Abstract
Background
Identification of genes that are differently expressed is a common approach used to analyze genetic mechanisms underlying cancer development. However, recent study results suggest that many such genes relate to a small number of biological functions. We hypothesized that analysis of these functions provides a better understanding of tumor biology than does actual identification of these genes does.
Materials and Methods
We re-analyzed publicly available gene expression data for paired samples of prostate tumor and adjacent normal tissue from the same patients to identify genes differently expressed in individual tumors and then used them to identify the functions.
Results
We found significant interindividual variation in the type and the number of functions. After adjusting for redundancy and nonspecificity of the functional terms, we identified seven functions. Several of them showed a strong association with clinical traits, e.g. age at diagnosis, preoperative prostate-specific antigen concentration, Gleason grade, and biochemical recurrence. Actin cytoskeleton was the function most frequently associated with clinical traits. Of note, the association between function and clinical traits was much stronger than that between the genes differently expressed and those traits.
Conclusion
Different prostate tumors differ in their functional profiles. Functions of differently expressed genes are strongly associated with clinical traits. This suggests that analysis of functions of differently expressed genes may provide a better description of tumor biology than does analysis of the respective genes.
Keywords: Gene expression, prostate cancer, in silico, functional profiling, functionality index
Identification of molecular mechanisms underlying cancer development and progression remains a challenge. Whole-genome profiling of gene expression is widely used to identify cancer-related genes. However, that method usually identifies too many genes that are differently expressed genes (DEGs), and it is hard to decide which ones are ‘drivers’ and which are ‘passengers’ of cancer progression. There is a growing appreciation that carcinogenesis may be driven by the modulation of a relatively small number of biological functions.
Two approaches are possible for studying carcinogenesis: gene-based and function-based approaches. The goals of these approaches are to identify cancer-related genes and cancer-related functions, respectively. At first glance, the differences between these approaches seem unimportant since differences in function are driven by different expression of the genes associated with the function. However, a given function may be modulated by different genes. One can expect to find, therefore, less variation between tumors on the functional level than there is on the gene level. Consistent with this expectation, the results of gene expression profiling of prostate cancer only weakly overlap the results of prostate genome-wide association studies on the gene level, whereas the overlap at the functional level is strong (1).
In this study, we undertook in silico functional profiling of individual prostate tumors. We identified a number of functions associated with the transition from the normal to the tumorous state and detected significant variation between individual tumors at the functional level.
Materials and Methods
We used gene expression data from the Gene Expression Omnibus (GEO) database (2) in two datasets: GSE21034 and GSE6919. GSE21034 was used as the primary dataset because i) it provided a relatively large sample size (n=29) and ii) a detailed clinical description was available for all patients. Samples for that dataset had been obtained from radical prostatectomies performed at the Memorial Sloan-Kettering Cancer Center (3). The patients had been followed up with history, physical examination, and serum prostate-specific antigen (PSA) concentration testing every three months for the first year, every six months for the second year, and annually thereafter. Biochemical recurrence (BCR) had been defined as a PSA ≥0.2 ng/ml on at least two occasions.
For validation, we used GEO dataset GSE6919. Tissue samples for GSE6919 had been acquired from the Health Sciences Tissue Bank of the University of Pittsburgh Medical Center (4,5). The tumor samples consisted of 58 cases of primary prostatic adenocarcinoma of Gleason grades 5 to 9.
To identify DEGs for a given tumor, we used the fold change (FC): FC = LOG2(T/N), in which T is the gene expression value in the tumor and N that in the adjacent normal prostate tissue. We separately ranked genes by using the absolute values of FC from that with the largest to that with the smallest change for each patient. The top 5% of the ranked genes were then functionally annotated by using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (6). To control for selection biases, all genes in the dataset were used as a background.
Biological processes from the Gene Ontology (GO) database (7) were used for the functional description of individual prostate cancer tumors. The functional profile of a tumor was described by using a vector length, m, in which the number of GO-defined biological processes with vector elements equals 0 when a specific function is not affected in a given tumor and equals 1 when that function is affected. Each tumor was categorized as ‘function positive’ or ‘function negative,’ depending on whether the corresponding unadjusted p-value from DAVID exceeded the threshold for a significance which was assumed to be 0.01. Functional profiles of a tumor were assessed by using Student's t testing to identify any association with age at prostate cancer diagnosis, pretreatment PSA concentration, pathological Gleason grade, and BCR.
Results
Biological functions affected in prostate tumors
The lists of the gene datasets with corresponding expression values can be found in the GEO database (2). Probes that are not linked to known genes were excluded. For each tumor, probes were ranked according to the absolute values of the FC. We found that the average pairwise correlation coefficient between ranks from different tumors was 0.22 ± 0.04, indicating a weak similarity in gene expression profiles across the tumors.
We assessed the distribution of the top 5% of DEGs across 2580 GO-defined terms for biological processes to identify those in which DEGs clustered more frequently than one would expect by chance. An unadjusted p-value of ≤0.01 was used as the threshold for significance. This threshold was selected to ensure that most of the true findings would pass the first step of analysis. We identified 264 terms that were significant in at least one tumor. A substantial percentage of the terms were nonspecific, e.g. “developmental process,” “regulation of biological process.” Such nonspecific terms were excluded from further analysis; this left 136 terms (see Additional file 1: Specific biological functions affected in individual prostate tumors.).
Validation of DE functions
As noted above, GEO dataset GSE6919 was used for validation. The data were treated in the same way as the data from GSE21034 were. We detected 311 biological processes that were significant in at least one tumor. There was significant overlap in functions between datasets GSE6919 and GSE21034: 121 terms were significant in both datasets, which is much larger than the number expected by chance (namely 4). Strong overlap between the two datasets was also observed for specific functions (57/145=0.39), whereas the expected proportion of overlapping functions was ~0.3%. Most importantly, the proportions of functions were similar in the two datasets: Pearson's r=0.68; n=454; p<<10–6.
Redundancy of the GO terms
There was significant redundancy of functions, with many GO terms apparently describing the same functions. For example, “biological adhesion,” “cell adhesion,” “cell–matrix adhesion,” “cell–substrate adhesion,” “cell junction organization,” “regulation of cell adhesion,” “cell–cell adhesion,” and “cell–substrate junction assembly” are all obviously linked to adhesion. To account for this redundancy, we combined synonymous terms to form a joint functional term. Only terms occurring at least five times were included in the analysis. If at least one of the terms from the joint group was significant, a tumor was considered positive. In total, 33 joint functional terms were identified. [Additional file 2: Specific and joint (after correction for term redundancy) functional terms from GSE21034.]
Association between functions of DEGs and clinical traits
We also assessed the data to identify any association between functions of DEGs and the patients’ clinical traits. Only specific functions were used. We applied the Bonferoni correction to correct for multiple comparisons. p-Values of ≤0.1 after correction for multiple testing were considered statistically significant. Thirteen functions significantly associated with clinical traits were thus identified (Table I).
Table I.
Specific terms associated with clinical traits.
| Function | DxAge | PreTxPSA | PathGGS | BCREvent |
|---|---|---|---|---|
| Antigen processing and presentation | 1.33 (0.1628) | 0.57 (0.3348) | 0.55 (0.3387) | 3.85 (0.0008) |
| Response to wounding | −0.11 (0.3932) | 0.56 (0.3367) | 0.27 (0.3810) | 3.30 (0.0033) |
| Immune system process | 0.03 (0.3954) | 3.37 (0.0027) | 0.70 (0.3078) | 2.55 (0.0189) |
| Vascular process in circulatory system | −0.46 (0.3548) | 1.92 (0.0656) | 4.17 (0.0003) | 2.32 (0.0307) |
| Positive regulation of cell adhesion | 0.39 (0.3658) | 4.16 (0.0003) | 2.91 (0.0084) | 2.32 (0.0307) |
| Positive regulation of cell-substrate adhesion | 0.39 (0.3658) | 4.16 (0.0003) | 2.91 (0.0084) | 2.32 (0.0307) |
| Regulation of muscle contraction | 0.39 (0.3658) | 4.16 (0.0003) | 2.91 (0.0084) | 2.32 (0.0307) |
| Response to estradiol stimulus | 0.39 (0.3658) | 4.16 (0.0003) | 2.91 (0.0084) | 2.32 (0.0307) |
| Extracellular matrix organization | 3.35 (0.0029) | 0.94 (0.2523) | 1.71 (0.0936) | 1.36 (0.1565) |
| Skeletal system development | 3.28 (0.0034) | 0.76 (0.2944) | 0.99 (0.2404) | 1.36 (0.1565) |
| Muscle contraction | 3.06 (0.0059) | 2.19 (0.0397) | 2.54 (0.0193) | 0.97 (0.2451) |
| Collagen fibril organization | 3.10 (0.0053) | −0.34 (0.3727) | 0.70 (0.3078) | 0.64 (0.3206) |
DxAge, Age at diagnosis; PreTxPSA, pretreatment prostate-specific antigen concentration; Path GGS, pathological grade by Gleason score; BCR_Event, biochemical recurrence. Student's t-test values are shown, with p-values in parentheses. The numbers in bold indicate significant associations after adjustment for multiple testing.
The strongest association was found between vascular process and Gleason grade. Positive regulation of cell adhesion, regulation of muscle contraction, and response to estradiol stimulus were positively associated with pretreatment PSA level. “Antigen processing and presentation” was positively associated with BCR. “Calcium ion transport” and “cell projection organization” were negatively associated with survival. Functions related to extracellular matrix (ECM) organization, collagen, and skeletal system development were positively associated with patient age at diagnosis.
Functions of DEGs were more strongly associated with clinical traits than DEGs
To assess whether functions of DEGs had stronger associations with clinical traits than the DEGs themselves had, we estimated the association between the expression level of individual genes and the patients’ clinical traits by using the same significance level after correction for multiple testing. We found only one out of 13,645 genes, namely death ligand signal enhancer (KIAA0141), that was positively associated with BCR. For the level of associations detected for functions, the expected number of genes associated with clinical traits would be 13/136 × 13,654 = 1305. This suggests that the function-based approach is more efficient than the gene-based approach for identifying associations with clinical traits.
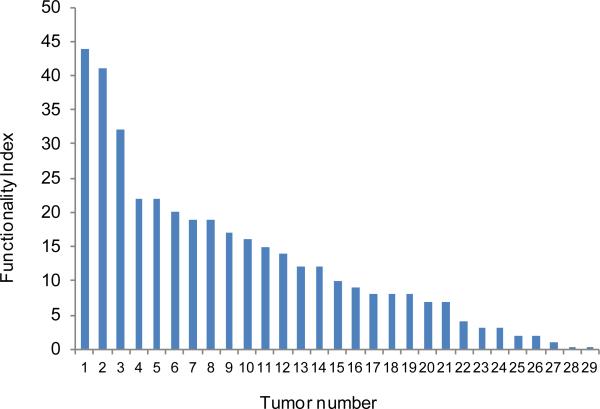
Functionality index
The total number of significant functions for each tumor was used as a functionality index (FI). The FI ranged from 0 to 44 (Figure 1). To test whether interindividual variation in FI is a result of interindividual variation in the number of DEGs, we estimated the number of genes with at least a twofold change in expression level in the tumors. We compared two groups of tumors, namely the five with the highest vs. the five with the lowest FI, and found no between-group statistically significant differences: 1589 ± 562 and 1761 ± 356 genes, respectively; df=10; p=0.88. The correlation between FI and the average absolute FC was also insignificant: Pearson's r=–0.14; n=29; p=0.47. These results suggest that tumors with low and high FIs have similar numbers of DEGs. However, in the low-FI group, the DEGs were randomly distributed across functional categories, whereas in the high-FI group, they were clustered into a small number of functional categories.
Figure 1. Functionality index distribution.
Each bar represents an individual tumor evaluated for the number of functions related to differently expressed genes.
We also assessed for any correlation between FI and clinical traits and found that the FI correlated positively with age at diagnosis and with BCR: Pearson's r=0.39; n=29; p=0.03 for both of these traits.
Correlation between functional terms
To identify common functional signatures for the transition from normal to tumorous prostate, we combined the data from the discovery and validation sets and used the functions that were affected in at least 10% of the tumors. Pairwise correlations were used to identify related functions. A Pearson's correlation coefficient of an absolute value of 0.38, which corresponds to a p-value of 0.01 after adjustment for multiple comparisons, was used as the threshold for statistical significance; 29 significant correlations were detected, all of them positive.
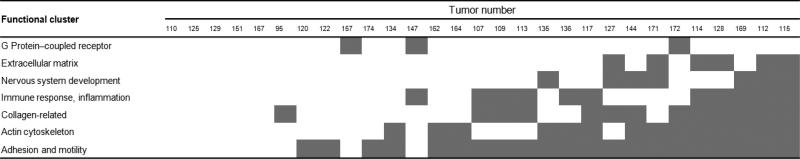
The correlated functions were then combined into functional clusters, seven of which were identified: i) adhesion and motility, ii) actin cytoskeleton, iii) collagen-related, iv) immune response and inflammation, v) nervous system development, vi) ECM and vii) G protein–coupled receptor. Figure 2 shows the functional profiles of the tumors from the dataset GSE21034. In some tumors (e.g. numbers 10, 25, 29, and 51) none of the functions were significantly correlated, whereas in others (e.g. numbers 112 and 115), six out of the seven functional clusters were affected.
Figure 2. Functional profile of individual tumors according to our analysis of functional clusters.
Shaded areas indicate that the corresponding functional cluster was significantly modified in a given tumor.
We also analyzed the association between the clusters and clinical traits (Table II). Overall, the results of the analysis were consistent with what we found for specific functions (Table I). A positive association between modulation of actin cytoskeleton and age at diagnosis was detected. ECM was also associated with age at diagnosis. Actin cytoskeleton was also positively associated with pretreatment PSA level, pathological Gleason grade, and BCR. The other function associated with BCR was immune response and inflammation. The FI was positively associated with Gleason grade and BCR.
Table II.
Association between functional clusters and clinical traits.
| Functional cluster | DxAge | PreTxPSA | PathGGS | BCR_Event |
|---|---|---|---|---|
| Adhesion and motility | 0.94 (0.2523) | 2.05 (0.0519) | 2.31 (0.0313) | 2.41 (0.0255) |
| Actin cytoskeleton | 3.61 (0.0015) | 3.98 (0.0006) | 3.60 (0.0015) | 4.04 (0.0005) |
| Collagen related | 0.62 (0.3248) | 1.70 (0.0951) | 1.88 (0.0704) | 2.76 (0.0119) |
| Immune response and inflammation | 0.98 (0.2428) | 0.77 (0.2921) | 0.47 (0.3531) | 3.38 (0.0027) |
| Nervous system development | 0.32 (0.3753) | 1.29 (0.1713) | 0.97 (0.2451) | −0.38 (0.3673) |
| Extracellular matrix | 3.61 (0.0015) | 1.39 (0.1504) | 2.93 (0.0080) | 1.86 (0.0729) |
| G Protein-coupled receptor | -2.93 (0.0080) | 0.87 (0.2689) | 1.34 (0.1607) | 1.24 (0.1823) |
| Functionality index | 1.89 (0.0692) | 2.98 (0.0071) | 3.20 (0.0042) | 3.83 (0.0008) |
DxAge, Age at diagnosis; PreTxPSA, pretreatment prostate-specific antigen concentration; Path GGS, pathological grade by Gleason score; BCR_Event, biochemical recurrence. Student's t-test values are shown, with p-values in parentheses. The numbers in bold indicate significant associations after adjustment for multiple testing.
Discussion
In their pivotal paper “The hallmarks of cancer,” Hanahan and Weinberg (8) identified self-sufficiency of growth signals, evasion of apoptosis, angiogenesis, and some other functions as hallmarks of cancer development. More recently, Pietras and Ostman (9) put these functions into the context of tumor–stroma interactions. In both studies, the number of cancer-associated functions was assumed to be rather small. The goal of our analysis was to identify cancer-associated functions. We thus focused on individual tumors to identify individual functional profiles and link them to the patients’ clinical characteristics.
A limitation of our approach is that it does not tell us whether the function is up-regulated or down-regulated. Genes associated with a given function form a complex network of interactions which is only partially understood, and it is difficult to predict the net result of modulated expression of the multiple genes forming the network; one of the few exceptions is the ECM. We estimated the FC for ECM-related genes, including collagens, elastins, fibrinogens, and laminins, and compared the FC estimates with those for other genes in the human genome. The average FC for the ECM genes was –0.29 ± 0.05, which is lower than the 0 FC expected under the null hypothesis (t-test=10.3; df=18426; p<10–22). For all other (i.e. not ECM-related) genes, the average FC was 0.02 ± 0.01, which is not statistically significantly different from 0. These results suggest that in prostate tumors, the ECM is less dense than it is in the surrounding normal prostate tissue. This lower ECM density may contribute to tumor growth and metastasis.
We also noted an association between positive regulation of cell adhesion and PSA. Some investigators, such as Romanov et al. (10), suggested that PSA is directly involved in the adhesion of prostate cancer cells to the bone marrow endothelium. We also found an association between the actin cytoskeleton and both vasculature and the Gleason grade (Tables I and II). A number of other investigators have also noted a link between genes for actin skeleton and Gleason grade, including positive associations for thymosin beta 15 (11), and for the actin filament-associated protein AFAP-110 (12). Other actin cytoskeleton-related proteins that are associated with Gleason grade include zinc finger protein 185 (ZNF185) (13), TAO kinase 2 (TAOK2, formerly known as PSK) (14), talin 1 (15), and beta-catenin (16,17). Taken together, these results support the idea that modulation of the actin cytoskeleton may underlie modulation of Gleason grade in the course of prostate cancer progression.
Functions associated with BCR included actin cytoskeleton and immune response. A number of actin cytoskeleton-related genes associated with prostate cancer progression have been identified recently (18-21), suggesting that modulation of the actin cytoskeleton is associated with progression. Although literature on the role of the immune response in prostate cancer progression is less extensive, the results of some studies also suggest a role for inflammation and immune response in progression (22,23).
We were surprised to find that classical cancer-associated functions, i.e. angiogenesis, apoptosis, and cell proliferation, were represented in only a small proportion of tumors: 0.11, 0.1, and 0.09, respectively. These functions included one or only a few genes that had very strong effects. For example, vascular endothelial growth factor A is a major player in angiogenesis, and modulating its expression has a dramatic effect on blood vessel development. When modulation of a function is driven by modulation of the expression of only one or a few genes, the resulting poor clustering signal may in turn result in a failure to detect any association. On the other hand, when multiple genes that have a relatively small effect contribute to a function, then simultaneous modulation of a number of those genes is required to modulate the function. This can easily be detected by the approach we have used.
Of note, the functions most frequently modulated in our analysis, namely cell adhesion and actin cytoskeleton, are controlled by multiple genes that have additive effects. Functions with polygenic control are not very suitable for experimental studies because their modulation requires simultaneously modulated expression of multiple genes. This may explain why modulation of these functions is not considered a hallmark of carcinogenesis.
We found that the number of significant functions in a given patient (i.e. the FI) were associated with age at diagnosis, Gleason grade, and BCR; these associations may result from multiple related weakly associated functions. However, when several weakly associated functions were pooled in the form of the FI, the overall associations became significant.
In summary, our analysis demonstrates that a function-centered approach to studying carcinogenesis more efficiently detects associations with clinical traits than does a gene-centered approach. In other words, profiling a panel of functions, e.g. by assessing overall and bone-specific adhesion and proliferation of tumor cells derived from biopsy specimens of primary tumors, may thus more efficiently predict prostate tumor progression than would an assessment of the expression of individual genes.
Supplementary Material
Acknowledgements
This research was supported in part by the National Institutes of Health through MD Anderson's Cancer Center Support Grant, CA016672. We thank Karen F. Phillips, ELS(D), of MD Anderson's Department of Genitourinary Medical Oncology, for editing the manuscript.
Footnotes
Authors’ contributions
IPG conceived of and designed the study and interpreted the results. JB performed the analyses under the supervision of IPG. CJL contributed to the study design and interpretation of results. IPG drafted the manuscript, and all authors read and approved the final manuscript.
Competing interests
The Authors have no relevant financial relationships with any commercial interests.
References
- 1.Gorlov IP, Gallick GE, Gorlova OY, Amos C, Logothetis CJ. GWAS meets microarray: Are the results of genome-wide association studies and gene-expression profiling consistent? Prostate cancer as an example. PLoS One. 2009;4:e6511. doi: 10.1371/journal.pone.0006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gene Expression Omnibus (GEO) database [ http://www.ncbi.nlm.nih.gov/geo/]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 6.Database for Annotation, Visualization, and Integrated Discovery (DAVID) [ http://david.abcc.ncifcrf.gov/] [PubMed]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.Pietras K, Ostman A. Hallmarks of cancer: Interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 10.Romanov VI, Whyard T, Adler HL, Waltzer WC, Zucker S. Prostate cancer cell adhesion to bone marrow endothelium: The role of prostate-specific antigen. Cancer Res. 2004;64:2083–2089. doi: 10.1158/0008-5472.can-03-3487. [DOI] [PubMed] [Google Scholar]
- 11.Bao L, Loda M, Janmey PA, Stewart R, Anand-Apte B, Zetter BR. Thymosin beta 15: A novel regulator of tumor cell motility upregulated in metastatic prostate cancer. Nat Med. 1996;2:1322–1328. doi: 10.1038/nm1296-1322. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Park SI, Artime MC, Summy JM, Shah AN, Bomser JA, Dorfleutner A, Flynn DC, Gallick GE. AFAP-110 is overexpressed in prostate cancer and contributes to tumorigenic growth by regulating focal contacts. J Clin Invest. 2007;117:2962–2973. doi: 10.1172/JCI30710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J- S, Gong A, Young CY. ZNF185, an actin–cytoskeleton-associated growth inhibitory LIM protein in prostate cancer. Oncogene. 2007;26:111–122. doi: 10.1038/sj.onc.1209769. [DOI] [PubMed] [Google Scholar]
- 14.Moore TM, Garg R, Johnson C, Coptcoat MJ, Ridley AJ, Morris JD. PSK, a novel STE20-like kinase derived from prostatic carcinoma that activates the c-Jun N-terminal kinase mitogen-activated protein kinase pathway and regulates actin cytoskeletal organization. J Biol Chem. 2000;275:4311–4322. doi: 10.1074/jbc.275.6.4311. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–1895. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitaker HC, Girling J, Warren AY, Leung H, Mills IG, Neal DE. Alterations in beta-catenin expression and localization in prostate cancer. Prostate. 2008;68:1196–1205. doi: 10.1002/pros.20780. [DOI] [PubMed] [Google Scholar]
- 17.Aaltomaa S, Karja V, Lipponen P, Isotalo T, Kankkunen JP, Talja M, Mokka R. Reduced alpha- and beta-catenin expression predicts shortened survival in local prostate cancer. Anticancer Res. 2005;25:4707–4712. [PubMed] [Google Scholar]
- 18.Assinder S, Cole N. Does TGF-beta induced formation of actin stress fibres reinforce Smad dependent TGF-beta signalling in the prostate? Med Hypotheses. 2011;76:802–804. doi: 10.1016/j.mehy.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Desiniotis A, Kyprianou N. Significance of talin in cancer progression and metastasis. Int Rev Cell Mol Biol. 2011;289:117–147. doi: 10.1016/B978-0-12-386039-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz WA, Ingenwerth M, Djuidje CE, Hader C, Rahnenfuhrer J, Engers R. Changes in cortical cytoskeletal and extracellular matrix gene expression in prostate cancer are related to oncogenic ERG deregulation. BMC Cancer. 2010;10:505. doi: 10.1186/1471-2407-10-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells CM, Whale AD, Parsons M, Masters JR, Jones GE. PAK4: A pluripotent kinase that regulates prostate cancer cell adhesion. J Cell Sci. 2010;123:1663–1673. doi: 10.1242/jcs.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elkord E. Immunology and immunotherapy approaches for prostate cancer. Prostate Cancer Prostatic Dis. 2007;10:224–236. doi: 10.1038/sj.pcan.4500964. [DOI] [PubMed] [Google Scholar]
- 23.Waugh DJ, Wilson C, Seaton A, Maxwell PJ. Multi-faceted roles for CXC-chemokines in prostate cancer progression. Front Biosci. 2008;13:4595–4604. doi: 10.2741/3025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.