radioresistance of CD133(+) glioblastoma stem-like cells. Neoplasia. 2012;14(2):150–158. doi: 10.1593/neo.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenblum ML, Wheeler KT, Wilson CB, Barker M, Knebel KD. In vitro evaluation of in vivo brain tumor chemotherapy with 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 1975;35(6):1387–1391. [PubMed] [Google Scholar]
- 46.Rosenblum MK, Knebel KD, Vasquez DA, Wilson CB. Brain-tumor therapy: quantitative analysis using a model system. J Neurosurg. 1977;46(2):145–154. doi: 10.3171/jns.1977.46.2.0145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.