Abstract
The aim of this study was to analyse various gene expression profiles of muscle tissue during normoxia, ischaemia and after reperfusion in human muscle free flaps, to gain an understanding of the occurring regulatory, inflammatory and apoptotic processes on a cellular and molecular basis. Eleven Caucasian patients with soft tissue defects needing coverage with microsurgical free muscle flaps were included in this study. In all patients, the muscle samples were taken from free myocutaneous flaps. The first sample was taken before induction of ischaemia in normoxia (I), another one after ischaemia (II), and the last one was taken after reperfusion (III). The samples were analysed using DNA-microarray, real-time-quantitative-PCR and immunohistochemistry. DNA-microarray analysis detected multiple, differentially regulated genes when comparing the different groups (I–III) with statistical significance. Comparing ischaemia (II) versus normoxia (I) educed 13 genes and comparing reperfusion (III) versus ischaemia (II) educed 19 genes. The comparison of reperfusion (III) versus normoxia (I) yielded 100 differentially regulated genes. Real-time-quantitative-PCR confirmed the results of the DNA-microarrays for a subset of four genes (CASP8, IL8, PLAUR and S100A8). This study shows that ischaemia and reperfusion induces alterations on the gene expression level in human muscle free flaps. Data may suggest that the four genes CASP8, IL8, PLAUR and S100A8 are of great importance in this context. We could not confirm the DNA-microarry and real-time-quantitative-PCR results on the protein level. Finally, these findings correspond with the surgeon’s clinical experience that the accepted times of ischaemia, generally up to 90 min., are not sufficient to induce pathophysiological processes, which can ultimately lead to flap loss. When inflammatory and apoptotic proteins are expressed at high levels, flap damage might occur and flap loss is likely. The sole expression on mRNA level might explain why flap loss is unlikely.
Keywords: gene expression, gene arrays, muscle free flaps, ischaemia, human tissue, hypoxia, reperfusion
Introduction
Microsurgical transfer of free myocutaneous flap-tissue has become a standardized technique in plastic and reconstructive surgery because of improvements of surgical techniques. Tissue engineering made impressive advances in the recent years. However, reconstructive microsurgery so far represents the most efficient approach to close large or complex tissue defects of the human body [1–7]. Indications for reconstructive procedures are multiple and have to be weighed on an individual basis. In spite of enormous advances in this field, the rate of flap failure is still 2–10% of all operations. Up to 25% of the transferred flaps even have to be revised because of complications [8–10]. According to Karsenti et al. the main reasons which lead to thrombotic closure of arterial and venous vessels are technical mistakes, such as, kinking of blood vessels and pairing disproportionate vessel diameters of donor and recipient [11].
For further increasing the success rates of free tissue transfer, a variety of drugs are applied to optimize rheology, ischaemic tolerance and prevent thrombosis of microsurgical anastomoses. However, there is no evidence-based treatment algorithm available at the moment [9] as in other medical fields [12, 13].
Further advances in the field of free myocutaneous tissue transfer and higher success rates require a fundamental knowledge of the molecular processes taking place in the flap tissue during surgery. Currently, the knowledge of these processes is still very limited or findings are often based on animal models only [14, 15].
Skeletal muscle tissue, in comparison with other body tissues, rarely tolerates longer periods of ischaemia and it has been found that apoptosis and even necrosis occur following ischaemia and reperfusion [16–18] (Fig. 1).
Fig 1.
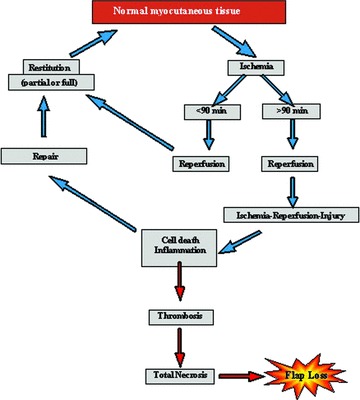
Potential pathways of tissue regeneration and necrosis in free muscle flap transfers.
The objective of this study is to examine the regulatory processes on a cellular and molecular basis that proceed in the free flap during the normoxic, ischaemic and reperfusion phases of surgery. Because of the fact that an essential part of ischaemia-reperfusion injury can be attributed to the processes at work during the reperfusion phase, this phase deserves particular interest [19]. Pro-inflammatory proteins such as Interleukin-1 or TNF-α, which are crucial for triggering systemic inflammatory reactions, are noticeably important. In terms of apoptosis, Caspase-3 is an important molecule as it connects the extrinsic and intrinsic signalling pathways (Fig. 2). Beyond that, the expression of proliferating cell nuclear antigen (PCNA) – which acts as processivity factor for DNA-polymerase – shall be analysed as an indicator of cell proliferation processes (Fig. 3). Moreover, it would be interesting and beneficial to further identify currently unconsidered genes and their protein products that are involved in regulating the adaptation of myocutaneous tissue towards ischaemia and reperfusion.
Fig 2.
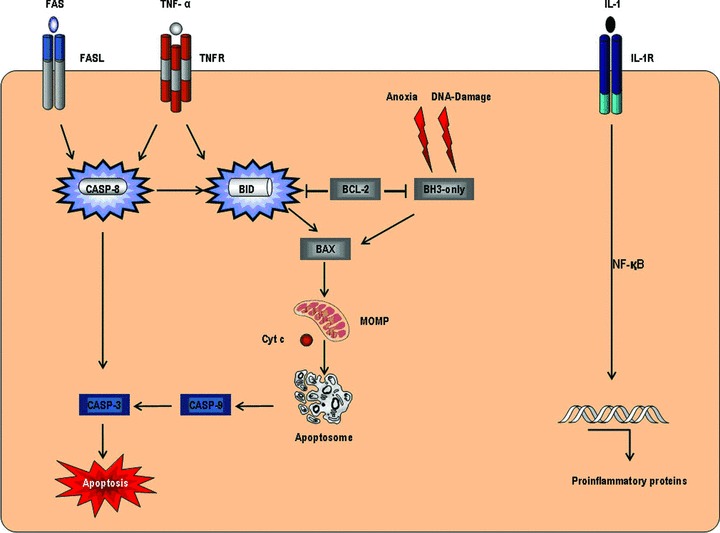
Extrinsic and intrinsic apoptotic signalling, inflammatory response via IL-1 and TNF-α.
Fig 3.
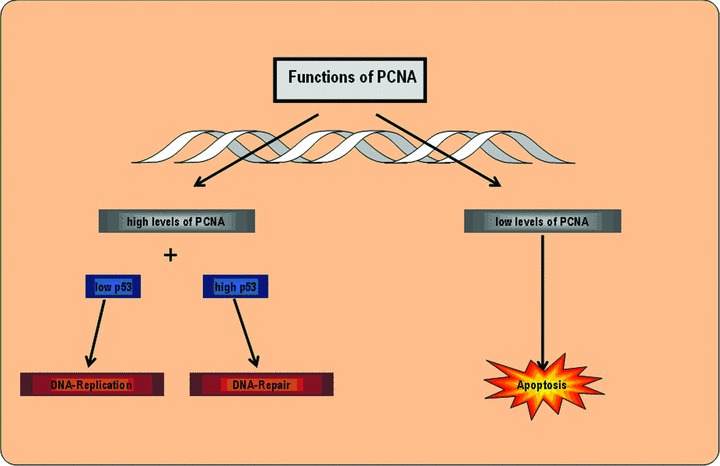
Functions of PCNA in DNA replication.
Materials and methods
Patients
Eleven Caucasian patients with a mean age of 56.6 years were included in this study (Table 1). Each patient had a considerable soft-tissue defect, which had to be covered by a free microsurgically anastomosed muscle flap. In all 11 patients, respectively, three muscle samples were retained intraoperatively in a standardized fashion from distal parts of the muscle flap after careful dissection of the dominant pedicle and surgical sectioning of all other minor pedicles. Special care was taken to ensure that the tissue samples were not taken from ischaemic- or venous-congested areas of the flap. All muscle samples had an average size of 1 cm3, which would have been otherwise discarded. This was approved by the scientific ELAN committee of the University Hospital Erlangen for 11 patients. In addition, each patient gave an informed consent. The recovering of tissue samples was uncomplicated, as tissue adaptation and shaping is performed routinely in all free tissue transplantations in order to perfectly adjust the transplanted tissue to the respective defect size. The first sample was taken in normoxia prior to the transection of the sustentative vessel pedicle (I), the second sample following a maximum ischaemia time of 72 ± 11 minutes (II), and the third sample was taken after successfully reanastomosing the vessels and reperfusion of 77 ± 22 min. (III). Each sample was then divided into halves. The first half of each sample was then put into Eppendorf-cups and shock frozen in liquid nitrogen at −196°C and in the following preserved in an −80°C freezer. The second half was conserved in 4% formaldehyde for histochemical analysis.
Table 1.
Patient data, Flap type, applied ischaemia and reperfusion times, applied methods
| Patient | Gender | Age | Flap type | Ischaemia (min.) | Reperfusion (min.) | Immunohistochemistry | DNA-microarray | RT-PCR | Flap survival |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 76 | M. latissimus | 76 | 60 | X | Yes | ||
| 2 | Female | 50 | M. gracilis | 79 | 56 | X | X | X | Yes |
| 3 | Female | 72 | M. latissimus | 91 | 96 | X | X | X | Yes |
| 4 | Male | 61 | M. latissimus | 62 | 71 | X | X | Yes | |
| 5 | Male | 61 | M. latissimus | 90 | 30 | X | X | Yes | |
| 6 | Female | 69 | M. gracilis | 81 | 53 | X | Yes | ||
| 7 | Male | 54 | M. gracilis | 62 | 69 | X | Yes | ||
| 8 | Male | 33 | M. gracilis | 69 | 119 | X | X | Yes | |
| 9 | Male | 33 | M. gracilis | 69 | 119 | X | Yes | ||
| 10 | Male | 50 | M. latissimus | 60 | 90 | X | Yes | ||
| 11 | Female | 64 | M. latissimus | 50 | 80 | X | Yes |
The samples were blinded and subsequently analysed corresponding to Table 1, either via DNA-microarray, real-time-quantitative-PCR or immunohistochemistry.
DNA-microarray
Isolation of RNA for microarray
The isolation of RNA from the muscle samples was performed by employing the TRIZOL-method corresponding to the instructions of Chomczynski and Sacchi 1987 [20].
RNA quality control
The isolated RNA was tested to ensure purity and subsequently underwent quality control measures. RNA-yield and concentration were assessed by photometry. Overall quality was adequate for further processing, which was ascertained by using Agilent 2100 Bioanalyzer® (Agilent, Santa Clara, CA, USA).
DNA-microarray
The sample-RNA was transcribed into complementary cDNA. For DNA-hybridization, the GeneChip® Human Genome U133A 2.0 Array (Affymetrix, Santa Clara, CA, USA) was used. This chip represents more than 18,500 transcriptional variants, and, among these 14,500 well-characterized genes, which were chosen from the GenBank®-Database.
Interpretation of microarray analysis
The interpretation was performed by the GeneChip® Scanner 3000 using the analysis software GeneChip® Operating Software 1.4 (GCOS 1.4). The analysis of the hybridization experiments was performed individually for each tissue sample. Each sample was then assigned to one group (n= 5) according to the point of time it was retained: normoxia (I), ischaemia (II) and reperfusion (III). The results of the analysis of DNA-microarray data were then compared with each other in groups to detect differences in gene expression patterns during the different stages of surgery. This resulted in three comparisons based on groups: ischaemia (II) versus normoxia (I), reperfusion (III) versus ischaemia (II) and reperfusion (III) versus normoxia (I).
Statistical analysis
To reliably compare the data of the microarray analysis and to minimize changes because of non-biological influences, the data were normalized by forming quantils and by applying a paired Student’s t-test. The statistical significance was defined at P < 0.05. To obtain valid data, genes were only considered as differentially expressed when having fold-changes of >2 or <−2. Fold-change was defined as the relative change of gene expression when comparing two conditions.
Subsequently, the statistically significant up- or down-regulated genes were determined for comparison of each group and then sorted based on their absolute fold-change.
Isolation of RNA
The lysis of the 100 μg tissue samples was performed by using Qiazol (Qiagen, Hilden, Germany). Afterwards, the samples were homogenized mechanically. The additional steps required for RNA-isolation were performed according to the handbook of the RNeasy Fibrous Tissue Mini Kit (Qiagen) and as published previously [21].
Quality control
The integrity of the isolated RNA was tested with gel electrophoresis of RNA and afterwards, RNA-purity and RNA-yield were determined using photometry.
Synthesis of cDNA
The sample-RNA was transcribed into cDNA by using the protocol of the Omniscript RT Kit (Qiagen).
Real-time-quantitative-PCR
Real-time-quantitative-PCR was performed after having retrieved the results of the DNA-microarray to confirm the data thus obtained. Therefore, we exemplarily performed real-time PCR on the samples of eight patients (n= 8) for four different genes (CASP8, IL8, PLAUR and S100A8), with potentially high clinical relevance. We used the primers according to Table 2 with a length of 20 bases and an average GC content of 55%. The applied house-keeping gene was GAPDH. The reagents of the PCR-Mix for each well – including Absolute QPCR SYBR Green Fluorescein Mix (ABgene, Epsom, UK) – were mixed as published previously [22]. The PCR was performed by the Biorad C1000 Thermal Cycler and analysed with CFX Manager Software Version 1.1.
Table 2.
Primer for real-time-quantitative-PCR
| Gene: CASP8 (Caspase 8) |
| Ace. No.: NM_001228 |
| Forward primer: 5′-catccagtcactttgccaga |
| Reverse primer: 3′ -gcatctgtttccccatgttt |
| Product size: 241 nt |
| Gene: GAPDH (gIyceraldehyde-3-phosphate dehydrogenase) |
| Ace. No.: NM_002046 |
| Forward primer: 5′-ccaggtggtctcctctgact |
| Reverse primer: 3′-ggtggtccaggggtcttact |
| Product size: 183 nt |
| Gene: IL8 (interleukin 8) |
| Ace. No.: NM_000584 |
| Forward primer: 5′-aaggaaaactgggtgcagag |
| Reverse primer: 3′-catctggcaaccctacaaca |
| Product size: 174 nt |
| Gene: PLAUR (plasminogen activator, urokinase receptor) |
| Ace. No.: NM_001005376 |
| Forward primer: 5′-agctatcggactggcttgaa |
| Reverse primer: 3′-catgtctgatgagccacagg |
| Product size: 117 nt |
| Gene S100A8 (S100 calcium binding protein A8) |
| Ace. No.: NM_002964 |
| Forward primer: 5′-atttccatgccgtctacagg |
| Reverse primer: 3′-acgcccatctttatcaccag |
| Product size: 167 nt |
Immunohistochemistry
The biopsies were fixed for 3 days in 4% formaldehyde. Afterwards, the tissue was embedded in paraffin and cut into sections of 2 μm.
To study apoptotic processes on the protein level, a primary antibody – diluted 1:100 in 3% bovine serum albumine (BSA) – against activated Caspase-3 (Abcam, Cambridge, UK) was employed. The secondary antibody was goat anti-rabbit antibody (Dianova, Hamburg, Germany) was diluted 1:500 in Tris HCL.
Primary antibodies against TNF-α and Interleukin-1 (both from Abcam) were used at a dilution of 1:100 in 3% BSA. For both the secondary goat anti-rabbit antibodies were diluted 1:200 (Dianova).
In addition, the biopsies were examined for activities of cell division by using primary antibodies against PCNA as published previously [23] (Proliferating Cell Nuclear Angtigen) (Dako, Glostrup, Denmark). The dilution was 1:500 in 3% BSA. The secondary goat anti-mouse antibody (Dianova) was diluted 1:250.
The avidin–biotin complex alkaline phosphatase detection system (Linaris, Wertheim-Bettingen, Germany) was used according to manufacturer’s instructions.
All sections were counterstained with haematoxylin and fixed under glass covers with Aquatex (Merck, Darmstadt, Germany).
The immunohistochemical staining was examined under the microscope and positive cells were counted by a blinded observer.
Results
DNA-microarray
As can be seen in Table 3, all group wise comparisons resulted in different numbers of statistically significant (paired Student’s t-test: P < 0.05) up- and down-regulated genes. Ischaemia (II) compared to normoxia (I) resulted in 13, and reperfusion (III) compared to ischaemia (II) resulted in 19 differentially expressed genes. The comparison of reperfusion (III) to normoxia (I) provided 100 differentially expressed genes.
Table 3.
Number of statistically significant genes derived from groupwise comparison
| Comparison | Detected genes | Induced genes | Repressed genes |
|---|---|---|---|
| Ischaemia versus normoxia | 13 | 4 | 9 |
| Reperfusion versus ischaemia | 19 | 5 | 14 |
| Reperfusion versus normoxia | 100 | 24 | 76 |
The Volcano-Plot in Figure 4 exemplarily illustrates the distribution of P-values against the logarithmical fold-change for the comparison of reperfusion (III) versus normoxia (I). The logarithmical fold-change is plotted on the x-axis and the P-values are plotted logarithmically on the y-axis. The horizontal line represents the P-value cut-off of 0.05. Every spot beneath this line and outside the white box represents one gene, which is significantly up- or down-regulated in comparison with reperfusion (III) versus normoxia (I).
Fig 4.
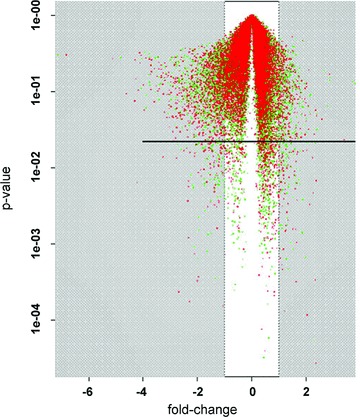
Volcan plot of the comparison between reperfusion and normoxia; x-axis: fold-change on logarithmical scale; y-axis: P-values on logarithmical scale; every spot represents one compared gene with corresponding fold-change and P-value; horizontal line: P-value cut-off of 0.05; all spots beneath this line and outside the white box represent statistically significant genes with at least a two-fold fold-change.
Tables 4–6 show – for each comparison in groups – 10 differentially expressed genes that were up- or down-regulated during various conditions. The genes were positioned based on their fold-change. Comparing ischaemia (II) versus normoxia (I) samples, a gene fold-change of +2 represents a double expression of the specific gene in the normoxic (I) sample. In contrast, a gene fold-change of −2 represents a double expression of the specific gene in the ischaemic (II) sample.
Table 4.
Comparison of ischaemia (II) to normoxia (I)
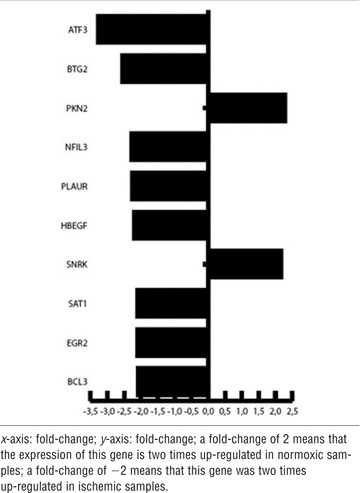 |
Table 5.
Comparison of reperfusion (III) to ischaemia (II)
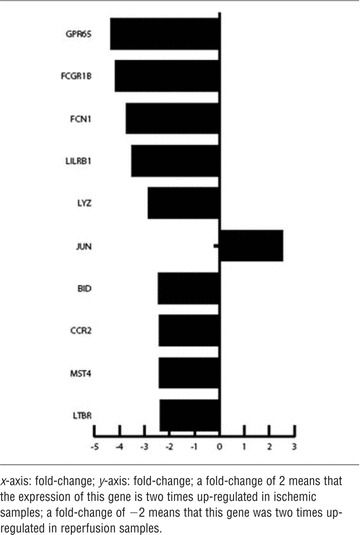 |
Table 6.
Comparison of reperfusion (III) to normoxia (I)
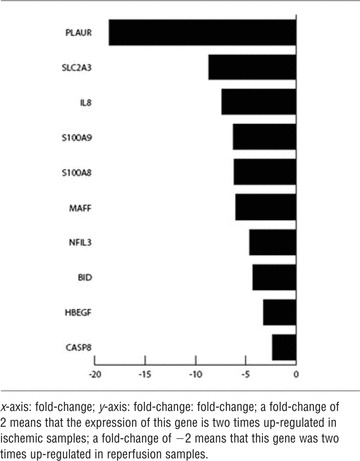 |
The comparison of ischaemia (II) versus normoxia (I) adduced an up-regulated expression of ATF 3, BTG 2, NFIL3, PLAUR, HBEGF, SAT1, EGR2, BCL3 in ischaemic (II) samples and of PKN2 and SNRK in the normoxic (I) samples.
Comparing reperfusion (III) versus ischaemia (II) GPR65, FCGR1B, FCN1, LILRB1, LYZ, BID, CCR2, MST4 and LTBR were up-regulated in reperfusion (III) samples, and JUN was up-regulated in ischemic (II) samples.
The highest number of differentially regulated genes could be seen in contrasting reperfusion (III) versus normoxia (I). The genes PLAUR, SLC2A3, IL-8, S100A9, S100A8, MAFF, NFIL3, BID, HBEGF and Caspase 8 were strongly up-regulated in the reperfusion (III) samples.
Table 7 gives an outline of those up-regulated genes that may be of great importance for adaptation of tissue towards ischaemia and reperfusion.
Table 7.
Summary of discussed genes, whose up-regulation had been detected via DNA-microarray and confirmed by real-time-quantitative-PCR (bold)
| Gene | Name | Function |
|---|---|---|
| ATF3 | Activating transcription factor 3 | Transcription factor, adaptation of tissue to anoxia |
| BID | BH3 interacting domain death agonist | Death agonist mediating mitochondrial damage |
| CASP8 | Caspase-8 | Apoptosis via extrinsic pathway |
| HBEGF | Heparin-binding EGF-like growth factor | Growth factor |
| IL8 | Interleukin-8 | Mediator of inflammatory response |
| PLAUR | Plasminogen activator, urokinase receptor | Fibrinolysis, reorganization of tissue |
| SI00A8 | S100 calcium binding protein A8 | Cell cycle regulation, pro inflammatory |
| S100A9 | S100 calcium binding protein A9 | Cell cycle regulation, pro-inflammatory |
| SLC2A3 | Solute carrier family 2, member 3 | Glucose transportation |
Real-time-quantitative-PCR
The gel electrophoresis of DNA in (Fig. 5) exemplarily illustrates the results of real-time-quantitative-PCR for patient one.
Fig 5.
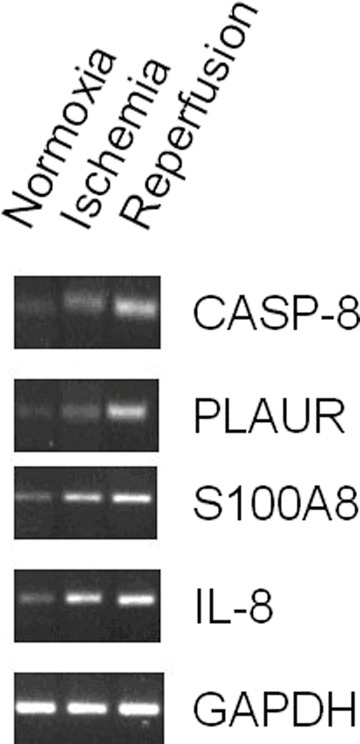
Exemplary DNA gel electrophoresis of real-time-quantitative-PCR product of patient 1.
In all tested genes, an up-regulation on RNA level from normoxia (I) over ischaemia (II) and following reperfusion (III) was detectable. Figure 6 shows the alterations of relative gene expression of Caspase-8 (A), Interleukin-8 (B), PLAUR (C) and S100A8 (D).
Fig 6.
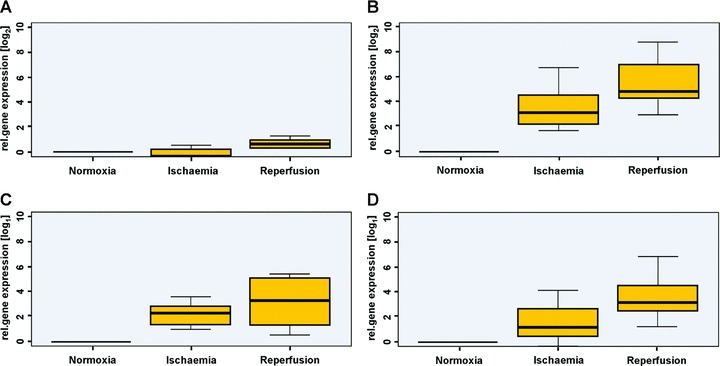
Real-time-quantitative-PCR reveals increased expression of Caspase-8. (A) Interleukin-8 (B), PLAUR (C) and S100A8 (D).
Immunohistochemistry
In normoxic (I), ischemic (II) and reperfusion (III) samples, Caspase-3, TNF-α, IL-1 and PCNA could not be detected. Figures 7 and 8 exemplarily demonstrate the results of immunohistochemic staining for Caspase-3 and PCNA.
Fig 7.
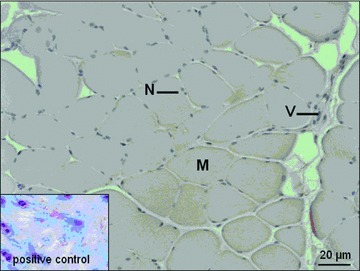
Immunohistochemical staining for Caspase-3. M: myofiber; N: nucleus; V: vessel.
Fig 8.
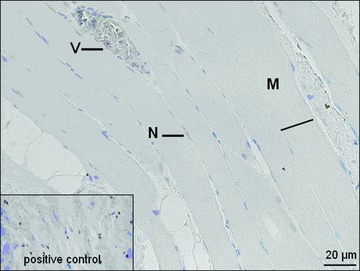
Immunohistochemical staining for PCNA. M: myofiber; N: nucleus; V: vessel.
Discussion
By performing quantitative real-time-quantitative-PCR, we could confirm the results of the DNA-microarray analysis in most instances. A noticeable up-regulation of plasmin-activator urokinase receptor (PLAUR) could be detected by DNA-microarray and confirmed by real-time-quantitative-PCR both for ischaemia (II) and for reperfusion (III) compared to normoxia (I). PLAUR is part of the plasmin-activating system, which is linked to the surface of cellular membranes (Fig. 9). It activates collagenases, beneath urokinase and plasminogen is converted into plasmin. Their proteolytic properties are necessary for fibrinolysis after thrombotic events and for the reorganization of tissues by degrading the extra cellular matrix. Because of the membrane attachment of PLAUR, the activation of these proteolytic systems happens in the vicinity of the cellular membrane [24–26]. The positive effects of the urokinase plasmin-activating systems on regeneration abilities of skeletal muscle tissue after injury have already been demonstrated in mice models. Thereby, the removal of excessively formed fibrin and the development of an adequate inflammatory reaction are elementary [27, 28]. The increased expression of PLAUR therefore appears to be one of the key processes that are necessary for the regeneration and reorganization of muscle tissue after longer periods of ischaemia.
Fig 9.
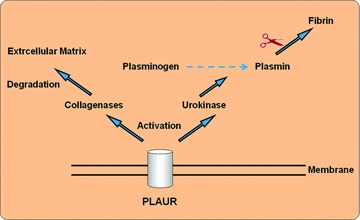
PLAUR – activation of urokinase, plasmin and collagenases.
The augmented expression of pro-apoptotic genes – such as Caspase-8, BID and Interleukin-8 – in reperfusion samples (III) is suggestive of apoptotic processes in muscle tissue after a period of ischaemia and reperfusion. Apoptosis can either be initiated by an extrinsic or an intrinsic signalling pathway [29, 30]. Among these pro-apoptotic genes – identified via DNA-microarray – was Caspase-8, which is essential for apoptosis via the extrinsic pathway (Fig. 2). This pathway is triggered by the binding of a ligand to a death receptor and is carried out by the subsequent activation of Caspase-8 or -9, which finally activate the effector caspases. The gene expression analysis of myocardial tissue following ischaemia reperfusion injury indicates that the up-regulation of Caspase-8 is specific for muscle cells, whereas Caspase-9 predominantly accumulates in vascular cells [31]. Further important physiologic functions of Caspase-8 in neo-vascularization procedures and terminal cellular differentiation are currently investigated [32–34].
In addition to increased expression of Caspase-8, the expression of BID (BH3 interacting death agonist) in reperfusion samples (III) was even more pronounced. The protein product of this gene – a member of the BCL-2 family – connects the extrinsic to the intrinsic apoptotic signalling pathway (Fig. 2) [35–37]. After cleavage by Caspase-8, it induces the mitochondrial liberation of pro-apoptotic factors like cytochrome c and thereby potentiates the apoptotic signal – initiated by Caspase-8, which leads to apoptotic cell death via controlled proteolysis [38]. In summary, the results of a modest increase of Caspase-8 expression and even greater increase of BID expression indicate that apoptosis in muscle cells after ischaemia and reperfusion is initiated by Caspase-8. However, the progression of apoptotic processes seems to be carried out via BID and the mitochondrial pathway, triggered by Caspase-8.
In addition, it could be demonstrated by this study that ischaemia and subsequent reperfusion trigger inflammatory processes in the line of free myocutaneous tissue transfer. This is indicated by the strong up-regulation of different markers of inflammation, as Interleukin-8, S100A8 and S100A9.
The pro-inflammatory cytokine IL-8 is produced by mesenchymal cells in the line of inflammatory processes. It serves as a chemotactic signal for neutrophile granulocytes [39]. Data provided by DNA-microarray and real-time-quantitative-PCR indicate that the production of pro-inflammatory cytokines – as IL-8 – is modestly increased during ischaemia (II) (Figs 5 and 6B), but strongly increased in reperfusion (III), as can be seen in Table 6[40]. Previously, it could be shown, that a significantly elevated concentration is measurable in blood plasma after 3–4 hrs of reperfusion [41]. Even in human myocardium elevated levels of IL-8 could be detected as markers of ischaemia causing inflammation [42].
The genes S100A8 and S100A9 are known to be indicators of inflammatory reactions [43]. The induction of these two genes by excessive physical exercise was described by Mortensen et al.[44]. This work is the first to demonstrate – using DNA-microarray (Table 6) and RT-PCR (Fig. 6D) – that these genes are induced in human muscle tissue after ischaemia and reperfusion in the line of free tissue transfer.
The increased expression during reperfusion (III) of the gene SLC2A3, which encodes a glucose transport protein (GLUT3), clearly displays the attempt of the tissue to compensate the lack of nutrition. This particular isoform of glucose transporters is – in comparison with the regularly, in muscle tissue expressed GLUT4 – independent of insulin levels. It has a high affinity for glucose molecules and thereby guarantees the supply of the tissue even in low glucose concentrations [45]. Recently the availability of GLUT-3 human skeletal muscle tissue was proven [46]. An increase of expression of GLUT-3 in hypoxic and ischaemic conditions has already been demonstrated in neuronal tissue [47].
The expression of activating transcription factor 3 (ATF3), a member of the family of ATF/CREB genes, was up-regulated in the ischemic samples (II) in comparison with normoxic samples (I). This pro-apoptotic gene is efficiently inducible under conditions of cellular stress, like hypoxia [48, 49]. Furthermore, it is being discussed whether it is involved in regulating the cell cycle arrest by binding to the promoter of Cyclin D1 [50].
The up-regulation of heparin-binding EGF-like growth factor (HBEGF) in ischaemia (II) and reperfusion (III) compared to normoxia corresponds to the results other surveys found in samples of rat brain [51, 52]. It is assumed that HBEGF has a protective effect on ischemic tissue [53]. Although HBEGF induces the expression of important angiogenic factors, no angiogenic effects on ischemic skeletal muscle could be detected [54, 55].
The immunohistochemical staining of normoxic-samples (I), ischemic-samples (II) and reperfusion-samples (III) could not detect Caspase-3, PCNA, TNF-α and Interleukin-10, and thus we could not confirm the DNA-microarry and real-time-quantitative-PCR results on the protein level. These results may implicate that the ischaemia (72 ± 11 min.) and reperfusion (77 ± 22 min.) times were too short to activate the generation and accumulation of these proteins in muscle tissue during free flap transfer. To understand the after transcriptional regulation in human muscle free flaps further studies have to be performed. These findings however correspond with the surgeon’s clinical experience that the accepted times of ischaemia, generally up to 90 min., are not sufficient to induce pathophysiological processes, which may lead to flap loss.
Although the relatively restricted sample size of 11 patients – because of the set-up and the high costs – needs further evaluation, the data on the behaviour of human tissue under realistic microsurgical conditions may valuably add to the pertinent literature and give rise for further investigations. This could include a larger number of patients. Based on the detected specific gene expression patterns, our results may be the beginning of further research. In summary, this study shows that ischaemia and reperfusion induces alterations on the gene expression level in human muscle free flaps. Data may suggest that the four genes CASP8, IL8, PLAUR and S100A8 are of great importance in this context. It may be hypothesized that by blocking the expression of these specific genes it may be possible to increase the ischaemic tolerance of muscle free flaps and prevent flap loss in reconstructive microsurgery.
Acknowledgments
This work was supported by ELAN grant of the Friedrich-Alexander-University Erlangen-Nürnberg (No. 08.01.07.1) and Xue Hong und Hans-Georg Geis Stiftung.
References
- 1.Arkudas A, Tjiawi J, Saumweber A, et al. Evaluation of blood vessel ingrowth in fibrin gel subject to type and concentration of growth factors. J Cell Mol Med. 2008;13:2864–74. doi: 10.1111/j.1582-4934.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleiziffer O, Horch RE, Hammon M, et al. T17b murine embryonal endothelial progenitor cells can be induced towards both proliferation and differentiation in a fibrin matrix. J Cell Mol Med. 2009;13:926–35. doi: 10.1111/j.1582-4934.2008.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutmacher DW, Horch RE, Loessner D, et al. Translating tissue engineering technology platforms into cancer research. J Cell Mol Med. 2009;13:1417–27. doi: 10.1111/j.1582-4934.2009.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polykandriotis E, Euler S, Arkudas A, et al. Regression and persistence: remodelling in a tissue engineered axial vascular assembly. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00828.x. 10.1111/j.1582-4934.2009.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallmichrath J, Stark GB, Kneser U, et al. Epidermal growth factor (EGF) transfection of human bone marrow stromal cells in bone tissue engineering. J Cell Mol Med. 2009;13:2593–601. doi: 10.1111/j.1582-4934.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beier JP, Klumpp D, Rudisile M, et al. Collagen matrices from sponge to nano: new perspectives for tissue engineering of skeletal muscle. BMC Biotechnol. 2009;9:34. doi: 10.1186/1472-6750-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkudas A, Tjiawi J, Saumweber A, et al. Evaluation of blood vessel ingrowth in fibrin gel subject to type and concentration of growth factors. J Cell Mol Med. 2009;13:2864–74. doi: 10.1111/j.1582-4934.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bizeau A, Guelfucci B, Giovanni A, et al. 15 years experience with microvascular free tissue transfer for repair of head and neck cancer defects. Ann Otolaryngol Chir Cervicofac. 2002;119:31–8. [PubMed] [Google Scholar]
- 9.Bui DT, Cordeiro PG, Hu QY, et al. Free flap reexploration: indications, treatment, and outcomes in 1193 free flaps. Plast Reconstr Surg. 2007;119:2092–100. doi: 10.1097/01.prs.0000260598.24376.e1. [DOI] [PubMed] [Google Scholar]
- 10.Vega S, Smartt JM, Jr, Jiang S, et al. 500 Consecutive patients with free TRAM flap breast reconstruction: a single surgeon’s experience. Plast Reconstr Surg. 2008;122:329–39. doi: 10.1097/PRS.0b013e31817f45cb. [DOI] [PubMed] [Google Scholar]
- 11.Karsenti G, Le Manach Y, Bouvier S, et al. Statins: a new pharmacological agent for free flap surgery. J Plast Reconstr Aesthet Surg. 2009 doi: 10.1016/j.bjps.2009.01.091. 10.1016/j.bjps.2009.01.091. [DOI] [PubMed] [Google Scholar]
- 12.Ashrafian H, Dwivedi G, Senior R. Assessing myocardial perfusion after myocardial infarction. PLoS Med. 2006;3:e131. doi: 10.1371/journal.pmed.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grenz A, Osswald H, Eckle T, et al. The reno-vascular A2B adenosine receptor protects the kidney from ischemia. PLoS Med. 2008;5:e137. doi: 10.1371/journal.pmed.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumeister S, Ofer N, Kleist C, et al. Reduction of skeletal muscle injury in composite tissue allotransplantation by heat stress preconditioning. Plast Reconstr Surg. 2004;114:1832–41. doi: 10.1097/01.prs.0000143577.36583.1b. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZG, Persons B, Lin L, et al. Vascular endothelial growth factor upregulates inducible nitric oxide synthase expression in the muscle flap ischemia model in the rat. J Reconstr Microsurg. 2009;25:219–25. doi: 10.1055/s-0028-1104550. [DOI] [PubMed] [Google Scholar]
- 16.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 17.Van Cruchten S, Van Den Broeck W. Morphological and biochemical aspects of apoptosis, oncosis and necrosis. Anat Histol Embryol. 2002;31:214–23. doi: 10.1046/j.1439-0264.2002.00398.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang WZ, Fang XH, Stephenson LL, et al. Ischemia/reperfusion-induced necrosis and apoptosis in the cells isolated from rat skeletal muscle. J Orthop Res. 2008;26:351–6. doi: 10.1002/jor.20493. [DOI] [PubMed] [Google Scholar]
- 19.Siemionow M, Arslan E. Ischemia/reperfusion injury: a review in relation to free tissue transfers. Microsurgery. 2004;24:468–75. doi: 10.1002/micr.20060. [DOI] [PubMed] [Google Scholar]
- 20.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 21.Surmann-Schmitt C, Widmann N, Mallein-Gerin F, et al. Stable subclones of the chondrogenic murine cell line MC615 mimic distinct stages of chondrocyte differentiation. J Cell Biochem. 2009;108:589–99. doi: 10.1002/jcb.22290. [DOI] [PubMed] [Google Scholar]
- 22.Surmann-Schmitt C, Widmann N, Dietz U, et al. Wif-1 is expressed at cartilage-mesenchyme interfaces and impedes Wnt3a-mediated inhibition of chondrogenesis. J Cell Sci. 2009;122:3627–37. doi: 10.1242/jcs.048926. [DOI] [PubMed] [Google Scholar]
- 23.Unglaub F, Thomas SB, Wolf MB, et al. Cartilage cell proliferation in degenerative TFCC wrist lesions. Arch Orthop Trauma Surg. 2009 doi: 10.1007/s00402-009-0883-z. 10.1007/s00402-009-0883-z. [DOI] [PubMed] [Google Scholar]
- 24.Hildenbrand R, Gandhari M, Stroebel P, et al. The urokinase-system—role of cell proliferation and apoptosis. Histol Histopathol. 2008;23:227–36. doi: 10.14670/HH-23.227. [DOI] [PubMed] [Google Scholar]
- 25.Hughes CC. Endothelial-stromal interactions in angiogenesis. Curr Opin Hematol. 2008;15:204–9. doi: 10.1097/MOH.0b013e3282f97dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rijken DC, Lijnen HR. New insights into the molecular mechanisms of the fibrinolytic system. J Thromb Haemost. 2009;7:4–13. doi: 10.1111/j.1538-7836.2008.03220.x. [DOI] [PubMed] [Google Scholar]
- 27.Bryer SC, Fantuzzi G, Van Rooijen N, et al. Urokinase-type plasminogen activator plays essential roles in macrophage chemotaxis and skeletal muscle regeneration. J Immunol. 2008;180:1179–88. doi: 10.4049/jimmunol.180.2.1179. [DOI] [PubMed] [Google Scholar]
- 28.Lluis F, Roma J, Suelves M, et al. Urokinase-dependent plasminogen activation is required for efficient skeletal muscle regeneration in vivo. Blood. 2001;97:1703–11. doi: 10.1182/blood.v97.6.1703. [DOI] [PubMed] [Google Scholar]
- 29.Kumar V, Abbas AK, Fausto N, Mitchell R. Robbins Basic Pathology. 8. New York: Saunders; 2007. [Google Scholar]
- 30.Unglaub F, Thomas SB, Kroeber MW, et al. Apoptotic pathways in degenerative disk lesions in the wrist. Arthroscopy. 2009;25:1380–6. doi: 10.1016/j.arthro.2009.04.071. [DOI] [PubMed] [Google Scholar]
- 31.Scarabelli TM, Stephanou A, Pasini E, et al. Different signaling pathways induce apoptosis in endothelial cells and cardiac myocytes during ischemia/reperfusion injury. Circ Res. 2002;90:745–8. doi: 10.1161/01.res.0000015224.07870.9a. [DOI] [PubMed] [Google Scholar]
- 32.Kang TB, Ben-Moshe T, Varfolomeev EE, et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J Immunol. 2004;173:2976–84. doi: 10.4049/jimmunol.173.5.2976. [DOI] [PubMed] [Google Scholar]
- 33.Scharner D, Rossig L, Carmona G, et al. Caspase-8 is involved in neovascularization-promoting progenitor cell functions. Arterioscler Thromb Vasc Biol. 2009;29:571–8. doi: 10.1161/ATVBAHA.108.182006. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Q, Xu Q. Caspase-8, a double-edged sword for EPC functioning. Arterioscler Thromb Vasc Biol. 2009;29:444–6. doi: 10.1161/ATVBAHA.108.183087. [DOI] [PubMed] [Google Scholar]
- 35.Fischer B, Coelho D, Dufour P, et al. Caspase 8-mediated cleavage of the pro-apoptotic BCL-2 family member BID in p53-dependent apoptosis. Biochem Biophys Res Commun. 2003;306:516–22. doi: 10.1016/s0006-291x(03)01004-0. [DOI] [PubMed] [Google Scholar]
- 36.Yin XM. Signal transduction mediated by Bid, a pro-death Bcl-2 family proteins, connects the death receptor and mitochondria apoptosis pathways. Cell Res. 2000;10:161–7. doi: 10.1038/sj.cr.7290045. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Ding WX, Qian T, et al. Bid activates multiple mitochondrial apoptotic mechanisms in primary hepatocytes after death receptor engagement. Gastroenterology. 2003;125:854–67. doi: 10.1016/s0016-5085(03)01066-7. [DOI] [PubMed] [Google Scholar]
- 38.Broughton BR, Reutens DC, Sobey CG. Apoptotic mechanisms after cerebral ischemia. Stroke. 2009;40:e331–9. doi: 10.1161/STROKEAHA.108.531632. [DOI] [PubMed] [Google Scholar]
- 39.Baggiolini M, Clark-Lewis I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 1992;307:97–101. doi: 10.1016/0014-5793(92)80909-z. [DOI] [PubMed] [Google Scholar]
- 40.Hua HT, Al-Badawi H, Entabi F, et al. CXC chemokine expression and synthesis in skeletal muscle during ischemia/reperfusion. J Vasc Surg. 2005;42:337–43. doi: 10.1016/j.jvs.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 41.Huda R, Solanki DR, Mathru M. Inflammatory and redox responses to ischaemia/reperfusion in human skeletal muscle. Clin Sci. 2004;107:497–503. doi: 10.1042/CS20040179. [DOI] [PubMed] [Google Scholar]
- 42.Gabrielsen A, Lawler PR, Yongzhong W, et al. Gene expression signals involved in ischemic injury, extracellular matrix composition and fibrosis defined by global mRNA profiling of the human left ventricular myocardium. J Mol Cell Cardiol. 2007;42:870–83. doi: 10.1016/j.yjmcc.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 43.Gebhardt C, Nemeth J, Angel P, et al. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72:1622–31. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Mortensen OH, Andersen K, Fischer C, et al. Calprotectin is released from human skeletal muscle tissue during exercise. J Physiol. 2008;586:3551–62. doi: 10.1113/jphysiol.2008.153551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dragu A. Kontraktilität, Kontraktur und BNP-Expression unter hypoxischen Bedingungen und Glukoseentzug an isolierten menschlichen myokardialen Trabekeln 95 Bl : Ill, graph Darst; (ger / dt) Freiburg im Breisgau, Univ, Diss. 2000.
- 46.Copland JA, Pardini AW, Wood TG, et al. IGF-1 controls GLUT3 expression in muscle via the transcriptional factor Sp1. Biochim Biophys Acta. 2007;1769:631–40. doi: 10.1016/j.bbaexp.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Vannucci SJ, Reinhart R, Maher F, et al. Alterations in GLUT1 and GLUT3 glucose transporter gene expression following unilateral hypoxia-ischemia in the immature rat brain. Brain Res Dev Brain Res. 1998;107:255–64. doi: 10.1016/s0165-3806(98)00021-2. [DOI] [PubMed] [Google Scholar]
- 48.Hartman MG, Lu D, Kim ML, et al. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol Cell Biol. 2004;24:5721–32. doi: 10.1128/MCB.24.13.5721-5732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X, Li X, Guo B. KLF6 induces apoptosis in prostate cancer cells through up-regulation of ATF3. J Biol Chem. 2008;283:29795–801. doi: 10.1074/jbc.M802515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J Biol Chem. 2006;281:10473–81. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 51.Jin K, Mao XO, Sun Y, et al. Heparin-binding epidermal growth factor-like growth factor: hypoxia-inducible expression in vitro and stimulation of neurogenesis in vitro and in vivo. J Neurosci. 2002;22:5365–73. doi: 10.1523/JNEUROSCI.22-13-05365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka N, Sasahara M, Ohno M, et al. Heparin-binding epidermal growth factor-like growth factor mRNA expression in neonatal rat brain with hypoxic/ischemic injury. Brain Res. 1999;827:130–8. doi: 10.1016/s0006-8993(99)01319-0. [DOI] [PubMed] [Google Scholar]
- 53.Weuste M, Wurm A, Iandiev I, et al. HB-EGF: increase in the ischemic rat retina and inhibition of osmotic glial cell swelling. Biochem Biophys Res Commun. 2006;347:310–8. doi: 10.1016/j.bbrc.2006.06.077. [DOI] [PubMed] [Google Scholar]
- 54.Chalothorn D, Moore SM, Zhang H, et al. Heparin-binding epidermal growth factor-like growth factor, collateral vessel development, and angiogenesis in skeletal muscle ischemia. Arterioscler Thromb Vasc Biol. 2005;25:1884–90. doi: 10.1161/01.ATV.0000175761.59602.16. [DOI] [PubMed] [Google Scholar]
- 55.Mehta VB, Zhou Y, Radulescu A, et al. HB-EGF stimulates eNOS expression and nitric oxide production and promotes eNOS dependent angiogenesis. Growth Factors. 2008;26:301–15. doi: 10.1080/08977190802393596. [DOI] [PubMed] [Google Scholar]


