Abstract
The genes of the homeotic complex (HOX) encode DNA binding homeodomain proteins that control developmental fates by differentially regulating the transcription of downstream target genes. Despite their unique in vivo functions, disparate HOX proteins often bind to very similar DNA sequences in vitro. Thus, a critical question is how HOX proteins select the correct sets of target genes in vivo. The homeodomain proteins encoded by the Drosophila extradenticle gene and its mammalian homologues, the pbx genes, contribute to HOX specificity by cooperatively binding to DNA with HOX proteins. For example, the HOX protein labial cooperatively binds with extradenticle protein to a 20-bp oligonucleotide that is sufficient to direct a labial-like expression pattern in Drosophila embryos. Here we have analyzed the protein-DNA interactions that are important for forming the labial-extradenticle-DNA complex. The data suggest a model in which labial and extradenticle, separated by only 4 bp, bind this DNA as a heterodimer in a head-to-tail orientation. We have confirmed several aspects of this model by characterizing extradenticle-HOX binding to mutant oligonucleotides. Most importantly, mutations in base pairs predicted to contact the HOX N-terminal arm resulted in a change in HOX preference in the heterodimer, from labial to Ultrabithorax. These results demonstrate that extradenticle prefers to bind cooperatively with different HOX proteins depending on subtle differences in the heterodimer binding site.
Full text
PDF
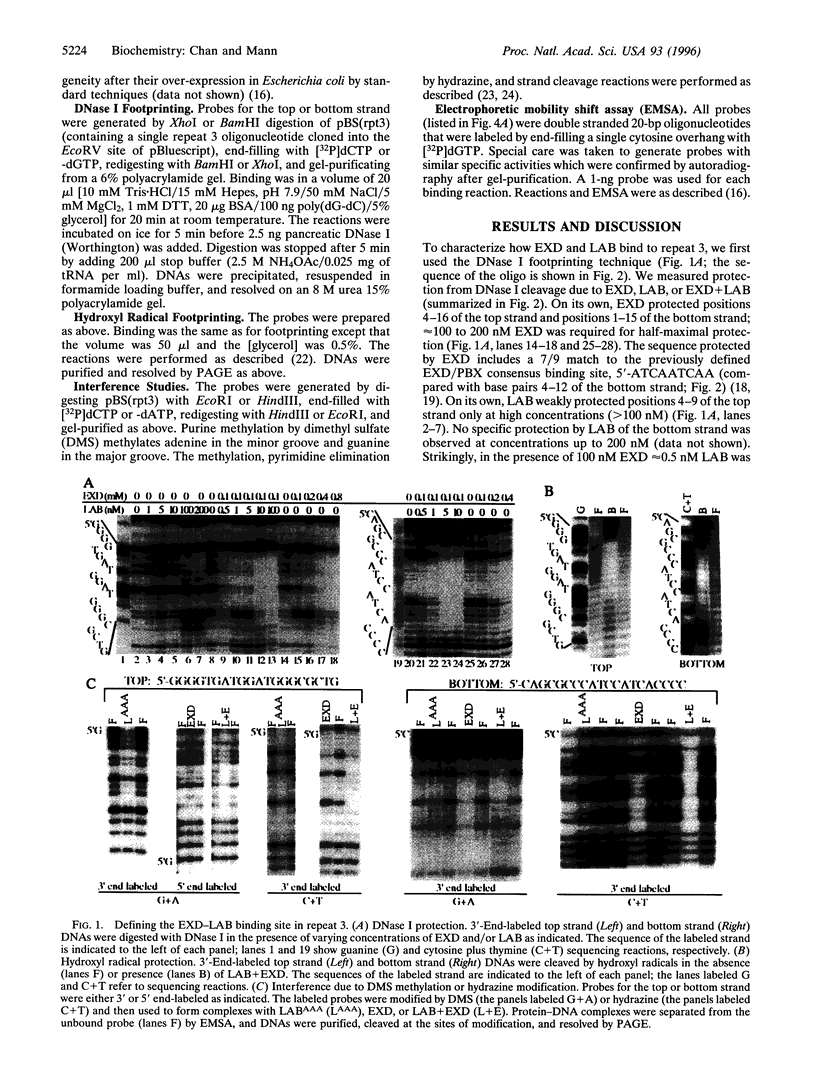

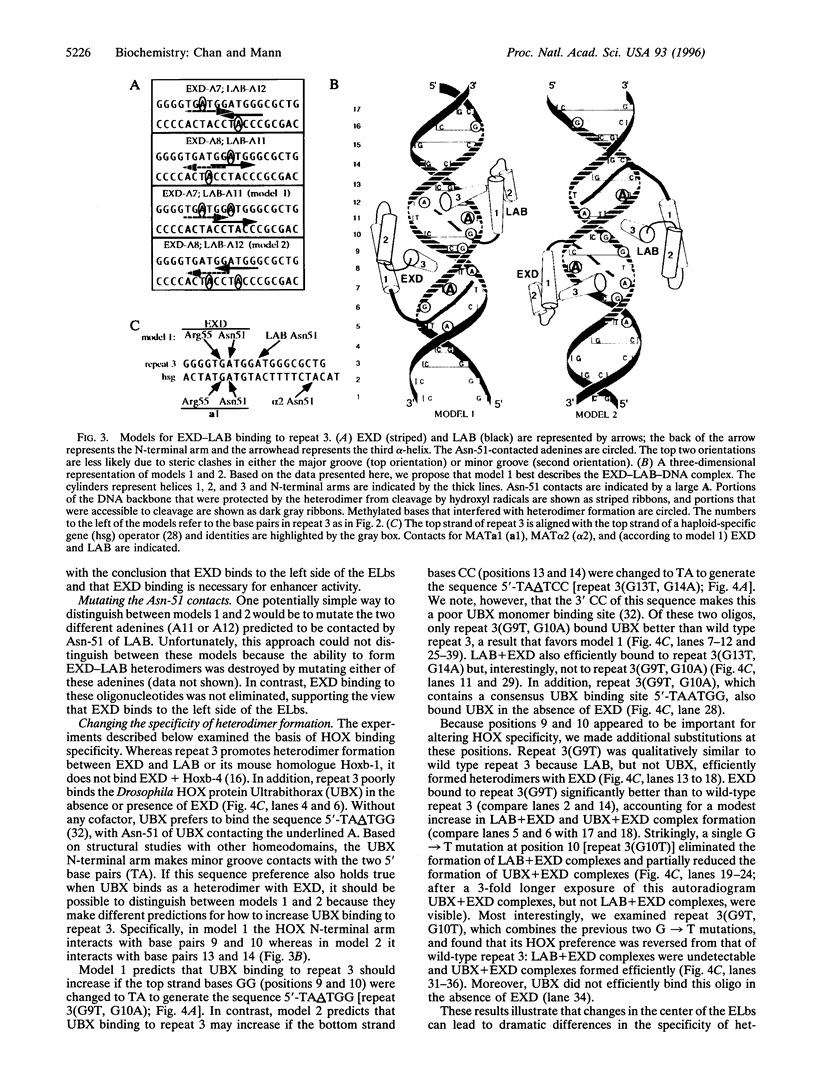


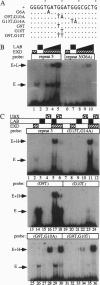
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Billeter M., Qian Y. Q., Otting G., Müller M., Gehring W., Wüthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993 Dec 20;234(4):1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. K., Jaffe L., Capovilla M., Botas J., Mann R. S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994 Aug 26;78(4):603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chang C. P., Shen W. F., Rozenfeld S., Lawrence H. J., Largman C., Cleary M. L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995 Mar 15;9(6):663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Dixon W. J., Hayes J. J., Levin J. R., Weidner M. F., Dombroski B. A., Tullius T. D. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- Ekker S. C., Jackson D. G., von Kessler D. P., Sun B. I., Young K. E., Beachy P. A. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994 Aug 1;13(15):3551–3560. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., Young K. E., von Kessler D. P., Beachy P. A. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991 May;10(5):1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel W. A., Singson A. W., Margolis J. S., Bang A. G., Posakony J. W., Murre C. Dpbx, a new homeobox gene closely related to the human proto-oncogene pbx1 molecular structure and developmental expression. Mech Dev. 1993 May;41(2-3):155–161. doi: 10.1016/0925-4773(93)90045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J., Qian Y. Q., Billeter M., Furukubo-Tokunaga K., Schier A. F., Resendez-Perez D., Affolter M., Otting G., Wüthrich K. Homeodomain-DNA recognition. Cell. 1994 Jul 29;78(2):211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Goutte C., Johnson A. D. Recognition of a DNA operator by a dimer composed of two different homeodomain proteins. EMBO J. 1994 Mar 15;13(6):1434–1442. doi: 10.1002/j.1460-2075.1994.tb06397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Kaelin W. G., Jr, Krek W., Sellers W. R., DeCaprio J. A., Ajchenbaum F., Fuchs C. S., Chittenden T., Li Y., Farnham P. J., Blanar M. A. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992 Jul 24;70(2):351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- Kalionis B., O'Farrell P. H. A universal target sequence is bound in vitro by diverse homeodomains. Mech Dev. 1993 Sep;43(1):57–70. doi: 10.1016/0925-4773(93)90023-q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps M. P., Murre C., Sun X. H., Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990 Feb 23;60(4):547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991 Dec 3;30(48):11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- LeBrun D. P., Cleary M. L. Fusion with E2A alters the transcriptional properties of the homeodomain protein PBX1 in t(1;19) leukemias. Oncogene. 1994 Jun;9(6):1641–1647. [PubMed] [Google Scholar]
- Li T., Stark M. R., Johnson A. D., Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995 Oct 13;270(5234):262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- Lis J., Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993 Jul 16;74(1):1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- Lu Q., Knoepfler P. S., Scheele J., Wright D. D., Kamps M. P. Both Pbx1 and E2A-Pbx1 bind the DNA motif ATCAATCAA cooperatively with the products of multiple murine Hox genes, some of which are themselves oncogenes. Mol Cell Biol. 1995 Jul;15(7):3786–3795. doi: 10.1128/mcb.15.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992 Jan 24;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Neuteboom S. T., Peltenburg L. T., van Dijk M. A., Murre C. The hexapeptide LFPWMR in Hoxb-8 is required for cooperative DNA binding with Pbx1 and Pbx2 proteins. Proc Natl Acad Sci U S A. 1995 Sep 26;92(20):9166–9170. doi: 10.1073/pnas.92.20.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourse J., Mellentin J. D., Galili N., Wilkinson J., Stanbridge E., Smith S. D., Cleary M. L. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990 Feb 23;60(4):535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- Peifer M., Wieschaus E. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 1990 Jul;4(7):1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- Phelan M. L., Rambaldi I., Featherstone M. S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995 Aug;15(8):3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöpperl H., Bienz M., Studer M., Chan S. K., Aparicio S., Brenner S., Mann R. S., Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995 Jun 30;81(7):1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Van Dijk M. A., Voorhoeve P. M., Murre C. Pbx1 is converted into a transcriptional activator upon acquiring the N-terminal region of E2A in pre-B-cell acute lymphoblastoid leukemia. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6061–6065. doi: 10.1073/pnas.90.13.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- van Dijk M. A., Murre C. extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994 Aug 26;78(4):617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- van Dijk M. A., Peltenburg L. T., Murre C. Hox gene products modulate the DNA binding activity of Pbx1 and Pbx2. Mech Dev. 1995 Jul;52(1):99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]