Abstract
Background
Diffuse low-grade gliomas (LGGs) form a heterogeneous subgroup of gliomas in adults. Chromosome (chr) arms 1p/19q codeletion and IDH mutation have been shown to be closely associated with oligodendroglial phenotype and better prognosis. We sought to identify relevant biomarkers in non 1p/19q codeleted LGGs.
Methods
We characterized a retrospective series of 126 LGGs using genomic arrays, microsatellite analysis, IDH sequencing, MGMT promoter methylation assay, and p53 expression analysis.
Results
Our study confirms that 1p/19q codeletion, mutually exclusive with p53 overexpression, was associated with: (i) better prognosis, (ii) oligodendroglial phenotype, (iii) MGMT promoter methylation, and (iv) IDH mutation. Interestingly, 1p/19q codeleted tumors occur in older patients at diagnosis. Our study shows that non 1p/19q codeleted LGGs can be divided in 5 main genomic subgroups: (i) 11p loss, (ii) 19q loss (iii) 7 gain, (iv) 19 gain, and (v) unclassified. In non 1p/19q codeleted LGGs, we demonstrated that (i) 11p loss is associated with astrocytoma phenotype and has an independent negative prognostic value, and (ii) 19q loss diminished the favorable prognostic value of IDH mutation. Our findings were validated in an independent cohort of 98 LGGs.
Conclusion
Novel genomic entities and biomarkers have been identified in non 1p/19q codeleted LGGs. Our findings may help to stratify non 1p/19q codeleted LGGs, facilitating future individualization of treatment. Further prospective studies are warranted to support our findings.
Keywords: biomarker; genomic array; low-grade gliomas; IDH; MGMT, TP53
Diffuse low-grade gliomas (LGGs, WHO grade II) in adults form a very heterogeneous group of neoplasms in terms of pathological and clinical features.1,2 They include diffuse WHO grade II astrocytomas, oligodendrogliomas, and oligoastrocytomas. Most often, LGGs are slow growing tumors. However, they inevitably recur and progress to higher-grade tumors (ie, anaplastic gliomas/WHO grade III and glioblastomas/WHO grade IV).2,3 The current classification of LGGs is based on morphological criteria edited by the WHO.2 In practice, diagnosis of LGGs remains challenging, with significant intra- and interobserver variability.4 In addition, these tumors have a broad range of survival that varies from 1 to 30 years.5
Over the past decades, several molecular markers with clinical relevance have been identified in LGGs. They might help to establish a histomolecular classification of LGGs:6–8 (i) chr arms 1p/19q codeletion;6,9,10 (ii) isocitrate dehydrogenase (IDH) 1 and 2 mutations,8,10,11 (iii) TP53 mutation/p53 overexpression,11,12 and (iv) MGMT promoter methylation.13,14 Non 1p/19q codeleted LGGs have not been perfectly characterized so far but their characterization is likely to include several genomic groups. Triple-negative tumors (ie, without p53 overexpression, 1p/19q codeletion, and IDH mutation) have recently been described as a tumor group with worse prognosis.12,15 In order to decipher non 1p/19q codeleted LGGs at a molecular level, we performed a genome-wide analysis in a cohort of 126 LGGs and validated our results in an independent cohort of 98 samples.
Material and Methods
Patients and Tumors
The following criteria were used to include cases and tumors in the present study: (i) aged 18 years or older at pathological diagnosis, (ii) diagnosis of supratentorial LGG (astrocytoma, oligodendroglioma, or oligoastrocytoma) according to the WHO classification at first surgery,2 (iii) detailed clinical information at diagnosis and during follow-up, (iv) availability of paired blood and fresh frozen tumor samples, (v) informed consent from participants for molecular analysis, and (vi) genomic profiling of the tumor DNA by bacterial artificial chromosome (BAC)-array based comparative genomic hybridization (BAC-aCGH). All tumors were centrally reviewed by 2 neuropathologists (K.M. and H.A.), who were blinded for molecular and clinical data.
All patients received conventional therapy consisting of surgical resection as extensive as clinically and technically possible. Surgery was followed by radiotherapy and/or alkylating-based chemotherapy (nitrosourea or temozolomide) at unequivocal clinical and/or radiological tumor progression.
DNA Extraction and BAC-aCGH
DNA extraction was performed using DNeasy Mini kit (Qiagen) according to the manufacturer's recommendations. DNA concentration and quality were determined by spectrophotometry (NanoDrop®). All samples had high-quality genomic DNA with an A260-A280 ratio purity of 1.8:2. One-megabase resolution BAC-aCGH experiments were conducted as described previously.16
MGMT Promoter Methylation Analysis
The EZ DNA Methylation Gold Kit (Zymo Research) for bisulfite conversion of DNA was used for epigenomic analyses. Starting amount of DNA was 300 ng, and all modification reactions were performed according to manufacturer's instructions. PCR amplification and high-resolution melting analysis were carried out in LighCycler480 (Roche®) using the following primer sequences: 5′-CGTTTGCGATTTGGTGAGTGTT-3′ and 5′-CCTACAAAACCACTCGAAACTACCA-3′. Amplification consisted of 10 minutes at 95°C, followed by 50 cycles of 15 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. After amplification, a postamplification melting curve program was initiated by heating to 95°C for 1 minute, cooling to 70°C for 30 seconds, and increasing the temperature to 95°C (heating rate 0.01°C/s) while continuously measuring fluorescence. Artificial methylation using CpG Methylase M.ssI M0226S (New England Biolabs®) was performed according manufacturer's instructions and used as a positive control. Peripheral blood DNA was considered negative control. When the peak corresponding to methylated DNA was >50% of the corresponding unmethylated DNA peak, the sample was considered methylated, as previously described.17 All reactions were performed in duplicate.
p53 Expression
Paraffin-embedded tissue slices, 10 μm in thickness, were immunostained. Monoclonal antibody anti-p53 (clone DO-7) from DAKO® was used at a titer of 1:50. A tumor was scored positive if >10% of tumor cells showed strong nuclear staining, as reported previously.15
IDH1 and IDH2 Mutation Analysis
IDH1 and IDH2 were assessed by Sanger sequencing, as previously described.10
Microsatellite Markers Analysis
Microsatellite polymorphisms analysis was performed. as previously described.18 Chr arms 1p/19q status was identified using BAC-aCGH and validated by microsatellite polymorphisms analysis using the following markers: D1S450, D1S2667, D1S234, D1S2890, D1S2841, D19S425, D19S219, D19S412, and D19S418. In addition, loss of 11p detected by BAC-aCGH was validated using D11S922, D11S4088, D11S4096, D11S4200, and D11S4083.
Biostatistical and Bio-informatics Analysis
Statistical analysis was done using the statistical software R (http://www.r-project.org/). Mann-Whitney U, χ2, and Fisher' exact tests were used to test for association between clinical variables and molecular alterations. False discovery rate method19 was used to adjust P values for multiple testing.
BAC-aCGH was normalized with the R package MANOR20 and analyzed with the R package GLAD21 using default parameters and visualized using Nexus® Copy Number software. Data matrices were generated, containing the imbalance status (“0”, “1”, and “-1” indicates copy neutral, gain, and loss, respectively) for each BAC. From this data matrix, the frequency of gains and losses was represented by histograms. Minimal common regions (MCR) of chr imbalances were analyzed using significance testing for aberrant copy number (STAC) with default parameters. Footprint P value of STAC algorithm was selected to report nonrandom genomic copy number aberrations.22 Hierarchical clustering of non 1p/19q codeleted LGGs was performed with a modification of the R package gplots using Ward agglomeration and Euclidean distance. Genomic aberrations observed in ≥ 5% of tumors were selected as recurrent. Agreement between BAC-aCGH and microsatellite markers analysis was assessed using Fleiss κ test. A κ value of <0 indicated poor agreement, while a κ value of 0.81–1.00 indicated good agreement.23
Log-rank test was used for survival comparisons. Progression-free survival (PFS) was calculated from surgery to tumor progression, death, or end of follow-up. Overall survival (OS) was calculated from surgery to death or end of follow-up. The effect of each individual molecular marker and genomic group on OS was investigated using the Cox proportional hazard model (P < .1 was considered to be the inclusion criterion for factors that could be included into the multivariate analysis): surgery (resection vs biopsy), phenotype (oligodendroglial vs astrocytoma), MGMT promoter methylation status, and IDH mutation. The model was designed to take each of the factors into account without considering interaction terms. A 2-sided P value of <.05 was considered statistically significant.
The Validation Cohort
The validation cohort included 98 LGGs (40 astrocytomas, 15 oligoastrocytomas, and 43 oligodendrogliomas) and was provided by van Thuijl, HF et al (unpublished data). All patients were >18 years old, treatment naïve, deceased, or when still alive, a follow-up of more than 6 years. The pathological diagnosis was confirmed by an experienced pathologist (P.W.). Formalin-fixed paraffin-embedded material was used for copy number aberrations using Massively Parallel Sequencing analysis (Illumina HiSeq2000). Shallow whole genome sequencing was performed with a sequence depth of 0.4X of the genome, as previously described.24 The IDH1 and IDH2 mutation status, obtained by direct sequencing, was available for all samples with the exception of one (technical failure).
Results
Clinical Data
The initial population included 150 LGG samples according to local diagnosis. All patients were treated and followed in Pitié-Salpêtrière Hospital and were diagnosed between 2000 and 2010. After central review by 2 neuropathologists (H.A. and K.M.), 126 neoplasms (53 oligodendrogliomas, 50 oligoastrocytomas, and 23 astrocytomas) exhibiting LGG histology were confirmed and included in the present study. Clinical characteristics of patients are summarized in Table 1.
Table 1.
Progression-free survival and overall survival according to clinical, pathological, and molecular features of tumors and patients
| Variable | Category | Number | Median PFS (95% CI) years | Log-rank P value | Median OS (95% CI) years | Log-rank P value |
|---|---|---|---|---|---|---|
| Age | ≤ 40 | 68 | 3.6 (2.9–4.2) | .8 | 6.5 (6–7) | .4 |
| > 40 | 58 | 2.9 (2.1–3.6) | 5.8 (3.4–8.2) | |||
| Sex | Male | 67 | 3 (2–4) | .13 | 6.9 (5–8.8) | .25 |
| Female | 59 | 3.5 (2.6–4.5) | 6.4 (5.2–7.6) | |||
| KPS | ≥ 80 | 117 | 3.5 (2.9–4.1) | .09 | 6.5 (5.3–7.7) | .7 |
| < 80 | 6 | 2.7 (0.9–4.5) | NR | |||
| Surgery | Resection | 92 | 3.5 (2.9–4.2) | .2 | 6.5 (5.5–7.5) | .056 |
| Biopsy | 34 | 2.7 (2.1–3.3) | 6.9 | |||
| Phenotype | Oligodendroglial | 103 | 3.5 (2.9–4.1) | .2 | 6.9 (6.2–7.5) | .06 |
| Astrocytic | 23 | 2 (1.8–2.3) | 5 (3.7–6.4) | |||
| IDH mutation | No | 14 | 2.3 (0.9–3.8) | .029 | 4.9 (0.6–9.1) | .006 |
| Yes | 103 | 3.6 (2.9–4.2) | 6.9 (5.9–7.9) | |||
| 1p/19q codeletion | No | 94 | 3 (2.2–3.9) | .024 | 5.5 (4.4–6.5) | .0002 |
| Yes | 32 | 4 (1.5–6.4) | NR | |||
| p53 overexpression | No | 86 | 3.2 (2.5–3.9) | .5 | 6.5 (5.7–7.4) | .7 |
| Yes | 34 | 2 (0.3–3.8) | NR | |||
| MGMT promoter methylation | No | 29 | 2.3 (1.2–3.5) | .1 | 4.5 (1.4–7.6) | .004 |
| Yes | 88 | 3.4 (2.9–3.8) | 6.9 (5.6–7.6) |
Abbreviations: KPS, Karnosfky performance status; N, number; NR, not reached; OS, overall survival; PFS, progression-free survival
Median age and median Karnofsky Performance Status (KPS) of patients at diagnosis was 38.8 years (interquartile range [IQR] 30.6%–46.3%) and 90 (IQR 90%–100%) respectively. The sex ratio male-to-female was 1:1. The median PFS and OS for all cases was 3.2 years (95% CI, 2.6–3.8) and 6.5 years (95 CI%, 5.6–7.5), respectively. The median follow-up was 4.5 years (95% CI, 3.4–5.2).
The prognostic significance of clinical features of patients and pathological characteristics of tumors is summarized in Table 1. Sex, age categorized (≤ or >40 years), and KPS did not have statistically significant prognostic value (Table 1). It should be noted that using a cut-off of 50 years yielded the same results in univariate analysis (data not shown).25,26 Oligodendroglial tumors (oligodendrogliomas and oligoastrocytomas) had a trend towards better prognosis (Table 1). Distribution of age, sex, KPS, and surgical procedure among the different histological subgroups was similar (data not shown). Surgical procedure (subtotal or partial resection) showed a trend towards better prognosis in terms of OS compared with biopsy (Table 1).
Molecular Data
The molecular alterations detected in the training cohort of LGG were as follows: (i) 28.3% (34/120) overexpressed p53, (ii) 88% (103/117) IDH (IDH1 or IDH2) mutated, and (iii) 75.2% (88/117) MGMT promoter methylated. Molecular alterations observed within histological subgroups are presented in Table 2. None of these aberrations was associated with age, KPS, or extent of surgical resection (data not shown).
Table 2.
Molecular characteristics according to histological phenotype
| Oligodendroglioma n = 53 (%) | Oligoastrocytoma n = 50 (%) | Astrocytoma n = 23 (%) | P value | |
|---|---|---|---|---|
| 1p/19q codeletion | 25/53 (47.2) | 5/50 (10) | 2/23 (8.7) | 1.06e-05 |
| P53 overexpression | 8/51 (15.7) | 19/49 (38.8) | 7/20 (35) | .02 |
| IDH mutation | 48/53 (90.6) | 37/43 (86) | 18/21 (85.7) | .7 |
| MGMT promoter methylation | 36/50 (72) | 37/47 (78.7) | 5/20 (75) | .7 |
Chr arms 1p/19q codeletion was associated with (i) better prognosis in terms of PFS and OS, (ii) oligodendroglial phenotype, (iii) IDH mutation, and (iv) MGMT promoter methylation (Tables 1–3). p53 overexpression was associated with astrocytic phenotype (P = .02) but did not impact prognosis (Tables 1–3). MGMT promoter methylation was associated with prolonged OS only in univariate analysis (P = .004, 4.5 years vs 6.9 years) and with IDH mutation (P = .005) (Tables 1–3). IDH mutation was positively associated with favorable OS (P = .006) and better PFS (P = .029) in univariate analysis (Tables 1–3).
Table 3.
Pairwise associations between molecular alterations
| Variable | 1p/19q codeletion | P53 overexpression | IDH mutation | MGMT promoter methylation |
|---|---|---|---|---|
| 1p/19q codeletion | −0.03 | +++0.01* | 0.1 | |
| P53 overexpression | −0.03 | 0.1 | 0.6 | |
| IDH mutation | +++0.01* | 0.1 | ++0.005* | |
| MGMT promoter methylation | 0.1 | 0.6 | ++0.005* |
*Significant at P value <.05. Positive association: + (OR 1.01-3); ++ (OR 3.01-5); +++ (OR 5.01-infinity). Negative association: - (OR 0.99–0.25).
Genome-wide Analysis
Recurrent gains were identified on chr arms 7q (19%); 7p (14%); 19p (12%); 22q (11%); 8p (9%); 11q and 20q (8% each); 8q (7%); 19q (6.3%); 16p (6%); and 11p (8.8%). Recurrent losses were found on chr arms 19q (48%); 1p (30%); 17p (12%); 4p (13%); 10p, 19p, and 22q (10% each); 10q (9%); 17q (8%); 18p (7%); 9p, 13q, and 16p (6% each); 4p (6%); 15q (6%); and 11p (7.2%) (Fig. 1).
Fig. 1.
Summary of frequencies of genomic imbalances detected by BAC-aCGH in the overall population of 126 low-grade gliomas. Losses are depicted in red on the left and gains in blue on the right.
Hierarchical Cluster Analysis of Low-grade Gliomas
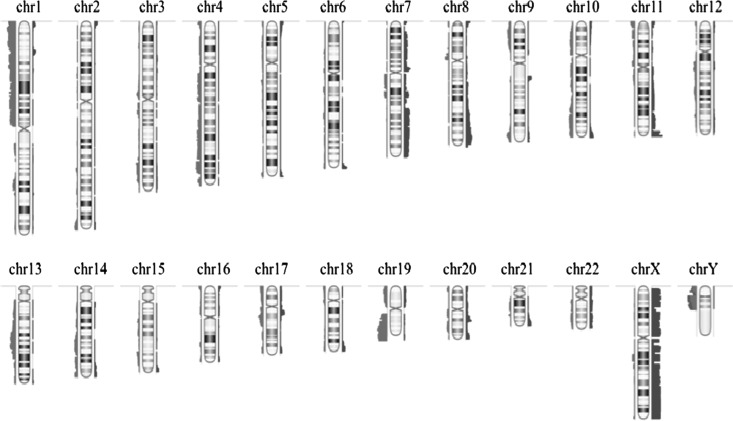
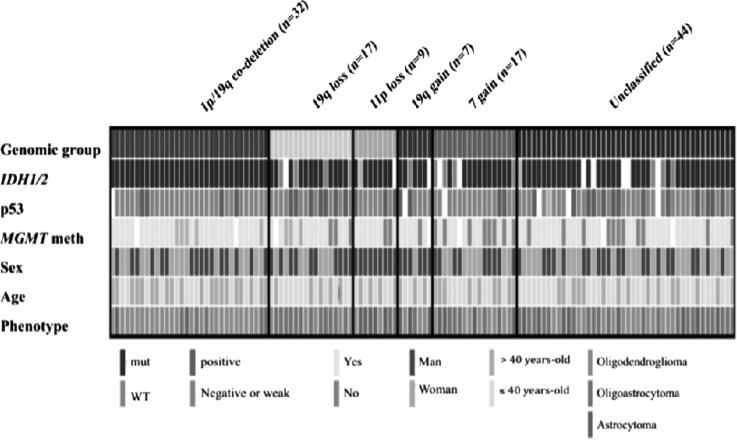
The overall population was classified using supervised hierarchical clustering (Fig. 2). In order to identify genomic groups with clinical relevance, genomic aberrations were correlated with clinical (survival), pathological, and molecular parameters (p53 overexpression, IDH mutation, and MGMT promoter methylation). Since 1p/19q codeletion is a well-established chr aberration, we considered this genomic aberration as a group. The most frequent genomic aberrations (observed in ≥5% of LGGs) were selected. Chr copy number aberrations that clustered closely on the unsupervised clustering (Supplementary Fig. S1) were chosen and, after visual inspection of each genomic profile, genomic profiles were reclassified in supervised manner to build homogeneous genomic groups (Fig. 2). The selected genomic aberrations were (i) 1p/19q codeletion with pericentromeric breakpoints (25.4%), (ii) 19q loss (13.5%), (iii) 11p loss (7.2%), (iv) 7 gain (13.5%), and (v) 19 gain (5.5%). Tumors with neither 1p/19q codeletion nor aforementioned genomic aberrations were termed unclassified (34.9%). Then, these genomic aberrations were also selected in the validation cohort. Copy number aberrations detected using BAC-aCGH were validated by microsatellite analysis with a good agreement for 1p/19q codeletion and 11p loss (κ = 0.92 and 0.9, respectively). The distribution of molecular characteristics within genomic groups is presented in Fig. 3. The clinical characteristics according to the genomic groups are reported in Table 4. Age, KPS, surgical procedure, and sex distribution were equally distributed among genomic subgroups identified in the group of non 1p/19q codeleted LGGs (Table 4).
Fig. 2.
Supervised genomic subgroups in the entire series. Each row indicates a genomic locus grouped within chromosome arms, and each column indicates a sample. The first row indicates the genomic group.
Fig. 3.
Summary of molecular and histological characteristics of each genomic group. Abbreviations: meth, methylation; mut, mutation; WT, wild-type.
Table 4.
Clinical and molecular features for each genomic group
| Variable | 1p/19q Codeletion | 19q Loss | 11p Loss | 7 Gain | 19q Gain | Un classified | P value* |
|---|---|---|---|---|---|---|---|
| n = 126 (%) | 32 | 17 | 9 | 17 | 7 | 44 | |
| Type (O/M/A) (%) | 78/16/6 | 29/70/0 | 22/22/56 | 48/29/23 | 29/29/42 | 25/54/21 | .00001* |
| Age, median (years) | 42 | 35 | 43 | 40 | 41 | 33 | .02 |
| Sex (M/F) | 0.8 | 1.4 | 2 | 1.4 | 6 | 0.9 | NS |
| KPS, median (%) | 90 | 90 | 90 | 90 | 90 | 90 | NS |
| Resection/Biopsy (%) | 75/25 | 65/35 | 77/21 | 65/35 | 75/25 | 77/23 | NS |
| OS, years median (years) | 12.1 | 3.5 | 5.2 | 3.9 | 8.4 | 5.3 | .0002* |
| PFS, years median (years) | 6.6 | 2.2 | 2.7 | 2.2 | 3 | 3.5 | .02 |
| IDH mutation (%) | 100 | 75 | 89 | 77 | 83 | 90 | NS |
| p53 overexpression (%) | 13 | 47 | 35 | 19 | 33 | 37 | .0001* |
| MGMT promoter methylation (%) | 87 | 69 | 100 | 50 | 83 | 75 | .06 |
*Significant at P <.05.
Abbreviations: F, female; KPS, Karnofsky Performance Status;M, male; NS, not significant; O/M/A, oligodendroglioma/oligoastrocytoma or mixed/astrocytoma; OS, overall survival; PFS, progression-free survival
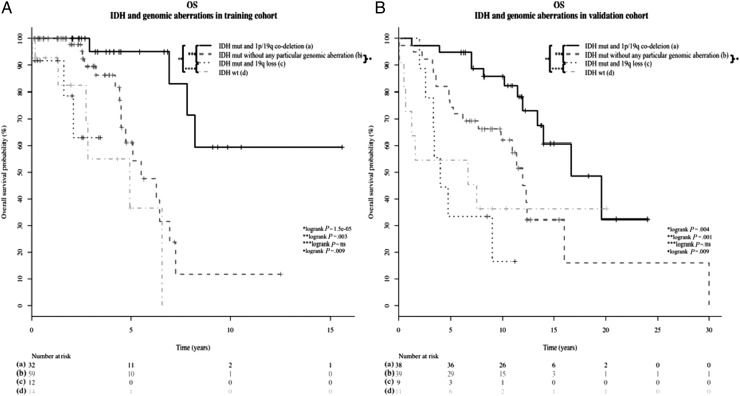
1p/19q codeleted LGGs had oligodendroglial phenotype (30/32, 94%), good prognosis, and tended to be older at diagnosis (42 vs 37 years, P = .03) compared with LGGs harboring other genomic patterns (Tables 2 and 4, Supplementary Fig. S2). LGGs with 19q loss exhibited oligodendroglial phenotype (oligodendroglioma or oligoastrocytoma) (17/17, 100%; among them, 12/17 were oligoastrocytomas) and IDH mutation (12/16; 75%) (Table 4). Interestingly, 11p loss was predominantly found in astrocytomas (56%, 5/9 were astrocytomas in 11p loss vs 15%, 18/117 in LGGs harboring other genomic aberrations; P = .01; Table 4). The MCR of loss in 11p-deleted LGG samples was localized between 11p15.5 and 11p15.4 cytobands (region 11:442.341–5.376.935, 5Mbp), STAC footprint P value = .009. This region included 13 BAC probes and 175 genes (eg, IGF2, CD81, CDKN1C, NAP1L4, CARS, and NUP98, Supplementary Table S1). Finally, LGGs with chr 7 gain had poor prognosis (Table 4). Within non 1p/19q codeleted tumors, IDH mutation was associated with better prognosis compared with IDH wild-type. Interestingly, tumors in our cohort with co-occurrence of IDH mutation and 19q deletion (75%, 12/16) had similar prognosis as IDH wild-type LGG (Fig. 4, Panels A and B).
Fig. 4.
Kaplan-Meier curves comparing overall survival (OS) in (a) IDH mutated and 1p/19q codeleted low-grade glioma (LGG) versus (b) IDH mutated without 1p/19q codeletion LGG versus (c) IDH mutated and 19q lost LGG versus (d) IDH wild-type LGG The comparison was conducted in the training cohort (Panel a) and in the validation cohort (Panel b). Abbreviations: mut, mutation; ns, non-significant ; WT, wild-type.
LGGs classified as triple negative (negative for p53 expression, IDH mutation, and 1p/19q codeletion)12,15 (n = 12) had a significantly worse prognosis when analyzing OS (P = .03), and a trend was shown for PFS (P = .07) (Supplementary Fig. S3, Panels A and B). However, from the genomic point of view, these tumors were highly heterogeneous and harbored several genomic aberrations (Fig. 3).
Two gene amplifications were detected (2/126; 1.6%): (i) MDM4 amplification in the chr 19 gain group (1/7; 14.3%) and (ii) EGFR amplification in the unclassified group (1/44; 2.2%).
Multivariate Analysis for Outcome
After adjusting for the confounding factors described in the biostatistical and bio-informatics analysis section, 19q loss without concurrent 1p loss on the one hand and 11p loss on the other hand had statistically significant unfavorable prognostic value (Table 5).
Table 5.
Cox proportional hazards ratio for overall survival
| Factor | Hazard ratio |
Death | |
|---|---|---|---|
| 95% CI | P value | ||
| Genomic group | |||
| 1p/19q Codeletion | 1 | ||
| 19q loss | 20.8 | 3.2–136 | 0.001 |
| 11p loss | 5.7 | 1.1–30.4 | 0.04 |
| 7 gain | 6.2 | .9–40.1 | 0.054 |
| 19q gain | 9.8 | 1.6–60.3 | 0.013 |
| Unclassified | 7.5 | 3.5–126 | 0.006 |
| Histological phenotype | |||
| Astrocytoma | 1 | ||
| Oligodendroglial | 0.4 | .2–1.003 | 0.051 |
| IDH mutation | |||
| Yes | 1 | ||
| No | 0.5 | .2–1.3 | 0.4 |
| Surgery | |||
| Biopsy | 1 | ||
| Resection | 0.27 | .1–0.8 | 0.04 |
| MGMT promoter methylation | |||
| No | 1 | ||
| Yes | 0.4 | .1 to 1.3 | 0.1 |
In bold, P values of variables that have been confirmed in the validation cohort.
Validation Cohort
The selected genomic aberrations with clinical value in the training cohort of LGGs were confirmed in the independent validation cohort of 98 LGGs. The distribution of genomic aberrations in the validation cohort was similar to the training cohort as shown in Supplementary Fig. S4. For example, the 1p/19q codeletion distribution according to the histological phenotype was similar in both cohorts of LGGs. Indeed, in the validation cohort of LGGs, 2 of 38 were astrocytomas, 3 of 38 were oligoastrocytomas, and 33 of 38 were oligodendrogliomas. As mentioned previously in the training cohort, cases with 1p/19q codeleted LGGs were also significantly older at diagnosis in the validation cohort (41 vs 35 years; P = .01; Supplementary Fig. S2, panel B). Similarly, the frequencies of IDH mutations were virtually the same in both cohorts of LGGs (89.7%; 87/97 of LGGs in validation cohort were IDH-mutated; Supplementary Fig. S4). In summary, the validation cohort and the training cohort shared similar characteristics.
We have also identified the same genomic groups within the validation cohort: (i) 1p/19q codeletion (38.8%) with centromeric breakpoints, (ii) 19q loss (9.2%), (iii) 11p loss (10.2%), (iv) 7 gain (9.2%), (v) 19 gain (9.2%), and (vi) 23.4% of tumors that were unclassified.
Overall, 10 LGGs from the validation cohort were 11p-deleted, and 9 of these samples were astrocytomas. Therefore, 11p loss was associated with astrocytoma phenotype (90%; in 11p-deleted LGGs vs 35% in non 11p-deleted LGGs, P = .002; Supplementary Fig S1, panel B). In addition, 11p loss had independent prognostic significance in multivariate analysis (Supplementary Table S2). Finally, 9 LGGs from the validation cohort exhibited 19q loss without 1p loss (3 astrocytomas, 3 oligoastrocytomas, and 3 oligodendrogliomas; Supplementary Fig S3). Interestingly, IDH-mutated and 19q-deleted LGGs (100%; 9/9) also had a similar prognosis compared with IDH-mutated LGGs (Fig. 4, panel B).
Discussion
LGG is a complex subgroup of glial tumors with a heterogeneous prognosis. Molecular markers, including protein expression and genetic imbalances, have enabled us to identify 3 major molecular subgroups of LGG with clinical significance: (i) IDH mutated and 1p/19q codeleted, (ii) IDH mutated/non 1p/19q codeleted, and (iii) IDH wild-type.8,15
Our study was focused on the clinical significance of molecular markers (ie, p53 expression, MGMT promoter methylation, and IDH1/IDH2 mutations) and genomic aberrations in non 1p/19q codeleted LGG patients. We have identified several genomic groups providing novel insights into the clinicomolecular portrait of these tumors.
Our study confirmed that LGGs are heterogeneous from a genomic point of view and provided a detailed picture of the most frequent molecular alterations reported in LGGs, notably IDH mutation (87%), MGMT promoter methylation (75%), and p53 overexpression (43%). It should be noted that a wide range in frequency of these alterations has been reported in previous studies: IDH mutation from 68%–87%,8,12,27 MGMT promoter methylation from 58%–93%, and p53 overexpression from 13%–88%.5,13,17 This variability may be due to the distribution of tumor phenotype within the studies (ie, p53 overexpression is more frequent in astrocytomas),12,25 and the different analytical and interpretation procedures (ie, regarding MGMT promoter methylation status).17 In addition, we also confirmed that 1p/19q codeletion and IDH mutation are associated with favorable prognosis in LGG.1,6,8,10 Conversely, MGMT promoter methylation was not associated with favorable survival. Indeed, there are conflicting data regarding the impact of this possible marker in LGG.5,8,11,13 Our study also confirms that triple-negative LGGs (p53 not overexpressed, IDH wt) have dismal prognosis.12,15 It should be noted that these triple-negative tumors, even though being described as relatively homogeneous from a radiological point of view (ie, located in the insula),15 have highly heterogeneous genomic profiling. Thus, further studies focusing on this rare subtype (12/126 in our series) are needed to better characterize their driving molecular alterations.
Our study also characterized LGGs without 1p/19q codeletion. Based on BAC-aCGH results, LGG could be subdivided into 5 homogeneous genomic groups using a supervised analysis: (i) 11p loss, (ii) 19q loss, (iii) 7 gain, (iv) 19 gain, and (v) unclassified. Interestingly, tumors harboring 11p loss were astrocytomas. This clinicomolecular correlation has been previously suggested.26 It should be noted that 11p-deleted LGGs form a small subgroup of LGGs accounting for ∼9% of LGGs (7.2% in the discovery cohort and 10.2% in the validation cohort). Both series of tumors investigated in the present work are retrospective, and biases related to retrospective studies, including sampling biases and treatment heterogeneity, might impact our results. The MCR of loss on 11p in 11p-deleted LGGs was identified between 11p15.5 and 11p15.4 cytobands, Supplementary Table S1. Interestingly, the 11p15.5 cytoband includes several genes that may act as tumor suppressors (eg, CDKN1C, NUP98, and SLC22A18).28–30 Further studies are warranted to investigate these candidate genes in oncogenesis of 11p-deleted LGGs. Our study has shown that chr 19q loss suppresses the favorable prognostic value of IDH mutation in non 1p/19q codeleted LGGs.8 Indeed, IDH mutated LGGs harboring 19q loss have similar prognosis to IDH wild-type tumors.
Interestingly, tumors with 1p/19q codeletion (all IDH mutated) occur in significantly older patients (median age = 42 years) than non 1p/19q codeleted tumors (median age = 36.9 years; P = .03, Supplementary Fig. S2).
Novel recurrent mutations have been identified in WHO grade II and grade III gliomas. In 1p/19q codeleted oligodendrogliomas, recurrent mutations have described CIC (homologue of Capicua Drosophila) being localized in 19q and in FUBP1 (far upstream element binding protein 1) being localized in 1p.31 In non 1p/19q codeleted gliomas, ATRX (alpha thalassemia/mental retardation syndrome X-linked) is mutated in about half of generally astrocytoma phenotype gliomas harboring IDH1/IDH2 mutations.32,33 ATRX inactivation has been associated with alternative lengthening of telomeres (ALT).32 Very recently, mutations in TERT promoter in virtually all 1p/19q codeleted tumors and 80% of glioblastomas have been described.34 The impact of these recurrent somatic mutations in the oncogenesis and prognosis of LGGs remains to be established.
Although, our study is monocentric with homogeneous therapeutic guidelines, its retrospective nature limits investigations of the impacts of medical treatments in prognosis.
In summary, our study provides a comprehensive overview of genomic aberrations in LGGs. We have designated 5 genomic groups within non 1p/19q codeleted LGGs. Interestingly, these genomic groups have clinical value, particularly 11p loss and 19q loss. Finally, we demonstrated that genomic changes commonly associated with the pathogenesis of LGGs may help to stratify these tumors regarding prognosis and, in the near future, may facilitate the individualization of treatment.
Supplementary Material
Funding
This work is part of the national program Cartes d'Identité des Tumeurs ® (CIT) (http://cit.ligue-cancer.net/). The research leading to these results received funding from the program “Investissements d'avenir” ANR-10-IAIHU-06. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Agusti Alentorn has been granted a fellowship by Obra Social la Caixa and ARTC.
Supplementary Material
Acknowledgments
Conflict of interest statement. None declared.
References
- 1.Smith JS, Perry A, Borell TJ, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18(3):636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 2.Louis D, Ohgaki H, Wiestler O, Cavenee W. WHO Classification of Tumors of the Central Nervous System. 4th ed. Ogdensburg: Renouf Publishing Co. Ltd; 2007. [Google Scholar]
- 3.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64(6):479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 4.Coons SW, Johnson PC, Scheithauer BW, et al. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79(7):1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 5.Bourne TD, Schiff D. Update on molecular findings, management and outcome in low-grade gliomas. Nat Rev Neurol. 2010;6(12):695–701. doi: 10.1038/nrneurol.2010.159. [DOI] [PubMed] [Google Scholar]
- 6.van den Bent MJ, Looijenga LH, Langenberg K, et al. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97(5):1276–1284. doi: 10.1002/cncr.11187. [DOI] [PubMed] [Google Scholar]
- 7.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 10.Labussière M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann C, Hentschel B, Tatagiba M, et al. German Glioma Network. Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res. 2011;17(13):4588–4599. doi: 10.1158/1078-0432.CCR-10-3194. [DOI] [PubMed] [Google Scholar]
- 12.Kim YH, Nobusawa S, Mittelbronn M, et al. Molecular classification of low-grade diffuse gliomas. Am J Pathol. 2010;177(6):2708–2714. doi: 10.2353/ajpath.2010.100680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everhard S, Kaloshi G, Crinière E, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60(6):740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 14.Komine C, Watanabe T, Katayama Y, et al. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13(2):176–184. doi: 10.1111/j.1750-3639.2003.tb00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metellus P, Coulibaly B, Colin C, et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010;120(6):719–729. doi: 10.1007/s00401-010-0777-8. [DOI] [PubMed] [Google Scholar]
- 16.Idbaih A, Marie Y, Lucchesi C, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122(8):1778–1786. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 17.Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, methylight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118(17):4201–4211. doi: 10.1002/cncr.27392. [DOI] [PubMed] [Google Scholar]
- 18.Hoang-Xuan K, Capelle L, Kujas M, et al. Temozolomide as initial treatment for adults with low-grade oligodendrogliomas or oligoastrocytomas and correlation with chromosome 1p deletions. J Clin Oncol. 2004;22(15):3133–3138. doi: 10.1200/JCO.2004.10.169. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statist Soc Series B. 1995;57(1):289–300. [Google Scholar]
- 20.Neuvial P, Hupé P, Brito I, et al. Spatial normalization of array-CGH data. BMC Bioinformatics. 2006;7:264. doi: 10.1186/1471-2105-7-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hupé P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20(18):3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- 22.Diskin SJ, Eck T, Greshock J, et al. STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 2006;16(9):1149–1158. doi: 10.1101/gr.5076506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(2):159–174. [PubMed] [Google Scholar]
- 24.van Thuijl HF, Ylstra B, Würdinger T, et al. Genetics and pharmacogenetics of diffuse gliomas. Pharmacol Ther. 2013;137(1):78–88. doi: 10.1016/j.pharmthera.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Ständer M, Peraud A, Leroch B, Kreth FW. Prognostic impact of TP53 mutation status for adult patients with supratentorial World Health Organization Grade II astrocytoma or oligoastrocytoma: a long-term analysis. Cancer. 2004;101(5):1028–1035. doi: 10.1002/cncr.20432. [DOI] [PubMed] [Google Scholar]
- 26.Dahlback HS, Gorunova L, Brandal P, et al. Genomic aberrations in diffuse low-grade gliomas. Genes Chromosomes Cancer. 2011;50(6):409–420. doi: 10.1002/gcc.20866. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 28.Borriello A, Caldarelli I, Bencivenga D, et al. p57(Kip2) and cancer: time for a critical appraisal. Mol Cancer Res. 2011;9(10):1269–1284. doi: 10.1158/1541-7786.MCR-11-0220. [DOI] [PubMed] [Google Scholar]
- 29.Singer R, Zhao R, Barsotti AM, et al. Nuclear pore component Nup98 is a potent tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol Cell. 2012;48(5):799–810. doi: 10.1016/j.molcel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu SH, Feng DF, Ma YB, et al. Promoter methylation and downregulation of SLC22A18 are associated with the development and progression of human glioma. J Transl Med. 2011;9:156. doi: 10.1186/1479-5876-9-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannan K, Inagaki A, Silber J, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3(10):1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.