Abstract
Background
Gracilaria tenuistipitata is an agarophyte with substantial economic potential because of its high growth rate and tolerance to a wide range of environment factors. This red seaweed is intensively cultured in China for the production of agar and fodder for abalone. Microsatellite markers were developed from the chloroplast genome of G. tenuistipitata var. liui to differentiate G. tenuistipitata obtained from six different localities: four from Peninsular Malaysia, one from Thailand and one from Vietnam. Eighty G. tenuistipitata specimens were analyzed using eight simple sequence repeat (SSR) primer-pairs that we developed for polymerase chain reaction (PCR) amplification.
Findings
Five mononucleotide primer-pairs and one trinucleotide primer-pair exhibited monomorphic alleles, whereas the other two primer-pairs separated the G. tenuistipitata specimens into two main clades. G. tenuistipitata from Thailand and Vietnam were grouped into one clade, and the populations from Batu Laut, Middle Banks and Kuah (Malaysia) were grouped into another clade. The combined dataset of these two primer-pairs separated G. tenuistipitata obtained from Kelantan, Malaysia from that obtained from other localities.
Conclusions
Based on the variations in repeated nucleotides of microsatellite markers, our results suggested that the populations of G. tenuistipitata were distributed into two main geographical regions: (i) populations in the west coast of Peninsular Malaysia and (ii) populations facing the South China Sea. The correct identification of G. tenuistipitata strains with traits of high economic potential will be advantageous for the mass cultivation of seaweeds.
Keywords: cpSSRs, Genetic analysis, Haplotype, Rhodophyta
Findings
Background
Gracilaria tenuistipitata Chang and Xia is a red seaweed that is distributed not only throughout the tropical and subtropical regions in the Western Pacific [1], but also in the Indian Ocean [2]. This seaweed is cultured intensively in China for the production of agar and fodder for abalone [3,4]. This species’ wide tolerance to cultivation environment, high growth rate and high agar yield make it suitable for cultivation [5-7]. G. tenuistipitata was first described by Chang and Xia in 1976, and a new variety, G. tenuistipitata var. liui, was reported in 1988 [1]. G. tenuistipitata is found in Vietnam [8-10], Thailand [11], Philippines [12] as well as Kuah, Pulau Langkawi [2] and Batu Laut, Selangor [13,14] in Malaysia.
In seaweeds, the morphological species concept remains a basis for species-level and intraspecific studies [15]. However, seaweeds are well-known for their high morphological plasticity, and the same species of seaweeds can have a wide range of morphological features when grown under different environmental conditions. Correct identification is vital for commercial exploitation, such as with species of Kappaphycus, Gracilaria, Sargassum, and Caulerpa. Despite being chosen from the same clone and cultivated under identical conditions, these species may still vary in terms of yield and morphological features. Hence, molecular markers are pivotal for the observation and exploitation of genetic variations, which include genome structure, organization, and evolution throughout the entire genome [16]. Moreover, molecular markers provide an important tool for evaluating the levels and patterns of genetic diversity and have previously been utilized successfully in a variety of plant species [17-19].
Microsatellites, or simple sequence repeats (SSRs), are tandem repeats of 1–6 nucleotides that are distributed throughout the genomes of most eukaryotic species [18]. SSRs are the markers of choice for a variety of applications in plant genetics and breeding because of their multiallelic nature, higher levels of polymorphisms, codominant inheritance and relatively abundant [20]. Microsatellite loci have been successfully identified in a number of seaweeds, such as the red Gracilariopsis lemaneiformis [21], the green Ulva intestinalis [22] and the brown Saccharina (Laminaria) japonica [23]. The conventional method of developing SSR markers by generating genomic libraries after sequencing large numbers of clones to search for the SSR-containing DNA regions [24] is labor-intensive, time-consuming and costly [25-27]. An abundance of DNA sequence information has now been generated and deposited in online databases with the establishment of genome and expressed sequence tag (EST) sequencing projects in many algal species. These highly polymorphic SSR markers are useful in germplasm characterization, marker-assisted selection, cultivar identification, genetic diversity and phylogenetic relationship studies [28].
To date, the potential usage of microsatellite markers in seaweeds has been tested in Gracilaria gracilis [29] and Porphyra haitanensis [30] for assessing genetic variability, Undaria pinnatifida for exploring genetic structure [31], Porphyra sp. for germplasm identification [32], Asparagopsis taxiformis for determining ploidy level and sexual reproduction [33], and Gracilariopsis lemaneiformis [21] and Chondrus crispus for performing genetic diversity studies [34], among others. In Malaysia, molecular research has focused on Gracilaria species to investigate genetic relationships using random amplified polymorphic DNA (RAPD) [35,36] and genetic diversity using the mitochondrial cox1 gene [37]. cDNA libraries have also been generated for the Malaysian Gracilaria changii [38] and Sargassum binderi [39].
Chloroplast DNA (cpDNA) varies in terms of haploid nature, uniparental inheritance and lack of recombination compared with the nuclear genome [15,40]. The complete plastid genome sequences available in Rhodophyta include the chloroplast [41] and mitochondrial genomes [42] of Porphyra purpurea and the mitochondrial genome of Chondrus crispus [43]. In 2004, Hagopian et al. sequenced the chloroplast genome of G. tenuistipitata var. liui, which was the first plastid genome successfully completed from the subclass Florideophycidae (Rhodophyta) [44]. The life history of G. tenuistipitata var. liui was completed in vitro by Barufi et al. [45], and the mitochondria genome was successfully sequenced in 2010 [46]. Recently, expressed sequence tags of G. tenuistipitata var. liui were established and deposited in a public database, the National Center for Biotechnology Information (NCBI) [47]. This finding shows that there is increasing interest in G. tenuistipitata as a model organism for functional genomics studies.
Chloroplast SSRs (cpSSRs) have been used mostly in plant studies, such as the design of cpSSRs primers to amplify targeted regions in a diverse array of plant species [48], the development of universal primers to amplify SSRs in grasses by Provan et al. (2004a) [49] and also the development of a set of universal cpSSR primers to explore cpDNA diversity among sub-tropical and tropical fruit crops [50]. However, few studies of cpSSRs have been conducted in algae, such as the development of universal primers for the amplification of chloroplast coding and non-coding regions in Chlorophyta and Rhodophyta [51].
The aims of this study were to develop a set of microsatellite markers from the chloroplast genome of G. tenuistipitata using available bioinformatics tools and to test the derived primers on eighty specimens of G. tenuistipitata from six different localities to examine the genetic variation of G. tenuistipitata from different populations.
Methods
Ethics statement
The Gracilaria tenuistipitata specimens were not collected from any national parks or protected areas. No specific permissions were required to collect the specimens.
Sample collection and DNA extraction
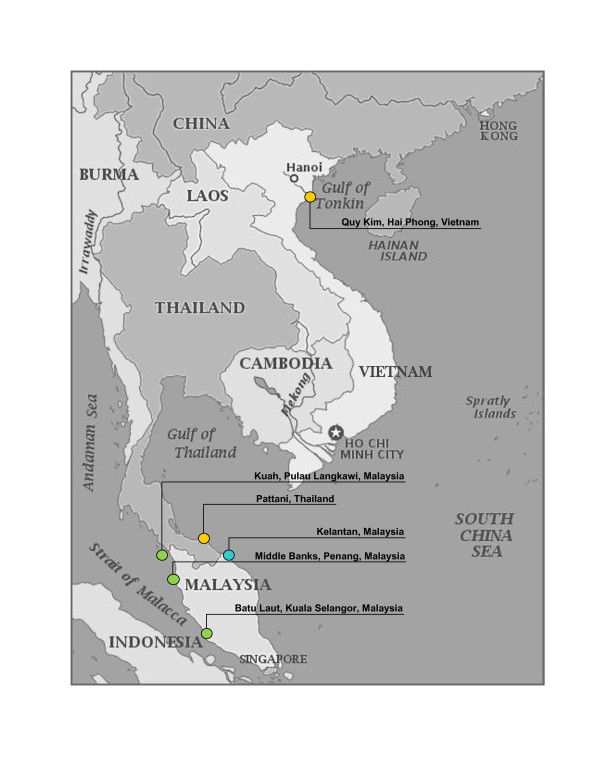
A total of 80 Gracilaria tenuistipitata specimens were collected from six different localities: i) Batu Laut, Selangor, Malaysia ii) Middle Banks, Penang, Malaysia iii) Kuah, Pulau Langkawi, Malaysia iv) Kelantan, Malaysia v) Quy Kim, Hai Phong, Vietnam and vi) Pattani, Thailand (Figure 1). Female gametophytes (haploid) and tetrasporophytes (diploid) were identified by their reproductive organs. The samples were grounded into powder in liquid nitrogen and DNA extraction was performed using the DNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions with minor modifications. The extracted genomic DNA was kept at -20°C for further analysis.
Figure 1.
Map displaying the six different locations where the G. tenuistipitata were collected. The haplotypes of various localities are represented by different colors. Samples from Batu Laut, Middle Banks and Kuah (Malaysia) correspond to Clade A; Samples from Thailand and Vietnam correspond to Clade B1 and samples from Kelantan, Malaysia correspond to Clade B2. (adapted from http://www.yourchildlearns.com/online-atlas/Asia/vietnam-map.htm).
SSR development
The 183,883 bp G. tenuistipitata var. liui chloroplast genome DNA sequence was downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) and saved in FASTA format. Perl scripts were developed to search for microsatellite tandem repeats in the genome sequence data using the MIcroSAtellite (MISA) search module (http://pgrc.Ipk-gatersleben.de/misa/). To avoid homoplasy, only perfect SSRs (with no substitutions interrupting the core motif) and nucleotides with repeated patterns of length two or more were used in this study. Primers were designed using Primer3 [52]. The parameters used for the design of SSRs were defined in such a way that the primer annealing temperatures varied from 48 to 55°C with primer length within 20–24 bp, GC content between 50 - 70% and the expected product size between 280 and 350 bp.
SSR analysis
Polymerase chain reactions (PCRs) were performed in a total volume of 15 μl containing 1.5 μl 10x PCR buffer (Takara Biotechnology, Dalian, China), 0.2 mM dNTP mix (Takara), 0.5 U Taq polymerase (Takara), 0.3 mM of each primer pairs, and 25–50 ng genomic DNA with UHQ water added to a total volume of 15 μl. The amplification reaction conditions were as follows: 5 min denaturation step at 94°C, followed by thirty-five cycles of 1 min at 94°C, 1 min at 52°C, 2 min elongation at 72°C, and a final extension at 72°C for 5 min. The forward primer of each primer pair was fluorescently [69-carboxy-fluorecine (FAM)] labelled. The optimized annealing temperature for PCR amplifications was 52°C and this optimum temperature was used to test all the specimens. The amplification products were separated on a 3.0% MetaPhor® agarose gel (FMC or Cambrex Corporation, USA) at 75 V for 75 min and visualized by SYBR SAFE staining (Invitrogen, USA). Amplification products were sent for fragment analysis to detect alleles using an automated DNA sequencer.
Data analysis
Primer-pairs that exhibited good amplification in all the eighty G. tenuistipitata samples were considered usable. SSR products were scored using a binary matrix method with “1” (presence) and “0” (absence) based on the SSR pattern that was amplified by each primer-pair designed. These DNA fingerprints of G. tenuistipitata specimens from different locations were constructed using GelQuest software [53] based on the size standard template ABI GeneScan 50–500 and followed with similarity matrix cluster analysis based on the UPGMA (unweighted pair group method using arithmetic means) using ClusterVis software version 1.4.2 [53].
The height of the peak greater than 50% the height of the highest peak was selected to avoid background noise, and the amplified fragment size analyzed was set in a range that included the expected amplified fragment size. Cluster analysis was performed based on the amplified fragment sizes of the samples, and the dendrogram was generated with the hyperbin width set to a value that will resulted in a group of defined amplified fragment sizes. The resulting UPGMA dendrogram was visualized and edited using the TreeMe software [54].
To verify that the polymorphisms were not because of indels in the regions flanking the cpSSRs, three PCR products that were randomly selected from each locality were purified and sequenced for each primer pair. The sequencing data were analyzed and edited using Chromas 1.45 (Technelysium Pty Ltd., Australia) and BioEdit 7.0.9.0 [55] software. Edited sequences were aligned by the CLUSTAL X program [56].
Results
Eight perfect SSRs with designed primer-pairs (Table 1) were obtained from the 183,883 bp Gracilaria tenuistipitata var. liui chloroplast genome. Of the eight primer-pairs, five (62.5%) were mononucleotide repeats, two (25%) were dinucleotide repeats and only one (12.5%) was a trinucleotide repeat. All cpSSRs obtained were A, T or AT repeats. No CG microsatellites were identified. All primer-pairs were tested on 80 specimens of G. tenuistipitata from different localities. The eight primer-pairs demonstrated good amplification in 3% Metaphor agarose gel electrophoresis.
Table 1.
Primer pairs derived from the chloroplast genome of G. tenuistipitata var. liui
| Name | Forward primer (5′-3′) | Reverse primer (5′-3′) | Repeat motif | Predicted product size (bp) | Genomic coordinates of the SSR loci | Ta (°C) | Coding/non-coding region |
|---|---|---|---|---|---|---|---|
| GT1 |
TTTATCAACGATCCCTGTAG |
AATGGACTGTAATTCACCAA |
(T)10 |
336 |
4314 – 4323 |
52 |
Non-coding |
| GT2 |
TTTTTGAGCGATATTTTGAC |
AGAATAAGACCACCTGAACC |
(T)10 |
310 |
78590 – 78599 |
53 |
Coding |
| GT3 |
CCATAATGGAGATCTGTTTG |
CTGGCAACATAGTTAGCATT |
(T)10 |
306 |
84904 – 84913 |
53 |
Coding |
| GT4 |
AGCAATCCTAAATTGACAAC |
AAGGTAGACCAGGAGAAAAA |
(ATT)5 |
295 |
99538 – 99552 |
53 |
Coding |
| GT5 |
AGAAATTGATCAAGCTGTT |
TTTTCAGCAATTGGAGTATC |
(AT)8 |
329 |
105129 – 105144 |
52 |
Non-coding |
| GT6 |
CCTACAATCAGAATGGAATG |
AGCTTCCAAGAAAAATGAGT |
(A)10 |
303 |
143312 – 143321 |
53 |
Coding |
| GT7 |
ATCCTTCTTTTAAGCCGTAG |
TCTTCCATGAAGTCTTCTTTT |
(T)10 |
309 |
161988 – 161997 |
53 |
Non-coding |
| GT8 | CTCCTGACATGATAAACACC | CTTCGATTTGTTTAATGAGC | (AT)7 | 288 | 179473 – 179486 | 52 | Non-coding |
Ta Annealing temperature.
Mononucleotide primer-pairs (GT1, GT2, GT3, GT6 and GT7) and the trinucleotide primer-pair (GT4) demonstrated good amplification but were monomorphic on the samples tested. Only one defined amplified fragment size, 336 bp for primer-pair GT1, 312 bp for primer-pair GT2, 308 bp for primer-pair GT3, 295 bp for primer-pair GT4, 301 bp for primer-pair GT6 and 307 bp for primer-pair GT7 was derived and the fragment results indicated that all samples fell within the same defined amplified fragment size.
For the primer-pair GT5, three defined amplified fragment sizes, 327 bp, 329 bp and 333 bp, were derived and samples with similar base size peaks fell into the same defined amplified fragment size (Table 2). The dendrogram for primer-pair GT5 indicated that the G. tenuistipitata specimens were grouped into three main clades: (a) G. tenuistipitata from Kelantan, Malaysia (b) G. tenuistipitata from Quy Kim, Hai Phong, Vietnam and Pattani, Thailand and (c) G. tenuistipitata from Kuah, Batu Laut and Middle Banks (Malaysia). All three clades were supported with similarity coefficients of 0.5.
Table 2.
Alleles of each G. tenuistipitata from six different localities for primer-pairs GT5 and GT8
|
Location |
Number of samples tested |
Primer GT5 |
Primer GT8 |
|||
|---|---|---|---|---|---|---|
| Allele (327 bp) | Allele (329 bp) | Allele (333 bp) | Allele (284 bp) | Allele (294 bp) | ||
| Batu Laut, Selangor, Malaysia |
15 |
1 |
0 |
0 |
0 |
1 |
| Middle Banks, Penang, Malaysia |
15 |
1 |
0 |
0 |
0 |
1 |
| Quy Kim, Hai Phong, Vietnam |
15 |
0 |
1 |
0 |
1 |
0 |
| Pattani, Thailand |
15 |
0 |
1 |
0 |
1 |
0 |
| Kelantan, Malaysia |
10 |
0 |
0 |
1 |
1 |
0 |
| Kuah, Pulau Langkawi, Malaysia | 10 | 1 | 0 | 0 | 0 | 1 |
1, Presence of SSR product.
0, Absence of SSR product.
For the primer-pair GT8, two defined amplified fragment sizes, 284 bp and 294 bp were derived, and samples with similar amplified fragment sizes exhibited a cell value in that column. The dendrogram for primer-pair GT8 indicated that the G. tenuistipitata specimens were grouped into two main clades: (a) G. tenuistipitata from Batu Laut, Middle Banks and Kuah (Malaysia) and (b) G. tenuistipitata from Pattani (Thailand), Quy Kim, Hai Phong (Vietnam), and Kelantan (Malaysia). All two clades were also supported with similarity coefficients of 0.5.
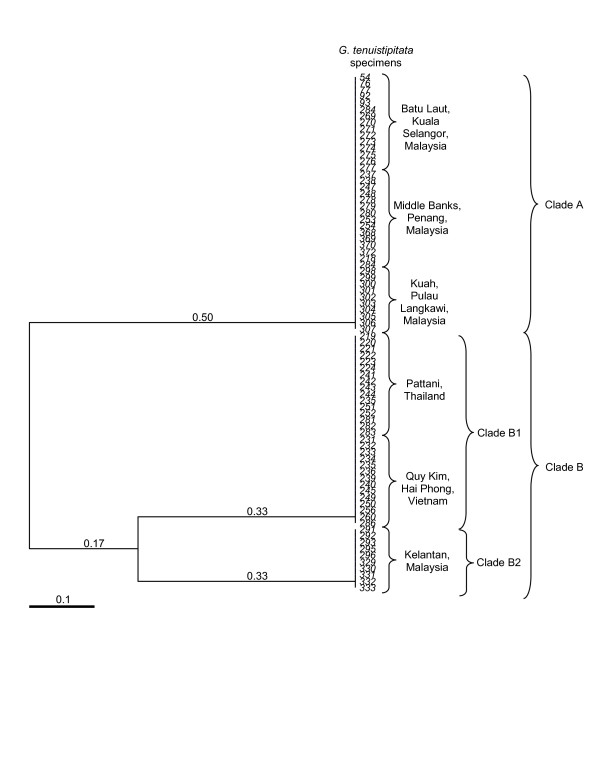
The combined dataset from the two primer-pairs produced a dendrogram (Figure 2) with similarity coefficients ranging from 0.17 to 0.5. From the generated dendrogram, the 80 specimens of G. tenuistipitata were grouped into two major clades: Clade A consisting of G. tenuistipitata from Batu Laut, MiddleBanks and Kuah (Malaysia), and Clade B consisting of G. tenuistipitata from Pattani (Thailand), Quy Kim, Hai Phong, (Vietnam), and Kelantan (Malaysia). Clade B is further divided into two sub-clades: Clade B1 consisting of G. tenuistipitata from Pattani (Thailand) and Quy Kim, Hai Phong, (Vietnam), and Clade B2 consisting of G. tenuistipitata from Kelantan (Malaysia). Both clades B1 and B2 were supported with a similarity coefficient of 0.33.
Figure 2.
Cluster analysis of eighty G. tenuistipitata specimens for the combined primer-pairs GT5 and GT8 from different localities using the unweighted pair group method with arithmetic average (UPGMA).
The sequencing results indicated that the amplification products of these primer-pairs were generated because of polymorphisms and not indels in the regions flanking the cpSSRs (the sequences of the amplified products can be provided upon request).
Discussion
In plants, the highest frequency of cpSSRs was 160 found in Arabidopsis thaliana, and the lower numbers of cpSSRs were found in Nymphaea alba, the water lilies with 35 and Nuphar advena with 39 [57]. Studies have suggested that the cpSSRs are abundant in most plants [58-60] but less abundant in algae. This finding is consistent with our analysis on SSRs identified from the 23 algae chloroplast genomes deposited at the National Center for Biotechnology Information database (data not shown here). The analysis indicated that approximately 50% of the chloroplast genome contains low numbers of repetitive sequences, particularly on SSRs. Ectocarpus siliculosus, Parachlorella kessleri, Monomastix sp., Scenedesmus obliquus, and Chlorella vulgaris were found to have one SSR; Bryopsis hypnoides, Micromonas sp., Cyanidioschyzon merolae and Cyanidium caldarium were found to have two SSRs; Gracilaria tenuistipitata var. liui and Porphyra yezoensis have three SSRs; and only Leptosira terrestris has more than 60 SSRs. These results demonstrate that G. tenuistipitata var. liui used in this study is in the category of low numbers of repetitive sequences.
Of the eight primer-pairs we designed, only two revealed genetic polymorphisms in the eighty specimens tested. Primer-pair GT5 differentiates the G. tenuistipitata into three main groups at the 0.5 similarity level by cluster analysis. This result demonstrated that the populations of G. tenuistipitata from Batu Laut, Middle Banks and Kuah that grow in the Straits of Malacca, on the west coast of Peninsular Malaysia, were separated from the populations of Thailand, Vietnam and Kelantan, Malaysia, which face the South China Sea (Figure 1).
Primer-pair GT8 was able to differentiate the populations of G. tenuistipitata from Vietnam and Thailand from the populations of G. tenuistipitata from Malaysia (Batu Laut, Middle Banks and Kuah), but not the population from Kelantan, Malaysia, as was achieved with the use of primer-pair GT5. Primer-pair GT5 indicated that there are three possible haplotypes of G. tenuistipitata from the samples collected, whereas primer-pair GT8 only indicated two haplotypes of G. tenuistipitata.
Using the combined dataset of the two primer-pairs (Figure 2), G. tenuistipitata from Kelantan, Malaysia was more closely related to the specimens from Quy Kim, Hai Phong (Vietnam) and Pattani (Thailand) and distantly separated from G. tenuistipitata from Batu Laut, Middle Banks and Kuah (Malaysia).
Two of the eight primer-pairs that were generated from the chloroplast genome of G. tenuistipitata were able to distinguish the populations of G. tenuistipitata from different geographical origins (west and east coast of Peninsular Malaysia) and the population from Kelantan, Malaysia was separated from other localities. The variation in the repeated nucleotides can be used not only in genetic mapping and marker-trait association studies [28,61] but also in genetic diversity and population studies. This information regarding genetic diversity and differentiation of strains of the same species is pivotal particularly in cultivation, where red algae have contributed to 24.5% of the total world seaweed production [62] for food, fodder, ingredients in cosmetics and fertilizers, and hydrocolloid production (e.g., agar and alginate) [63]. This study demonstrated that variation in SSR length and stability varies among loci within species [64,65], and microsatellites obtained from genomic libraries have been found to be more polymorphic [64,66]. However, whether are more polymorphic than ESTs of G. tenuistipitata must be proven by comparing SSRs developed from the two sources.
To obtain the optimal forward and reverse primers that flanked the SSR region and to ensure robust PCR amplification, such parameters as oligonucleotide melting temperature, primer size, GC content and PCR product size were designed to a defined set of constraints [67]. Our study indicated that the microsatellites mined from the chloroplast genome of G. tenuistipitata in both coding and non-coding regions are A/T rich, which also supports the findings that (AT)n sequences are commonly found in genomic libraries of plants [68-70]. This result may be observed because of the excess of AT repeats and deficiencies of AC/TG and CG repeats. C or G mononucleotide repeats were rare or absent [57].
There is limited gene flow and a weak geographic structuring pattern, because the number of haplotypes was rather low. However, this can only be verified with wider samples from more populations. For example, the analysis of the distribution of genetic diversity of the seaweed Chondrus crispus by Provan and Maggs (2012) [34] included 19 populations from both sides of the North Atlantic using mitochondrial single nucleotide polymorphisms (SNPs), sequence data from two single copy nuclear regions and microsatellite loci. These researchers’ results revealed unique genetic variation for all marker classes in the rear-edge populations in Iberia but not in the rear-edge populations in North America. Thus, more samples from different populations and also different techniques of molecular assays will likely help us understand the migration and dispersal ability of the species.
Conclusions
This study is consistent with the observation by Schaal et al. that the chloroplast genome is suitable for the design of simple sequence repeats because of its lower rate of mutation and therefore contains potentially more effective genetic markers of population subdivision and differentiation than the nuclear genome [71]. Indeed, there are a few studies of chloroplast markers in population genetic, biogeographic and hybridization [59,71-73]. Although the cpSSRs that we developed were not able to distinguish between different populations of seaweeds, the population from Kelantan, Malaysia was characterized by a specific haplotype and exhibited an interesting geographical pattern in which samples facing the Straits of Malacca have a different haplotype from those facing the South China Sea. It is important to choose and breed G. tenuistipitata with traits of high economic value and use them for cultivation. Additional studies on gel strength, gel contents and growth rates among the different populations of G. tenuistipitata will provide valuable information regarding strain selection for cultivation. Hence, molecular genetic studies, particularly on the development of molecular markers, are essential to effectively select such strains. Further studies concerning developing SSRs from the mitochondrial genome [46] and ESTs [47] of G. tenuistipitata may result in the development of molecular markers for differentiation between species (at the strain and population level) and among species, which will be important for aquaculture.
Abbreviations
cpSSRs: Chloroplast simple sequence repeats; RAPD: Random amplified polymorphic DNA; SSRs: Simple sequence repeats; UPGMA: Unweighted pair group method with arithmetic average
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PE and SM supervised the project. WW and SL mined the SSR primers. SL, PE, DH and AP provided the specimens. SL and PE analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Sze-Looi Song, Email: looi511@hotmail.com.
Phaik-Eem Lim, Email: phaikeem@um.edu.my.
Siew-Moi Phang, Email: phang@um.edu.my.
Weng-Wah Lee, Email: leewengwah@gmail.com.
Dang Diem Hong, Email: ddhong60vn@yahoo.com.
Anchana Prathep, Email: anchana.p@psu.ac.th.
Acknowledgements
This study was funded in part by the MoHE–HIR grant (H-50001-00-A000025), University of Malaya Research Grant Scheme (RG025-09SUS) and Postgraduate Research Fund (PS304-2009B, PS269-2010B and PV082-2011B), University of Malaya.
References
- Chang CF, Xia BM. On two new Gracilaria (Gigartinales, Rhodophyta) from South China. Taxonomy of economic seaweeds with reference to some Pacific and Caribbean species. 1988;2:127–129. [Google Scholar]
- Terada R, Kawaguchi S, Masuda M, Phang SM. Taxonomic notes on marine algae from Malaysia. III. Seven species of Rhodophyceae. Bot Mar. 2000;43:347–357. [Google Scholar]
- Chiang YM, Lin JL. Nitrate uptake by nitrogen-starved plants of the red alga Gracilaria tenuistipitata var. liui. Jpn J Phycol. 1989;37:187–193. [Google Scholar]
- Lin M-C, Liao C-M. 65Zn(II) accumulation in the soft tissue and shell of abalone Haliotis diversicolor supertexta via the alga Gracilaria tenuistipitata var. liui and the ambient water. Aquaculture. 1999;178:89–101. doi: 10.1016/S0044-8486(99)00128-3. [DOI] [Google Scholar]
- Collén J, Pinto E, Pedersén M, Colepicolo P. Induction of oxidative stress in the red macroalga Gracilaria tenuistipitata by pollutant metals. Arch Environ Contam Toxicol. 2003;45:337–342. doi: 10.1007/s00244-003-0196-0. [DOI] [PubMed] [Google Scholar]
- Lee TM, Chang YC, Lin YH. Differences in physiological responses between winter and summer Gracilaria tenuistipitata (Gigartinales, Rhodophyta) to varying temperature. Bot Bul Acad Sin. 1999;40:93–100. [Google Scholar]
- Oliveira EC, Alveal K. In: The mariculture of Gracilaria (Rhodophyta) for the production of agar. Akatsuka I, editor. The Hague: SPB Academic Publishing; 1990. Introduction to applied phycology; pp. 553–564. [Google Scholar]
- Nguyen HD. In: Taxonomy of economic Seaweeds. Abbott IA, editor. La Jolla, California: California Sea Grant College; 1992. Vietnamese species of Gracilaria and Gracilariopsis; pp. 207–210. vol. 3. [Google Scholar]
- Tseng CK, Xia BM. On the Gracilaria in the western Pacific and Southeastern Asia region. Bot Mar. 1999;42:209–217. [Google Scholar]
- Tsutsui I, Huynh QN, Nguyên HD, Arai S, Yoshida T. The common marine plants of southern Vietnam. Numerous colour photographs Usa: Japan Seaweed Association. 2005;251:1–250. [Google Scholar]
- Lewmanomont K. In: Taxonomy of Economic Seaweeds. Abbott IA, editor. La Jolla, California: California Sea Grant College Program; 1994. The species of Gracilaria from Thailand; pp. 135–148. vol. 4. [Google Scholar]
- Abbott IA. In: Taxonomy of Economic Seaweeds with Reference to some Pacific Species. Abbott IA, editor. California: California Sea Grant College; 1994. New Records and a Reassessment of Gracilaria (Rhodophyta) from the Philippines; pp. 111–118. [Google Scholar]
- Lim PE, Phang SM. Gracilaria species (Gracilariales, Rhodophyta) of Malaysia including two new records. Malays J Sci. 2004;23:71–80. [Google Scholar]
- Phang SM. Seaweed resources in Malaysia: current status and future prospects. Aquat Ecosyst Health Manag. 2006;9:185–202. doi: 10.1080/14634980600710576. [DOI] [Google Scholar]
- Wattier R, Maggs CA. Intraspecific variation in seaweeds: the application of new tools and approaches. Adv Bot Res. 2001;35:171–212. [Google Scholar]
- O’Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womack JE, Marshall Graves JA. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- Lim PE, Wong CL, Phang SM. In: Advances in Seaweed Cultivation and Utilization in Asia. Phang SM, Critchley AT, Ang POJr, editor. Kuala Lumpur: University of Malaya Maritime Research Center Publication; 2006. Molecular taxonomy of seaweeds with emphasis on Rhodophyta and Phaeophyta; pp. 105–142. [Google Scholar]
- Powell W, Machray GC, Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996;1:215–222. doi: 10.1016/1360-1385(96)86898-1. [DOI] [Google Scholar]
- Rafalski A, Vogel MJ, Morgante M, Powell W, Andre C, Tingey SV. In: Non-mammalian genome analysis: a practical guide. Biren B, Lai E, editor. London: Academic Press; 1996. Generating and using DNA markers in plants; pp. 75–134. [Google Scholar]
- Sun JW, Jin MD, Zhou CJ, Yang QK, Weng ML, Duan LD, Xu P, Ma JH, Wang B. Identification of Porphyra lines (Rhodophyta) by AFLP DNA fingerprinting and molecular markers. Plant Mol Biol Report. 2005;23:251–262. doi: 10.1007/BF02772755. [DOI] [Google Scholar]
- Pang Q, Sui Z, Kang KH, Kong F, Zhang X. Application of SSR and AFLP to the analysis of genetic diversity in Gracilariopsis lemaneiformis (Rhodophyta) J Appl Phycol. 2010;22:607–612. doi: 10.1007/s10811-009-9500-3. [DOI] [Google Scholar]
- Kostamo K, Blomster J, Korpelainen H, Kelly J, Maggs CA, Mineur F. New microsatellite markers for Ulva Intestinalis (Chlorophyta) and the transferability of markers across species of Ulvaceae. Phycologia. 2008;47:580–587. doi: 10.2216/08-16.1. [DOI] [Google Scholar]
- Liu F, Wang X, Yao J, Fu W, Duan D. Development of expressed sequence tag-derived microsatellite markers for Saccharina (Laminaria) japonica. J Appl Phycol. 2010;22:109–111. doi: 10.1007/s10811-009-9426-9. [DOI] [Google Scholar]
- Eujayl I, Sledge MK, Wang L, May GD, Chekhovskiy K, Zwonitzer JC, Mian MA. Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theor Appl Genet. 2004;108(3):414–422. doi: 10.1007/s00122-003-1450-6. [DOI] [PubMed] [Google Scholar]
- Kantety RV, la Rota M, Matthews DE, Sorrells ME. Data mining for simple-sequence repeats in expressed sequence tags from barley, maize, rice, sorghum, and wheat. Plant Mol Biol. 2002;48:501–510. doi: 10.1023/A:1014875206165. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Thiel T, Stein N, Langridge P, Graner A. In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett. 2002;7:537–546. [PubMed] [Google Scholar]
- Yu J-K, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME. Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome. 2004;47:805–818. doi: 10.1139/g04-057. [DOI] [PubMed] [Google Scholar]
- Wang ML, Mosjidis JA, Morris JB, Chen ZB, Barkley NA, Pederson GA. Evaluation of Lespedeza germplasm genetic diversity and its phylogenetic relationship with the genus Kummerowia. Conserv Genet. 2008;10:79–85. [Google Scholar]
- Wattier R, Dallas JF, Destombe C, Saumitou-Laprade P, Valero M. Single locus microsatellites in Gracilariales (Rhodophyta): high level of genetic variability within Gracilaria gracilis and conservation in related species. J Phycol. 1997;33:868–880. doi: 10.1111/j.0022-3646.1997.00868.x. [DOI] [Google Scholar]
- Xie C-T, Chen C-S, Ji D-H, Xu Y. Characterization, development and exploitation of EST-derived microsatellites in Porphyra haitanensis Chang and Zheng (Bangiales, Rhodophyta) J Appl Phycol. 2008;21:367–374. [Google Scholar]
- Voisin M, Engel CR, Viard F. Differential shuffling of native genetic diversity across introduced regions in a brown alga: aquaculture vs. maritime traffic effects. Proc Natl Acad Sci USA. 2005;102:5432–5437. doi: 10.1073/pnas.0501754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Liu T, Guo B, Jin D, Weng M, Feng Y, Xu P, Duan D, Wang B. Development of SSR primers from EST sequences and their application in germplasm identification of Porphyra lines (Rhodophyta) Eur J Phycol. 2006;41:329–336. doi: 10.1080/09670260600740906. [DOI] [Google Scholar]
- Andreakis N, Kooistra WHCF, Procaccini G. Microsatellite markers in an invasive strain of Asparagopsis taxiformis (Bonnemaisoniales, Rhodophyta): insights in ploidy level and sexual reproduction. Gene. 2007;406:144–151. doi: 10.1016/j.gene.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Provan J, Maggs CA. Unique genetic variation at a species’ rear edge is under threat from global climate change. Proc R Soc B. 2012;279:39–47. doi: 10.1098/rspb.2011.0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PE, Thong KL, Phang SM. Molecular differentiation of two morphological variants of Gracilaria salicornia. J Appl Phycol. 2001;13:335–342. doi: 10.1023/A:1017532624398. [DOI] [Google Scholar]
- Sim MC, Lim PE, Gan SY, Phang SM. Identification of random amplified polymorphic DNA (RAPD) marker for differentiating male from female and sporophytic thalli of Gracilaria changii (Rhodophyta) J Appl Phycol. 2007;19:763–769. doi: 10.1007/s10811-007-9224-1. [DOI] [Google Scholar]
- Yow Y-Y, Lim P-E, Phang S-M. Genetic diversity of Gracilaria changii (Gracilariaceae, Rhodophyta) from west coast, Peninsular Malaysia based on mitochondrial cox1 gene analysis. J Appl Phycol. 2011;23:219–226. doi: 10.1007/s10811-010-9535-5. [DOI] [Google Scholar]
- Teo SS, Ho CL, Teoh S, Lee WW, Tee JM, Rahim RA, Phang SM. Analyses of expressed sequence tags (ESTs) from an agarophyte, Gracilaria changii (Gracilariales, Rhodophyta) Eur J Phycol. 2007;42:41–46. doi: 10.1080/09670260601012461. [DOI] [Google Scholar]
- Wong TKM, Ho CL, Lee WW, Rahim RA, Phang SM. Analyses of expressed sequence tags (ESTs) from Sargassum binderi (Phaeophyta) J Phycol. 2007;43:528–534. doi: 10.1111/j.1529-8817.2007.00349.x. [DOI] [Google Scholar]
- Zuccarello GC, Burger G, West JA, King RJ. A mitochondrial marker for red algal intraspecific relationships. Mol Ecol. 1999;8:1443–1447. doi: 10.1046/j.1365-294x.1999.00710.x. [DOI] [PubMed] [Google Scholar]
- Reith M, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol Biol Report. 1995;13:333–335. doi: 10.1007/BF02669187. [DOI] [Google Scholar]
- Burger G, Saint-Louis D, Gray MW, Lang BF. Complete sequence of the Mitochondrial DNA of the red Alga Porphyra Purpurea: cyanobacterial Introns and shared ancestry of red and green Algae. Plant Cell. 1999;11:1675–1694. doi: 10.1105/tpc.11.9.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc C, Boyen C, Richard O, Bonnard G, Grienenberger J-M, Kloareg B. Complete sequence of the mitochondrial DNA of the rhodophyte Chondrus crispus (Gigartinales). Gene content and genome organization. J Mol Biol. 1995;250:484–495. doi: 10.1006/jmbi.1995.0392. [DOI] [PubMed] [Google Scholar]
- Hagopian JC, Reis M, Kitajima JP, Bhattacharya D, de Oliveira MC. Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastids. J Mol Evol. 2004;59:464–477. doi: 10.1007/s00239-004-2638-3. [DOI] [PubMed] [Google Scholar]
- Barufi JB, Cabral de Oliveira E, Plastino EM, Cabral de Oliveira M. Life history, morphological variability and growth rates of the life phases of Gracilaria tenuistipitata (Rhodophyta: Gracilariales) in vitro. Sci Mar. 2010;74:297–303. doi: 10.3989/scimar.2010.74n2297. [DOI] [Google Scholar]
- Takahashi MM. Sequênciamento e análise do genoma mitocondrial de Gracilaria tenuistipitata (Gracilariales, Rhodophyta) Brazil: University of São Paulo, Institute of Biosciences; 2010. (Master Thesis). [Google Scholar]
- Collén PN, Collén J, Reis MS, Pedersen M, Setubal JC, Varani AM, Colepicolo P, Oliveira MC. Analysis of expressed sequence tags from the agarophyte Gracilaria tenuistipitata (Rhodophyta) J Appl Phycol. 2011;24:1–7. [Google Scholar]
- Chung S-M, Staub JE. The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theor Appl Genet. 2003;107:757–767. doi: 10.1007/s00122-003-1311-3. [DOI] [PubMed] [Google Scholar]
- Provan J, Biss PM, McMeel D, Mathews S. Universal primers for the amplification of chloroplast microsatellites in grasses (Poaceae) Mol Ecol Notes. 2004;4:262–264. doi: 10.1111/j.1471-8286.2004.00636.x. [DOI] [Google Scholar]
- Cheng YJ, Meng HJ, Guo WW, Deng XX. Universal chloroplast primer pairs for simple sequence repeat analysis in diverse genera of fruit crops. J Hortic Sci Biotechnol. 2006;81:132–138. [Google Scholar]
- Provan J, Murphy S, Maggs C. Universal plastid primers for Chlorophyta and Rhodophyta. Eur J Phycol. 2004;39:43–50. doi: 10.1080/09670260310001636668. [DOI] [Google Scholar]
- Rozen S, Skaletsky HJ. Bioinformatics methods and protocols: methods in molecular biology: 2000. Totowa, NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Hepperle D. Fingerprinter©. a software for the analysis, bin-assignment and 01-matrix construction of molecular fingerprints (AFLPs) 2002. Win32-Version Distributed by the author via: http://science.do-mix.de/
- Hepperle D. TreeMe©. a software for visualization, manipulation, layouting and labelling of phylogenetic trees. 2004. Win32-Version Distributed by the author via: http://www.sequentix.de/
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible stratrgies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Peakall ROD. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol Ecol Resour. 2009;9:673–690. doi: 10.1111/j.1755-0998.2008.02319.x. [DOI] [PubMed] [Google Scholar]
- Powell W, Morgante M, Andre C, McNicol J, Machray G, Doyle J, Tingey S, Rafalski J. Hypervariable microsatellites provide a general source of polymorphic DNA markers for the Chloroplast genome. Curr Biol. 1995;5:1023–1029. doi: 10.1016/S0960-9822(95)00206-5. [DOI] [PubMed] [Google Scholar]
- Powell W, Morgante M, McDevitt R, Vendramin G, Rafalski J. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci USA. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Powell W, Hollingsworth P. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol. 2001;16:142–147. doi: 10.1016/S0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity. 2007;99:125–132. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
- Fao. The state of world fisheries and aquaculture 2000. 2001. Electronic edition, http://fao.org.
- Chan CX, Ho CL, Phang SM. Trends in seaweed research. Trends Plant Sci. 2006;11:165–166. doi: 10.1016/j.tplants.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:713–722. doi: 10.1007/s001220051343. [DOI] [Google Scholar]
- Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.) Theor Appl Genet. 1999;100:697–712. [Google Scholar]
- Eujayl I, Sorrells M, Baum M, Wolters P, Powell W. Assessment of genotypic variation among cultivated durum wheat based on EST-SSRS and genomic SSRS. Euphytica. 2001;119:39–43. doi: 10.1023/A:1017537720475. [DOI] [Google Scholar]
- Robinson AJ, Love CG, Batley J, Barker G, Edwards D. Simple sequence repeat marker loci discovery using SSR primer. Bioinformatics. 2004;20:1475–1476. doi: 10.1093/bioinformatics/bth104. [DOI] [PubMed] [Google Scholar]
- Morgante M, Olivieri AM. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993;3:175–182. doi: 10.1111/j.1365-313X.1993.tb00020.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Weber JL, Zhong G, Tanksley SD. Survey of plant short tandem DNA repeats. Theor Appl Genet. 1994;88:1–6. doi: 10.1007/BF00222386. [DOI] [PubMed] [Google Scholar]
- Cardlea L, Ramsaya L, Milbournea D, Macaulaya M, Marshalla D, Waugha R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA. Phylogeographic studies in plants: problems and prospects. Mol Ecol. 1998;7:465–474. doi: 10.1046/j.1365-294x.1998.00318.x. [DOI] [Google Scholar]
- McCauley DE. The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol Evol. 1995;10:198–202. doi: 10.1016/S0169-5347(00)89052-7. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]