Abstract
Background
Drug-resistant Providencia rettgeri producing metallo-β-lactamase and 16S rRNA methylase has been reported in several countries. We analyzed P. rettgeri clinical isolates with resistance to carbapenems and aminoglycosides in a hospital in Nepal.
Methods
Five clinical isolates of multidrug-resistant P. rettgeri were obtained in a hospital in Nepal. Antimicrobial susceptibilities were determined using the microdilution method and entire genomes were sequenced to determine drug-resistant genes. Epidemiological analysis was performed by pulsed-field gel electrophoresis.
Results
Four of the 5 isolates were resistant to carbapenems (imipenem and meropenem), with MICs ≥16 mg/L, with the remaining isolate showing intermediate resistance to imipenem, with an MIC of 2 mg/L and susceptibility to meropenem with an MIC ≤1 mg/L. All 5 isolates had blaVEB-1. Of the 4 carbapenem-resistant strains, 3 had blaNDM-1 and 1 had blaOXA-72. All isolates were highly resistant to aminoglycosides (MICs ≥1,024 mg/L) and harbored armA. As the result of pulsed-field gel electrophoresis pattern analysis in the 5 P. rettgeri isolates, 4 had identical PFGE patterns and the fifth showed 95.7% similarity.
Conclusions
This is the first report describing multidrug-resistant P. rettgeri strains harboring blaNDM-1 or blaOXA-72 and armA isolated from patients in Nepal.
Keywords: NDM-1, OXA-72, 16S rRNA methylase, Providencia rettgeri, Molecular epidemiology
Background
Providencia rettgeri has been associated with hospital acquired infections, including catheter-related urinary tract infections, bacteremia, skin infections, diarrhea, and gastroenteritis [1,2]. To date, there have been 5 reports of P. rettgeri isolates harboring metallo-β-lactamase (MBL) encoding genes, including IMP-type MBL producers in Japan [3,4]; VIM-type MBL, PER-1 extended-spectrum β-lactamase (ESBL) and 16S rRNA methylase ArmA in Korea [5]; and NDM-type MBL in Israel [6] and Brazil [7].
NDM-type MBL was initially identified in Klebsiella pneumoniae and Escherichia coli in 2009 in Sweden [8]. Since then, NDM-1-producing Enterobacteriaceae have been isolated in various parts of the world [9,10].
Exogenously acquired 16S rRNA methylase genes responsible for very high levels of resistance to various aminoglycosides are widely distributed among Enterobacteriaceae and glucose-nonfermentative microbes [11]. Gram-negative pathogens producing 16S rRNA methylase ArmA have been isolated in various countries [11].
Although co-production of several resistance determinants is not rare in Enterobacteriaceae[12-16], it is less common in P. rettgeri[5]. We describe here P. rettgeri clinical isolates from Nepal that produce carbapenemase (NDM-1 or OXA-72) and 16S rRNA methylase (ArmA).
Methods
Bacterial strains
Five P. rettgeri clinical isolates were obtained from May to July 2012 from 5 patients at Tribhuvan University Teaching Hospital in Kathmandu, Nepal. Three isolates were from sputum and 2 from pus at surgical sites. Samples were obtained as part of standard patient care. Phenotypical identification [17] was confirmed by API 32GN (BioMérieux, Mercy l'Etoile, France) and 16S rRNA sequencing (1,497 bp) [18,19].
Antimicrobial susceptibilities
MICs were determined using the microdilution method, according to the guidelines of the Clinical Laboratory Standards Institute (CLSI) [20]. Breakpoints to antibiotics were determined. The modified Hodge test, the meropenem-sodium mercaptoacetic acid double-disk synergy test (Eiken Chemical, Tokyo, Japan) and E-test (imipenem/EDTA) (AB Biodisk, Solna, Sweden) were performed.
Entire genome sequencing
The entire genomes of these isolates were extracted and sequenced by MiSeq (Illumina, San Diego, CA). CLC genomics workbench version 5.5 (CLC bio, Tokyo, Japan) was used for de novo assembly of reads and to search for 923 drug-resistance genes, including genes encoding β-lactamases, 16S rRNA methylases and aminoglycoside-acethyl/adenylyltransferases; point mutations in the gyrA, parC and pmrCAB operons; and point mutations in the fos genes, including fosA, fosA2, fosA3, fosC and fosC2.
Pulsed-field gel electrophoresis (PFGE) and southern hybridization
PFGE analysis was performed as described [3]. An 813 bp probe for blaNDM-1 was synthesized by PCR amplification using the primers 5′-atggaattgcccaatattatgcac-3′ (forward) and 5′-tcagcgcagcttgtcggccatgcggg-3′ (reverse), and a 780 bp probe for blaOXA-72 was synthesized using the primers 5′-agtttctctcagtgcatgttcatctat-3′ (forward) and 5′-agaaccagacattccttctttcatttc-3′ (reverse). Southern hybridization to detect blaNDM-1 and blaOXA-72 was performed using these probes, which were detected using DIG High Prime DNA labeling and detection starter kit II (Roche Diagnostics, Mannheim, Germany).
Nucleotide sequence accession numbers
The nucleotide sequences surrounding blaNDM-1 and blaOXA-72 have been deposited in GenBank with the accession number AB828598 and AB857844, respectively.
Ethical approval
The study protocol was reviewed and approved by the Institutional Review Board of the Institute of Medicine, Tribhuvan University (ref. 6-11-E) and the Biosafety Committee, National Center for Global Health and Medicine (approval number: 23-M-49).
Results
Antimicrobial susceptibilities
Four of the 5 isolates were resistant to carbapenems (doripenem, imipenem and meropenem) and piperacillin/tazobactam, whereas the fifth was susceptible to piperacillin/tazobactam, doripenem and meropenem and showed intermediate resistance to imipenem (Table 1). All 5 isolates were highly resistant to cephalosporins (ceftazidime and cefepime), aztreonam, aminoglycosides (arbekacin, amikacin and gentamicin), ciprofloxacin, colistin and fosfomycin, and all 5 showed intermediate resistance to tigecycline. The four isolates resistant to carbapenems were negative with the modified Hodge test, but three of the four isolates were positive with the meropenem-sodium mercaptoacetic acid double-disk synergy test and E-test/EDTA.
Table 1.
Summary of the characteristics of the 5 P. rettegeri strains, including antimicrobial resistance profiles and resistant genes
|
Strains |
Tissue sources |
Infection |
|
MIC (mg/L) |
Antibiotics resistant genes |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | CAZ | FEP | IPM | DPM | MEM | ATM | ABK | AMK | GEN | CIP | CST | FOF | TIG | ||||
| IOMTU1 |
Pus |
SSI |
1,024 |
512 |
>1,024 |
64 |
32 |
16 |
64 |
1,024 |
>1,024 |
>1,024 |
>1,024 |
128 |
>128 |
512 |
4 |
blaNDM-1, blaOXA-10, blaVEB-1, blaTEM-1, blaADC-67, armA, aadA1, aadA2 |
| IOMTU4 |
Sputum |
NLRTI |
1,024 |
128 |
>1,024 |
256 |
16 |
16 |
32 |
1,024 |
>1,024 |
>1,024 |
>1,024 |
>256 |
>128 |
512 |
4 |
blaOXA-72, blaOXA-10, blaVEB-1, blaTEM-1, blaADC-67, armA, aadA1 |
| IOMTU91 |
Sputum |
NLRTI |
>1,024 |
1,024 |
>1,024 |
1,024 |
64 |
32 |
64 |
1,024 |
>1,024 |
>1,024 |
>1,024 |
256 |
128 |
128 |
4 |
blaNDM-1, blaOXA-10, blaVEB-1, blaTEM-1, blaADC-67, armA, aadA1 |
| IOMTU94 |
Pus |
SSI |
1,024 |
4 |
>1,024 |
256 |
2 |
1 |
1 |
>1,024 |
1,024 |
1,024 |
>1,024 |
256 |
>128 |
1,024 |
4 |
blaOXA-10, blaVEB-1, blaTEM-1, blaADC-67, armA, aadA1 |
| IOMTU99 | Sputum | NLRTI | >1,024 | 512 | >1,024 | 128 | 64 | 32 | 64 | 1,024 | >1,024 | >1,024 | >1,024 | >256 | >128 | 1,024 | 4 | blaNDM-1, blaVEB-1, blaOXA-10, blaTEM-1, blaADC-67, armA, aadA1 |
SSI, surgical site infection; NLRTI, nosocomial lower respiratory tract infection PIP, piperacillin; TZP, piperacillin/tazobactam; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; DPM, doripenem; MEM, meropenem; ATM, aztreonam; ABK, arbekacin; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; CST, colistin; FOF, fosfomycin; TIG, tigecycline.
Drug-resistant genes
All 5 isolates tested had several genes associated with β-lactam and aminoglycoside-resistance (Table 1). These isolates had blaVEB-1, blaOXA-10, blaTEM-1, blaADC-67 (ampC), armA and aadA1; 3 had blaNDM-1; and 1 had blaOXA-72. None of these isolates had any other β-lactamase encoding genes, including the class A genes blaSHVs and blaCTX-Ms; the class B genes blaAIM, blaDIM, blaFIM, blaGIM, blaIMPs, blaINDs, blaKHM, blaSIM, blaSMB, blaSPM, blaTMBs, and blaVIMs; or the class D gene blaOXAs except for blaOXA-10 and blaOXA-72. None had other genes encoding 16S rRNA methylases or aminoglycoside acetyl/adenylyltransferases. All 5 isolates had point mutations in the quinolone-resistance-determining regions of gyrA and parC, with amino acid substitutions of S83I and D87E in GyrA and S80I in ParC, but none had any mutations in the pmrCAB operon and fos genes. All sequences of the drug-resistant genes tested were identical to those registered in GenBank.
PFGE and southern hybridyzation
Of the 5 P. rettgeri isolates, 4 had identical PFGE patterns and the fifth showed 95.7% similarity (Figure 1). Three of these isolates had a plasmid harboring blaNDM-1 and one had a plasmid harboring blaOXA-72, with plasmid sizes ranging from 9.42 to 23.1 kbp (data not shown).
Figure 1.
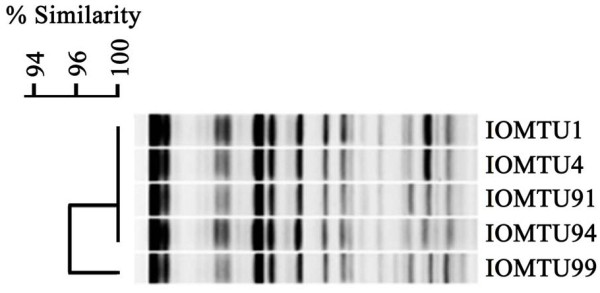
PFGE profiles obtained following Sfi I digestion of P. rettgeri chromosomes.
Genomic structures surrounding blaNDM-1 and blaOXA-72
The genetic environments surrounding blaNDM-1 (Accession no. AB828598) was blaNDM-1-bleMBL-trpF-dsbC-cutA1. All 3 isolates harboring blaNDM-1 (IOMTU1, 91 and 99) had the same genetic environments. The blaOXA-72 gene was flanked by conserved inverted repeats at the XerC/XerD binding sites [21], indicating mobilization by site-specific recombination mechanisms. The rep1 gene was located downstream of blaOXA-72 (Accession no. AB857844).
Discussion
The relatively high MICs to piperacillin/tazobactam and carbapenems of the five P. rettgeri isolates were likely due to the presence of blaNDM-1 or blaOXA-72. The enzymatic activities of metallo-β-lactamases, including NDM-1, were not inhibited by tazobactam [22], a β-lactamase inhibitor, in agreement with the MIC profiles of these isolates to piperacillin/tazobactam. The high MICs of all 5 isolates to ceftazidime, cefepime and aztreonam were likely due to the presence of blaVEB-1[23], and the presence of armA in these isolates was likely associated with their extremely high resistance to all aminoglycosides tested [11]. Point mutations in the quinolone-resistance-determining regions of gyrA and parC have been associated with high resistance to quinolones [24]. Point mutations in pmrCAB operon have been associated with the resistance of Acinetobacter spp. [25] and Pseudomonas aeruginosa[26] to polymxyin and colistin; and the presence of fos genes, including fosA, fosA2, fosA3, fosC and fosC2, has been associated with resistance to fosfomycin in Gram-negative bacteria [27-29].
Plasmids containing blaNDM-1 or blaOXA-72 may be disseminated among Gram-negative pathogens in Nepal. The genetic environments surrounding blaNDM-1 in our P. rettgeri strains (blaNDM-1-bleMBL-trpF-dsbC-cutA1) were also observed in other plasmids, including A. baumannii plasmid pAbNDM-1 from China (Accession no. JN377410), Citrobacter freundii plasmid pYE315203 from China (Accession no. JX254913), E. coli plasmid pNDM102337 from Canada (Accession no. JF714412), K. pneumoniae plasmid pKP-NCGM18-1 from Nepal (Accession no. AB824738) [30], K. pneumoniae plasmids pKPX-1, pKPN5047 and pNDM-HN380 from China (Accession nos. AP012055, KC311431 and JX104760, respectively), and P. rettgeri plasmid pFR90 (Accession no. JQ362415) from China. In addition, the genetic structures of OXA-72 producing Acinetobacter spp [31-34] and K. pneumoniae (Accession no. JX268653 and AB825955 deposited in 2012 and 2013, respectively) had the same genetic structure (blaOXA-72-rep1) as our strain of P. rettgeri.
Conclusions
To our knowledge, this is the first report describing P. rettgeri strains harboring blaNDM-1 or blaOXA-72 and armA isolated from patients in Nepal. These 5 strains were highly resistant to both β-lactams and aminoglycosides and expanded in a clonal manner in the hospital.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
TT: Performed PCR and sequencing, analyzed data and drafted the manuscript. TMA: Performed entire genome sequencing. RKD and MKS: Performed drug susceptibility tests. HO: Supervised this study. KS: Performed pulsed-field gel electrophoresis and its pattern analysis. TK and BMP: Designed protocols and supervised this study. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Tatsuya Tada, Email: ttada@ri.ncgm.go.jp.
Tohru Miyoshi-Akiyama, Email: takiyam@ri.ncgm.go.jp.
Rajan K Dahal, Email: dahalrajan2005@gmail.com.
Manoj K Sah, Email: yourmanoj_sah@yahoo.com.
Hiroshi Ohara, Email: ohara52.jp@gmail.com.
Kayo Shimada, Email: kshima@ri.ncgm.go.jp.
Teruo Kirikae, Email: tkirikae@ri.ncgm.go.jp.
Bharat M Pokhrel, Email: bmp268@hotmail.com.
Acknowledgements
The authors thank emeritus professor Masayasu Nakano, Jichi Medical University, for comments on the manuscript. This study was supported by grants [International Health Cooperation Research (23-A-301 and 24-S-5), a grant from the Ministry of Health, Labor and Welfare of Japan (H24-Shinko-Ippan-010), and JSPS KAKENHI Grant Number 24790432].
References
- Stock I, Wiedemann B. Natural antibiotic susceptibility of Providencia stuartii, P. rettgeri, P. alcalifaciens and P. rustigianii strains. J Med Microbiol. 1998;14:629–642. doi: 10.1099/00222615-47-7-629. [DOI] [PubMed] [Google Scholar]
- Yoh M, Matsuyama J, Ohnishi M, Takagi K, Miyagi H, Mori K, Park KS, Ono T, Honda T. Importance of Providencia species as a major cause of travellers' diarrhoea. J Med Microbiol. 2005;14:1077–1082. doi: 10.1099/jmm.0.45846-0. [DOI] [PubMed] [Google Scholar]
- Shiroto K, Ishii Y, Kimura S, Alba J, Watanabe K, Matsushima Y, Yamaguchi K. Metallo-beta-lactamase IMP-1 in Providencia rettgeri from two different hospitals in Japan. J Med Microbiol. 2005;14:1065–1070. doi: 10.1099/jmm.0.46194-0. [DOI] [PubMed] [Google Scholar]
- Nishio H, Komatsu M, Shibata N, Shimakawa K, Sueyoshi N, Ura T, Satoh K, Toyokawa M, Nakamura T, Wada Y, Orita T, Kofuku T, Yamasaki K, Sakamoto M, Kinoshita S, Aihara M, Arakawa Y. Metallo-beta-lactamase-producing Gram-negative bacilli: laboratory-based surveillance in cooperation with 13 clinical laboratories in the Kinki region of Japan. J Clin Microbiol. 2004;14:5256–5263. doi: 10.1128/JCM.42.11.5256-5263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kang HY, Shin KS, Kim J. Multidrug-resistant Providencia isolates carrying blaPER-1, blaVIM-2, and armA. J Microbiol. 2007;14:272–274. [PubMed] [Google Scholar]
- Gefen-Halevi S, Hindiyeh MY, Ben-David D, Smollan G, Gal-Mor O, Azar R, Castanheira M, Belausov N, Rahav G, Tal I, Mendelson E, Keller N. Isolation of genetically not related blaNDM-1 positive Providencia rettgeri in Israel. J Clin Microbiol. 2013;14:1642–1643. doi: 10.1128/JCM.00381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Assef AP, Pereira PS, Albano RM, Beriao GC, Chagas TP, Timm LN, Da Silva RC, Falci DR, Asensi MD. Isolation of NDM-producing Providencia rettgeri in Brazil. J Antimicrob Chemother. in press. [DOI] [PubMed]
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;14:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornaglia G, Giamarellou H, Rossolini GM. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis. 2011;14:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- Pillai DR, McGeer A, Low DE. New Delhi metallo-beta-lactamase-1 in Enterobacteriaceae: emerging resistance. CMAJ. 2011;14:59–64. doi: 10.1503/cmaj.101487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J, Arakawa Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat. 2012;14:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Sheng WH, Badal RE, Hsueh PR. SMART Program. Distribution of extended-spectrum beta-Lactamases, AmpC beta-Lactamases, and carbapenemases among enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the Study for Monitoring Antimicrobial Resistance Trends (SMART) Antimicrob Agents Chemother. 2013;14:2981–2988. doi: 10.1128/AAC.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MF, Francisco GR, O'Hara JA, de Oliveira Garcia D, Doi Y. Co-production of 16S Ribosomal RNA Methyltransferase RmtD and RmtG with KPC-2 and CTX-M-group ESBLs in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013;14:2397–2400. doi: 10.1128/AAC.02108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I, Souli M, Panagea T, Poulakou G, Kanellakopoulou K, Giamarellou H. Prevalence of 16S rRNA methylase genes in Enterobacteriaceae isolates from a Greek university hospital. Clin Microbiol Infect. 2012;14:E52–E54. doi: 10.1111/j.1469-0691.2011.03738.x. [DOI] [PubMed] [Google Scholar]
- Zacharczuk K, Piekarska K, Szych J, Zawidzka E, Sulikowska A, Wardak S, Jagielski M, Gierczynski R. Emergence of Klebsiella pneumoniae coproducing KPC-2 and 16S rRNA methylase ArmA in Poland. Antimicrob Agents Chemother. 2011;14:443–446. doi: 10.1128/AAC.00386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Liu Q, Han L, Sun J, Ni Y. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 and ArmA 16S rRNA methylase conferring high-level aminoglycoside resistance in carbapenem-resistant Enterobacter cloacae in China. Diagn Microbiol Infect Dis. 2010;14:326–328. doi: 10.1016/j.diagmicrobio.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Tang YW, Ellis NM, Hopkins MK, Smith DH, Dodge DE, Persing DH. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol. 1998;14:3674–3679. doi: 10.1128/jcm.36.12.3674-3679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymock D, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;14:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmon KE, Croft AC, Petti CA. Application of SmartGene IDNS software to partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J Clin Microbiol. 2006;14:4400–4406. doi: 10.1128/JCM.01364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards. Approved standard M07-A9. 8. Wayne, Pa: Clinical and Laboratory Standards Institute; 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed. [Google Scholar]
- D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;14:3528–3533. doi: 10.1128/AAC.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;14:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Naas T, Guibert M, Chaibi EB, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;14:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;14(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. 2009;14:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 2004;14:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beharry Z, Palzkill T. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J Biol Chem. 2005;14:17786–17791. doi: 10.1074/jbc.M501052200. [DOI] [PubMed] [Google Scholar]
- Xu H, Miao V, Kwong W, Xia R, Davies J. Identification of a novel fosfomycin resistance gene (fosA2) in Enterobacter cloacae from the Salmon River, Canada. Lett Appl Microbiol. 2011;14:427–429. doi: 10.1111/j.1472-765X.2011.03016.x. [DOI] [PubMed] [Google Scholar]
- Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother. 2010;14:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Miyoshi-Akiyama T, Dahal RK, Mishra SK, Ohara H, Shimada K, Kirikae T, Pokhrel BM. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int J Antimicrob Agents. 2013;14:372–374. doi: 10.1016/j.ijantimicag.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Werneck JS, Picao RC, Carvalhaes CG, Cardoso JP, Gales AC. OXA-72-producing Acinetobacter baumannii in Brazil: a case report. J Antimicrob Chemother. 2011;14:452–454. doi: 10.1093/jac/dkq462. [DOI] [PubMed] [Google Scholar]
- Wang H, Guo P, Sun H, Wang H, Yang Q, Chen M, Xu Y, Zhu Y. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob Agents Chemother. 2007;14:4022–4028. doi: 10.1128/AAC.01259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montealegre MC, Maya JJ, Correa A, Espinal P, Mojica MF, Ruiz SJ, Rosso F, Vila J, Quinn JP, Villegas MV. First identification of OXA-72 carbapenemase from Acinetobacter pittii in Colombia. Antimicrob Agents Chemother. 2012;14:3996–3998. doi: 10.1128/AAC.05628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goic-Barisic I, Towner KJ, Kovacic A, Sisko-Kraljevic K, Tonkic M, Novak A, Punda-Polic V. Outbreak in Croatia caused by a new carbapenem-resistant clone of Acinetobacter baumannii producing OXA-72 carbapenemase. J Hosp Infect. 2011;14:368–369. doi: 10.1016/j.jhin.2010.12.003. [DOI] [PubMed] [Google Scholar]


