Abstract
The transmembrane signaling mechanism of bacterial chemotaxis receptors is thought to involve changes in receptor conformation and dynamics. The receptors function in ternary complexes with two other proteins, CheA and CheW, that form extended membrane-bound arrays. Previous studies have shown that attractant binding induces a small (~2 Å) piston displacement of one helix of the periplasmic and transmembrane domains towards the cytoplasm, but it is not clear how this signal propagates through the cytoplasmic domain to control the kinase activity of the CheA bound at the membrane-distal tip, nearly 200 Å away. The cytoplasmic domain has been shown to be highly dynamic, which raises the question of how a small piston motion could propagate through a dynamic domain to control CheA kinase activity. To address this, we have developed a method for measuring dynamics of the receptor cytoplasmic fragment (CF) in functional complexes with CheA and CheW. Hydrogen exchange mass spectrometry (HDX-MS) measurements of global exchange of CF demonstrate that CF exhibits significantly slower exchange in functional complexes than in solution. Since the exchange rates in functional complexes are comparable to that of other proteins of similar structure, the CF appears to be a well-structured protein within these complexes, which is compatible with its role in propagating a signal that appears to be a tiny conformational change in the periplasmic and transmembrane domains of the receptor. We also demonstrate the feasibility of this protocol for local exchange measurements, by incorporating a pepsin digest step to produce peptides with 87% sequence coverage and only 20% back exchange. This method extends HDX-MS to membrane-bound functional complexes without detergents that may perturb the stability or structure of the system.
Introduction
Membrane proteins and their complexes are involved in many life processes including signal transduction, energy transduction, transmembrane transport, cell adhesion, and cell motility. Mechanistic understanding of these processes is hampered by the challenging nature of studies of membrane proteins. Not only is there very limited structural understanding of membrane proteins, which comprise less than 2% of known structures in the protein data bank, but methods for investigating mechanistic roles of dynamics have been historically difficult to extend to membrane proteins.
There has been recent success in measuring functionally important dynamics in membrane proteins. Solution NMR for example, which has provided key insights into the role of dynamics in catalysis by soluble proteins,1 has recently been used to measure ligand effects on conformational equilibria between inactive and activated states of detergent-solubilized beta-2 adrenergic receptor, providing insight into different types of activation.2–4 Unfortunately such approaches are difficult to apply to systems with a larger molecular weight, such as a membrane protein in a more native lipid bilayer environment or in a large complex with additional proteins, due to slow tumbling causing severe resonance broadening.
Although mass spectrometry does not have this limitation regarding molecular weight, membrane proteins and their complexes have long been assumed to be too challenging a target for MS, for example because of ionization suppression caused by some detergents.5–7 There has been remarkable recent progress in this area: multiple large membrane protein complexes have been analyzed by MS to reveal subunit stoichiometry and native lipid interactions.8,9 Hydrogen deuterium exchange mass spectrometry (HDX-MS) is a powerful approach for measuring protein dynamics, since amide proton exchange rates are sensitive to hydrogen bonding and solvent exposure.10–12 HDX-MS has been successfully applied to a wide range of soluble proteins to measure, for example, changes resulting from mutations, ligand binding, and protein-protein interactions13–16 as well as protein folding.17 Recently, HDX-MS has also been used to monitor structure and dynamics in a number of detergent-solubilized membrane proteins including G-protein coupled receptors and their complexes,18–20 an ADP/ATP carrier,21 and an ABC transporter,22 as well as in a nanodisc-inserted membrane protein.23 Thus HDX-MS is a promising tool for measuring functional dynamics of membrane proteins.
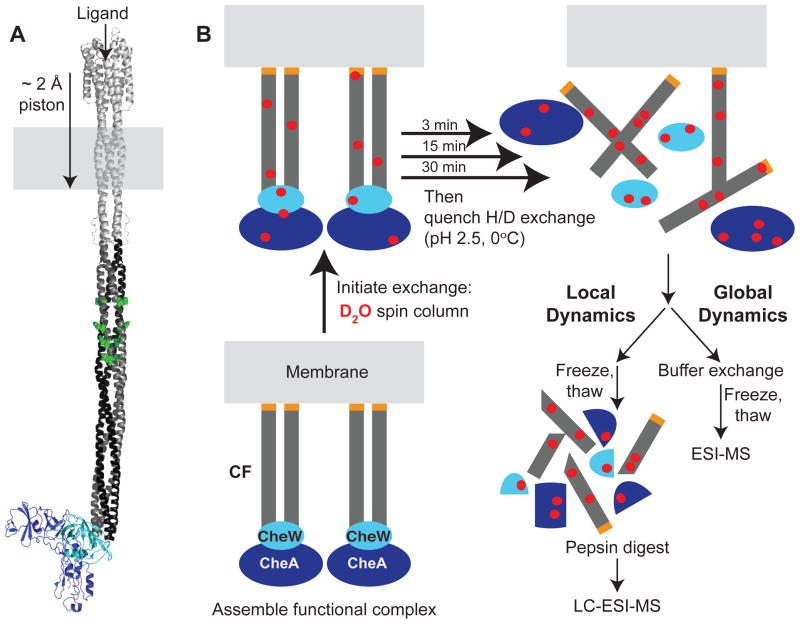
Bacterial chemotaxis receptors, which have been widely studied in an effort to understand the detailed mechanism of transmembrane signaling, display a paradoxically dynamic cytoplasmic domain.24,25 These receptors (Figure 1A) bind ligands in the periplasm and transmit a signal that controls activation of a kinase bound to the cytoplasmic tip of the receptor. Within the periplasmic and transmembrane regions, the ligand-induced signal is widely believed to consist of a tiny (~ 2 Å) piston motion of a transmembrane helix toward the cytoplasm.26 However, the cytoplasmic domain is known to be highly dynamic: a cytoplasmic domain fragment (CF) exchanges 90% of its amide protons to deuterium within 10 minutes,24 and the cytoplasmic domain of the intact receptor exhibits promiscuous cysteine crosslinking.25,27 How could a 2 Å piston motion be coupled through a dynamic cytoplasmic domain to control activation of the associated kinase? Chemotaxis receptors are known to operate in bacteria in large arrays (~ 200 nm dimensions) consisting of receptors and the associated CheW and CheA proteins,28–30 so perhaps the cytoplasmic domain dynamics are reduced in these complexes. Measurements of receptor dynamics in functional complexes are needed to resolve this paradox.
Figure 1.
Method for HDX-MS study of functional membrane-bound multi-protein complexes. (A) Model of an intact chemotaxis receptor with CheW (cyan) and CheA (blue) bound to the membrane-distal tip of the receptor cytoplasmic domain. The dimeric cytoplasmic fragment is represented in black/dark gray, with the methylation sites in green. (B) Such a membrane protein system can be simplified by assembling functional complexes on vesicles containing nickel-chelating lipids that bind the His-tag (orange) of the Asp receptor cytoplasmic fragment (CF), which in turn binds the other proteins of the complex (CheA and CheW).31 Deuterium exchange is initiated with a spin column to avoid dilution and dissociation of the complex. Exchange is quenched by lowering the temperature and pH. For global HDX measurements, the buffer is exchanged using spin columns and samples are flash-frozen and stored until ESI-MS analysis. For local HDX measurements, the quenched samples are flash-frozen. For LC-ESI-MS analysis, samples are thawed, digested with pepsin, injected onto a C18 column, and analyzed by mass spectrometry.
In order to study receptor dynamics in native-like arrays, we sought to avoid detergent solubilization which often perturbs the stability and function of membrane proteins. We developed an alternate approach that combines “template-directed assembly”31 with HDX-MS for measuring dynamics in functional protein complexes bound to membrane vesicles. Template directed assembly32 was developed by Weis and coworkers to assemble functional complexes of the cytoplasmic fragment (CF) of the aspartate receptor with CheA and CheW. The method employs vesicles containing a nickel-chelating lipid that binds to a histidine-tagged fragment of a membrane protein (CF), to which the other proteins bind to assemble a functional complex. This restores kinase activation that is lost in detergent-solubilized receptor preparations. Furthermore, both the kinase-activating (kinase-on) and methylation-activating (kinase-off) signaling states of the system can be produced by controlling the receptor density on the vesicles.33
The combined template-directed assembly HDX-MS method for measurement of dynamics in functional, membrane-bound multi-protein complexes in shown in Figure 1B. This approach is used for global exchange measurements on the Asp chemotaxis receptor CF, which demonstrate that formation of functional complexes significantly reduces its dynamics. Furthermore, we show that the method is compatible with local exchange measurements by identifying pepsin digest products covering 87% of the CF sequence, which will enable future measurements to test proposed models for how dynamics change during signaling. 34,35 Since other membrane protein systems are amenable to template-directed assembly,36–39 this provides a general HDX-MS approach for measuring functionally important dynamics in membrane protein complexes bound to vesicles, complementary to current applications of HDX-MS to membrane proteins in detergent micelles or nanodiscs.
Materials and Methods
Preparation and characterization of signaling complexes
CheA, CheW, CheY, and CF were prepared based on published protocols.40–43 Functional complexes of CF, CheA, and CheW bound to vesicles were prepared as previously described,33 using excess CheA and CheW to drive incorporation of all of the CF into complexes.42 These complexes were characterized using kinase and sedimentation assays.31 Details of these protocols are described in the Supplementary Information.
Global HDX-MS measurements
Spin columns were used both to initiate hydrogen exchange and to transfer samples into MS-compatible buffers. Unless otherwise specified, all centrifugation steps consisted of 2 min spins at 2000 rpm in a Beckman Coulter Allegra 6R Tabletop Centrifuge, at the specified temperatures. Hydrogen exchange was initiated using a 5 ml G10 Sephadex Zeba desalting column (Pierce Biotechnology), pre-equilibrated by four spins at 25°C, each with a 2 ml aliquot of D2O kinase buffer (75 mM Tris, 100 mM KCl, 5 mM MgCl2, 2 mM TCEP, 5% DMSO, pH electrode reading of 7.1, corresponding to a pDcorr=7.5). For the buffer exchange step, columns were prepared by hydrating 5 g of G25 Sephadex beads (Sigma Aldrich) overnight in 25 ml of water to obtain a suspension of swollen beads (sufficient for about thirty 1 ml columns). The next day, for each 1 ml syringe column, cotton wool was placed in the bottom of the syringe, the overnight hydrated G25 bead suspension was transferred onto the upright standing syringe to 1ml, and the water was allowed to drain completely. The column was equilibrated with 2 ml of desalting buffer (20 mM ammonium formate pH 2.5/10% acetonitrile) in sequential 200 μl aliquots and the solvent was allowed to drain through the column by gravity. To prevent the column from drying, 200μl of the desalting buffer was added to the closed column, which was then sealed with parafilm and kept on ice for use later in the day. Just before use, the excess solvent was removed from each desalting column by centrifugation at 4°C.
Preparation of the exchanged samples began with assembly of 2.5 ml of the relevant complex, which was checked for kinase activity, protein binding, and protein dissociation upon quenching, as described above. To initiate deuterium exchange, 2.0 ml of functional complexes were added to the 5 ml exchange column pre-equilibrated with room temperature D2O kinase buffer and the column was immediately centrifuged at 4°C. After the approximately 0.5 min deceleration of the centrifuge, the eluted exchanged complex was immediately placed in a 25°C water bath. For each time point, 100 μl of the exchanged complex was removed and exchange was quenched by addition of 10 μl of pre-chilled 50 mM potassium phosphate buffer (pH 1.1), bringing the final pH to 2.5. The quenched complex was immediately transferred to a pre-chilled desalting column and spun at 1000 rpm for 1 min at 4°C to exchange to a MS-compatible buffer (20 mM ammonium formate pH 2.5/10% acetonitrile). The flow through was transferred to a pre-chilled 1.5 ml eppendorf tube, flash-frozen in liquid nitrogen, and stored at −80°C until further analysis. Just before MS analysis, the sample was thawed in a 0°C ice water bath for 3 min.
Note that deuterium exchange occurred primarily at 25°C (except as noted below) and all steps post-quench were maintained at 0–4°C. The centrifugation of the exchange column was done in a 4°C centrifuge because of the need to immediately do the desalting of the first quenched sample at this temperature. Thus the first 2.5 min of exchange for all time points occurred between room temperature and 4°C, the temperatures of the exchange column and centrifuge, respectively.
Mass spectrometry was performed on an Esquire electrospray ion trap mass spectrometer (Bruker Daltonics) at the University of Massachusetts Mass Spectrometry Center. Before analysis, instrument performance was checked by injection of a 10 μM cytochrome c (Sigma Aldrich) control sample, then the line was cleaned thoroughly with three injections of 100 μL desalting buffer. Sample injections were done with a flow rate of 120 μL/hr. The source spray parameters were optimized for the lowest possible temperature to minimize back exchange, by increasing dry gas and nebulizer values to maintain good MS signals (dry temperature 150°C, dry gas 10 L/min, nebulizer 20.00 psi). MS experiments were conducted in positive ion mode, and spectra were obtained from m/z of 1100 to 1550 over a 1 minute acquisition time.
Bruker Daltonics Data Analysis software was used to convert the raw data to an average mass spectrum containing the series of m/z values for the protein. Data were processed with a smoothing value of 0.5 and a baseline subtraction of 0.8 to obtain symmetrical peaks with good signal to noise ratio, and deconvoluted to obtain the average molecular weight (MW) of the protein. The number of protons exchanged to deuterons at each time point was determined by subtracting the MW of un-exchanged protein from the MW of the exchanged protein. The number of exchanged protons was plotted vs. time, and fit to monoexponential {y = f1 − f1ek1t} and biexponential {y = f1 + f2 − [f1ek1t + f2ek2t]} curves using ProFit (Quantum Soft).
Two methods were used to determine the number of backbone amide protons that underwent back exchange to deuterium following the quench step into protonated buffers. In the first method, two 100 μl control samples of the non-exchanged soluble CF were each quenched with a 10 μL aliquot of 50 mM potassium phosphate buffer in D2O buffer (pD 1.4); one sample was loaded on a column previously equilibrated with 4°C H2O desalting buffer, and the other was loaded on a column previously equilibrated with 4°C D2O desalting buffer. The same control experiment was done on two samples of CF in functional complexes. Subsequent steps were identical to the handling of quenched experimental samples, so differences in deuterium incorporation in each sample pair should correspond to the extent of post-quench exchange that occurs under our experimental conditions. The second method involved preparation of a fully exchanged sample by denaturation of CF in D2O. Although CF has previously been shown to denature reversibly,44 protein aggregation was observed when CF4E was heated to 80°C in kinase buffer. Therefore 30 μM soluble CF4E was first exchanged into a buffer previously shown to support reversible thermal denaturation,45 using a 2 ml G10 Sephadex Zeba desalting column (Pierce Biotechnology) pre-equilibrated with a D2O buffer containing 20 mM potassium phosphate, pD 7.5, 50 mM NaCl, 5% DMSO. Following this spin column, the sample was heated at 80°C for 1 hour, cooled on ice for 30 min, quenched with 10 μl of potassium phosphate buffer in D2O (pD 1.4), and applied to a column previously equilibrated with 4 °C H2O desalting buffer. Subsequent steps were equivalent to the handling of quenched experimental samples, so the extent of proton incorporation in this fully deuterated sample should correspond to the extent of post-quench exchange that occurs under our experimental conditions.
Tandem MS identification of peptides
Since peptides produced for CF in solution and in functional complexes were the same, tandem MS was done only on CF in solution. A 30 μL volume of 30 μM CF was quenched with 15 μL of the quench buffer used for the local exchange experiments and digested by addition of 5 μL of 200 μM porcine pepsin (giving a 1:1 enzyme:protein ratio) for one minute in a 0°C ice water bath. The digest was injected at 200 μL/min into the C18 reversed phase column used for the local exchange experiments, which had been pre-equilibrated with Buffer A (0.1% formic acid in water) for 10 min. Peptides were eluted directly into the MS with a 30 min linear gradient from 0–95% Buffer B (0.1% formic acid in acetonitrile) at 100 μl/min. All of the peptides eluted by 60% Buffer B, but the standard 0–95% gradient used for most soluble proteins was used for this CF sample because no lipids were present. The strategy used for manual sequencing, aided by Analyst software, is described in the Supporting Information, and the identified peptides are listed in Table S1.
Local HDX-MS measurements
Preparation of the exchanged samples was similar to the global exchange experiments described above, with the following exceptions. A smaller amount (1 ml) of the relevant CF complexes were prepared and tested for activity and binding, and 700 μl was applied to a smaller (2 ml) exchange column that was pre-equilibrated and centrifuged at 25°C. For each time point, 30 μl of the exchanged complex was removed and added to 15 μl of the quench buffer (1% formic acid, 1M GuHCl, 20% glycerol, pH 1.6) in a 0°C ice water bath, yielding a final pH of 2.5. Each sample was immediately flash-frozen in liquid nitrogen and stored at −80°C until further analysis. Immediately prior to the MS experiment, the sample was thawed for 3 min in a 0°C ice water bath, porcine pepsin (Sigma Aldrich) was added in a 1:1 protein:enzyme molar ratio, and proteolysis proceeded for 1 min at 0°C. Subsequent injection into the HPLC column achieved both desalting of the sample and separation of the peptides.
To minimize back exchange, the injector, sample loops, HPLC columns, and lines connected both from the LC pump (Agilent 1100 G1312A) and the mass spectrometer were immersed in ice.46 The peptides were injected at 200μl/min into a 2.1 mm x 5 cm C18 reverse phase column (Higgins Analytical), pre-equilibrated in 95% Buffer A (0.1% formic acid in water). The sample was eluted directly into the mass spectrometer with a 5–40% gradient of Buffer B (0.1% formic acid in acetonitrile) over 7 min, followed by 40–60% Buffer B over 1 min. The 60% Buffer B maximum was chosen because this is sufficient to elute all the peptide, but avoids elution of the lipids (which elute at 80% Buffer B) into the MS instrument. The column was then re-equilibrated with 95% Buffer A for 8 min in preparation for the next sample.
After each sample injection, the syringe and injector valves were flushed with 500 μl kinase buffer, and a blank sample was injected to make sure that there were no carryover peptides from the previous run.47 After five sample injections, the column was separated from the ESI-MS and washed extensively for 30 min with 100% Buffer B to remove the lipids that accumulated in the column during previous sample injections.
Mass spectrometry was performed on a QSTAR-XL hybrid quadrupole/time-of-flight mass spectrometer (AB SCIEX) at the UMass Mass Spectrometry Center. Prior to MS analysis, the instrument was calibrated by direct injection of 30 μl of 10 μM renin substrate peptide (R-8129, Sigma-Aldrich), which should yield a spectrum consisting of two peptides with monoisotopic peak masses and charges of 586.9828, +2, and 879.9703, +3 respectively. In addition, prior to MS of exchanged CF samples, a CF4E pepsin digest was injected as a control to check performance of the column and MS. The MS instrument was set to positive ion mode, and the peptides were ionized with turbo ion spray ionization (ion source GS1=30 psi and GS2= 20 psi, curtain gas = 30 psi, ion spray voltage (IS) = 4500 V, heater temperature = 0°C). A 15 min gradient elution was used for peptide separation for the exchanged samples, to minimize back exchange of backbone amide protons, but the same peptides were identified based on the m/z and elution order.
Results
Template-directed assembly HDX-MS method for functional membrane-bound multi-protein complexes
With the goal of probing structure and dynamics in a functional complex of chemotaxis proteins, we combined the “template-directed assembly” method developed by Weis and coworkers31 with HDX-MS. As shown in Figure 1B, the method begins with the assembly of a membrane-bound complex of the Asp receptor cytoplasmic fragment (CF), CheA, and CheW. Assembly of the complex on vesicles involves using a cytoplasmic fragment of a membrane protein bearing a histidine tag, in this case the Asp receptor cytoplasmic fragment, that will bind to vesicles containing a nickel-chelating lipid, DGS-NTA-Ni2+. CheW and CheA bind to the cytoplasmic fragment assembled on vesicles to produce active complexes. This template-directed assembly approach can be used in general to create functional complexes of a membrane protein with its partners, as demonstrated by successful implementation for chemotaxis receptors,31,41 and for several eukaryotic receptor tyrosine kinases.36–39
Initiation of hydrogen exchange is performed using a spin column, to avoid dilution of the complex. Although this process is slower than the more typical 10-fold dilution into D2O buffer, it is an important choice for hydrogen exchange studies of complexes so that dilution does not lead to dissociation of the complex during the exchange time course. After each desired exchange time, an aliquot of the sample is removed and the exchange is quenched at pH 2.5 and 0°C. Finally, each exchanged sample is applied to another spin column to exchange from a buffer that supports assembly of a functional complex to a volatile buffer compatible with mass spectrometry. Each sample is immediately flash frozen in liquid nitrogen and stored at −80°C until mass spectrometry analysis.
Kinase assays were used to identify conditions that maximize the fraction of receptor in functional complexes. Since CheA in active complexes with chemoreceptors has 200-fold greater kinase activity than alone in solution,31 maximal kinase activity indicates maximum complex formation. We chose conditions under which all of the CF4E is bound to vesicles (30 μM CF4E and 580 μM lipid in extruded vesicles). In the presence of excess CheW (20 μM), kinase activity was measured as a function of CheA concentration and reached a maximum at 5 μM CheA (Figure S1a). Thus we chose 6 μM CheA, as it was sufficient to drive maximum formation of active complexes. With [CheA] fixed at 6 μM, kinase activity was measured as a function of CheW concentration and reached a maximum at 10 μM CheW (Figure S1b). Therefore, 12 μM CheW and 6 μM CheA were chosen as conditions favoring maximal incorporation of 30 μM CF into vesicle-bound, kinase-activating complexes. A sedimentation assay (Figure S2) confirms that all of the CF is bound to the vesicles. Comparison of the amounts of CheA and CheW present in the total and supernatant lanes indicates that some CheA and CheW are bound to the vesicle complexes (slightly greater amounts in the total lane), but a large excess remains in the supernatant. The use of excess CheA and CheW is consistent with the goal of having all of the CF4E in complexes with these proteins.
Quenching at acidic pH dissociates the majority of the receptor cytoplasmic fragments from the vesicles, which facilitates mass spectral analysis. We hypothesized that quenching to pH 2.5 (which is needed to minimize further exchange) would protonate the His tag on the CF4E, causing dissociation of the complex from the vesicles. The results of a sedimentation assay indicate that 75% of the CF4E does dissociate from the complex, based on the intensity of the CF4E band in the post-quench supernatant in Figure S2. Moreover, we observed that these acidic conditions also precipitate CheA and CheW, since their bands disappear from the post-quench supernatant. This may reduce the amount of CheA and CheW injected into the MS in subsequent steps (although the protocol does not involve a centrifugation step that would remove the precipitated proteins), which would be helpful for the current MS analysis of CF4E, but could hinder MS analysis of CheA and CheW in a future study.
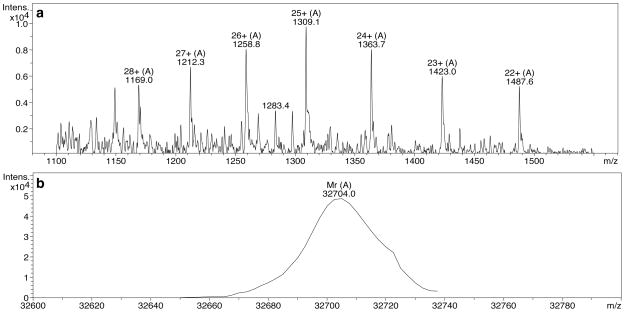
Despite the complexity of the sample, clear ESI-MS data are obtained for the CF4E from functional complexes on vesicles with CheA and CheW. Figure 2 shows representative MS results including (a) the charge state distribution, and (b) the results of the deconvolution of these charge states to obtain the average molecular mass of CF4E. The measured molecular weight of 32704±3 Da is in excellent agreement with the calculated 32707 Da molecular weight of the CF4E.
Figure 2. Mass spectrometry of CF from functional complexes.
(a) Mass spectrum showing charge state distribution of CF4E (+28 to +22 charge state peaks shown), (b) deconvolution of charge state distribution provides average molecular weight of CF4E (Note: although signal:noise is reduced in these samples that also contain vesicles and additional proteins, good mass accuracy and reproducibility is obtained. As reported in Table 2, the mass error estimated from independent measurements is ± 3 Da).
Control experiments were performed to measure the extent of amide backbone back exchange for this experimental protocol. After hydrogen to deuterium exchange is quenched, the buffer exchange (desalting spin column) introduces a protonated buffer, and then back exchange can occur during the freeze, thaw, and injection into the ESI-MS. Two independent methods were used to determine the extent of back exchange. One approach measured the mass difference between samples that were buffer-exchanged into a protonated vs. deuterated MS buffer. This difference, which was the same for CF4E alone and CF4E in vesicle-bound complexes, represents the total number of deuterons that back exchange due to the buffer exchange into protonated MS buffer. The other approach measured the mass of CF fully deuterated via thermal denaturation/renaturation in a deuterated buffer, followed by buffer exchange into the protonated MS buffer. The difference between this mass and the theoretical mass of deuterated CF4E again represents the total number of deuterons that back exchange due to the steps conducted in protonated MS buffer. As shown in Table 1, these methods gave consistent values for total back exchanged protons. Assuming the back exchange includes all sidechain sites, the calculated number of back exchanged backbone amide sites is 30–36. This relatively low extent of back exchange, 10–12% of the amide backbone sites, indicates that the protocol is well-optimized for rapid post-quench handling of the pH 2.5, chilled samples (each sample is unfrozen for approximately 3 min between quench and MS spectrum).
Table 1.
Back exchange for the experimental protocol.
| Buffer exchange H vs D buffer | Denatured & refolded in D2O | |
|---|---|---|
| Total sites back exchanged | 331±3a | 338±2b |
| Backbone amide sites back exchangedc (% of 300 total) | 331-302 = 29 (10%) | 338-302 = 36 (12%) |
Mass after D2O desalting column minus mass after H2O desalting column (mean ± standard deviation of two measurements).
Mass of fully deuterated protein (calculated) minus mass of D2O denatured protein after H2O desalting column (mean ± standard deviation of two measurements).
Assuming all 302 sidechain sites back exchange.
Global HDX-MS demonstrates a large-scale change in receptor dynamics upon formation of functional complexes
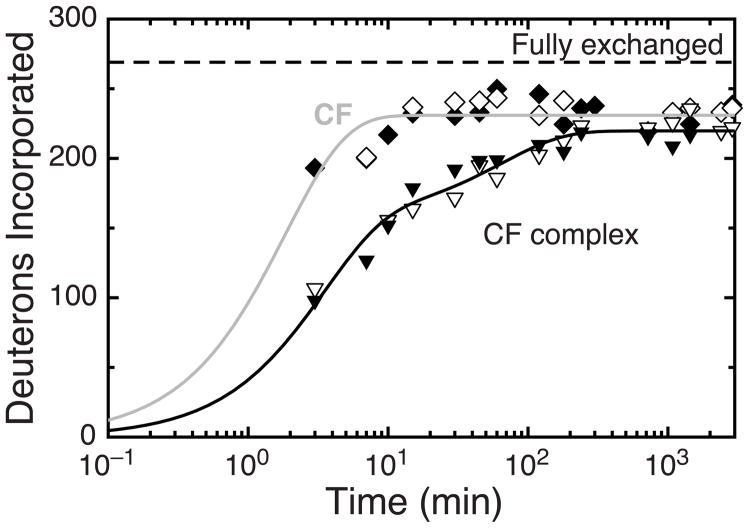
Using the protocol described above, global hydrogen exchange (exchange of amide protons of the entire protein) was measured for the Asp receptor CF4E in solution and for CF4E assembled in functional complexes with CheA and CheW on vesicles consisting of 50% DOPC/50% DGS-NTA-Ni2+. Figure 3 shows that hydrogen exchange of CF4E in functional complexes (triangles) is substantially slower than hydrogen exchange of CF4E alone in solution (diamonds), with exchange behavior consistent for independent experiments (white vs. black). The intermediate case, CF bound to vesicles in the absence of CheA and CheW, was not investigated because previous studies have shown that such samples are prone to CF-mediated vesicle aggregation that interferes with formation of functional complexes.41 The lines in Figure 3 represent monoexponential (gray) and biexponential (black) fits of the data for CF4E in solution and CF4E in functional complexes, respectively.
Figure 3.
Global hydrogen exchange of CF4E. Exchange occurs faster for CF in solution (diamonds) than in functional complexes assembled at high density on vesicles with CheA and CheW (triangles). Black and white symbols represent independent experiments. The estimated error of ± 3 Da is smaller than the data symbols. Dashed line shows maximum possible exchange of 269 protons obtained for CF4E denatured in D2O (corrected for back exchange). Lines are monoexponential (gray) and biexponential (black) fits to the data.
Table 2 compares the exchange rates and fractions for CF4E in solution and CF4E in kinase-on functional complexes. In both cases there is a very fast fraction f0 that back exchanges too quickly to be monitored by the experiment, estimated to be 32 amide protons (11%) based on controls described above. There is also a very slow fraction f3 of 37–42 protons (12–14%) that do not exchange in 16 hours. This was previously observed for CF in solution: in deuterium exchange NMR24 or tritium exchange45 experiments, approximately 10% of CF2Q2E does not exchange even after long times (3 days). The measured exchange time course for CF in solution shows a single fast exchange phase for all remaining 231 protons, with a rate constant of 0.54 min−1. In contrast, for CF in functional complexes, nearly a third of these protons (71 protons) exhibit a 50-fold slower exchange rate of 0.01 min−1, and a biexponential curve that includes “fast” and “slow” exchange rates is needed to achieve an adequate fit. A further slowing of hydrogen exchange for CF in complexes results in a fast exchange rate that is reduced from 0.54 to 0.37 min−1. A single isotopic cluster was observed in the mass spectra at each exchange time, consistent with a single population of CF rather than a fast exchanging (heavy) and slow exchanging (light) population. This indicates that the biphasic behavior is due to different exchange rates for protons within each CF rather than different conformations of CF. The overall change for CF upon formation of functional complexes is from a protein with nearly all (90%) fast-exchanging amide protons to one with only about 60% fast-exchanging protons.
Table 2.
Hydrogen exchange rates for CF4E in solution and in functional complexes.
| f0 very fasta | f1 fast | k1 (min−1) | f2 slow | k2 (min−1) | f3 very slowa | |
|---|---|---|---|---|---|---|
| Cytoplasmic fragmentb | 32 ± 3 11% |
231 ± 2 77% |
0.54 ± 0.06 | 0 | 0 | 37 12% |
| Kinase-active complexc | 32 ± 3 11% |
155 ± 6 51% |
0.37 ± 0.05 | 71± 6 24% |
0.01 ± 0.04 | 42 14% |
f0 based on back exchange controls (average of 3 measurements); errors in other parameters are errors of the fit. f3 is remainder to reach 300 amide protons.
Fit to y = f1 − f1ek1t
Fit to y = f1 + f2 − [f1ek1t + f2ek1t]
Receptor signaling states do not exhibit large-scale changes in dynamics
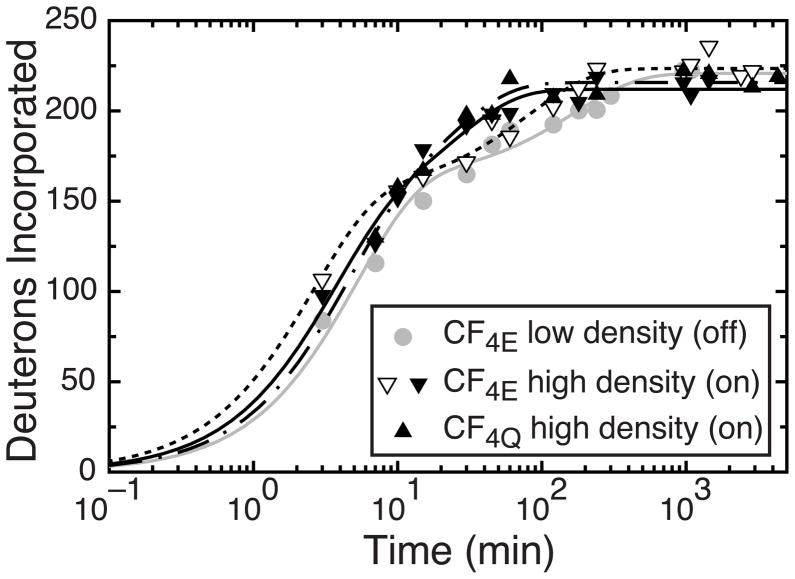
For intact chemotaxis receptors, ligand binding to the periplasmic domain and methylation of the cytoplasmic domain (at the green sites shown in Figure 1A) shift the signaling state in opposite directions between the kinase-on/methylation-off state (favored by methylation) and the kinase-off/methylation-on state (favored by attractant ligand binding). Although the CF lacks the ligand binding domain of the receptor, different methylation state constructs can be compared to investigate differences between signaling states. Furthermore, Weis and coworkers demonstrated that complexes of CF4E, CheA, and CheW assembled on vesicles at high and low density (50% or 10% nickel-chelating lipid, respectively) exhibit kinase-on/methylation-off and kinase-off/methylation-on activities, respectively.33 These activity measurements suggested that CF4E assembled in functional complexes on vesicles at low density (10% nickel-chelating lipid) mimics the ligand-bound signaling state of the intact receptor, and all other preparations (CF4E at high density and CF4Q at low or high density) mimic the ligand-free signaling state. Therefore comparisons of hydrogen exchange of high density complexes (50% nickel-chelating lipid) of CF4E and CF4Q investigate the effect of methylation and comparisons of high and low density complexes of CF4E investigate the effect of signaling state.
Close inspection of Figure 4 and Table 3 reveals no significant differences in measured global hydrogen exchange for high density complexes of CF4Q, high density complexes of CF4E, or low density complexes of CF4E. Any differences between exchange of different states are no greater than the difference between replicates of the high density CF4E state (white triangles/dash line and black triangles/black line in Figure 4). This is also clear in the fit parameters reported in Table 3, which are all largely within the range found for the replicates of the high density CF4E state. Therefore we conclude that there are no significant measurable differences in hydrogen exchange between these signaling states of functional CF complexes.
Figure 4.
Global hydrogen exchange of CF complexes in different signaling states. Kinase-off state (gray circles and line) is prepared by assembly of CF4E at low density (10% nickel-chelating lipid). Kinase-on state is prepared by either assembly of CF4E at high density (50% nickel-chelating lipid, black and white inverted triangles, which are the same data as shown in Figure 3, fit by black and dashed lines, respectively), or by assembly of CF4Q at high density (black upright triangles and dash-dot line). The estimated error of ± 3 Da is smaller than the data symbols.
Table 3.
Hydrogen exchange rates for CF complexes in different signaling states.a
| f1 | k1 (min−1) | f2 | k2 (min−1) | f3 (very slow)b | |
|---|---|---|---|---|---|
CF4E low density (kinase-off state)

|
160±7 | 0.20±0.02 | 60±8 | 0.006±0.002 | 48 |
| CF4E high density (kinase-on state) ■ | 154±6 | 0.37±0.05 | 71±6 | 0.012±0.003 | 43 |
| CF4E high density (kinase-on state) ▼ | 133±23 | 0.31±0.08 | 79±22 | 0.042±0.015 | 56 |
| CF4Q high density (kinase-on state) ▲ | 134±41 | 0.25±0.12 | 81±41 | 0.049±0.02 | 53 |
Reported errors are errors of the fitting. Errors are substantially greater, as estimated from averaging the two replicate samples of CF4E high density to yield k2 = 0.027 ± 0.021.
Assuming f0 = 32 back exchanged amide protons, f3 is the remainder to reach 300 amide protons.
Local exchange measurements are feasible in functional complexes
Local exchange measurements, which involve analysis of exchange properties of individual peptides derived from the complex, would make it possible to determine if there are small-scale changes in hydrogen exchange and to localize those changes within the protein. Furthermore, proposals for the role of dynamics in chemoreceptor signaling suggest that increased dynamics in one region may be counterbalanced by decreased dynamics of another region,34,35,48 which might not be detectable in a global exchange measurement unless the fast and slow exchange rates differed by at least 10-fold so they could be resolved by the global exchange experiment. Therefore we developed a local exchange protocol for template-assembled functional complexes (see Figure 1B), involving a pepsin digest to generate peptides that were identified by tandem MS (MS/MS). As for the global exchange experiment, a series of exchanged samples was prepared and frozen for later MS analysis. In this case, samples were flash-frozen immediately after quenching hydrogen exchange. However, sample thawing was very slow and resulted in protein aggregation, both of which were resolved by modifying the quench buffer to 1% formic acid, 1M GuHCl, 20% glycerol, pH 1.6. This prevented protein aggregation and decreased the thawing time to 3 min.
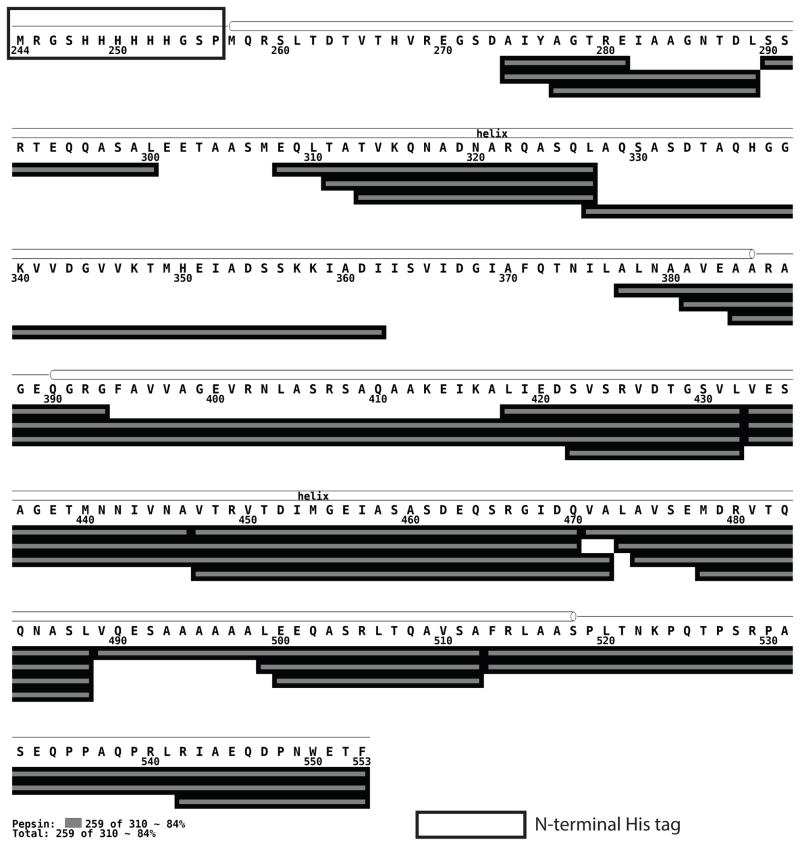
The peptide map shown in Figure 5 displays the peptic peptides identified by tandem MS. The same peptides, which cover 84% of the sequence of this construct, were observed following pepsin digestion of CF4E both in solution and in vesicle-bound complexes, in the original potassium phosphate quench buffer and in the modified GuHCl buffer. Tandem MS analysis to identify the peptides was performed in guanidine hydrochloride (GuHCl) buffer. The CF sequence begins at residue 257, after the N terminal His tag sequence, resulting in coverage of the actual CF sequence to 87%. A CF sample denatured in D2O was used as a control to determine the extent of back exchange for each peptide under the HDX protocol. As shown in Table S1, the CF peptides exhibited between 13 and 30% back exchange, with an average back exchange of 20%. The 87% peptide coverage and 20% average back exchange indicates that local exchange measurements should be possible for this and other template-assembled complexes. These measurements are underway and, while beyond the scope of this manuscript, will be the subject of a future publication.
Figure 5.
Pepsin digest peptides identified by tandem MS cover 84% of the sequence. Pepsin digest fragments are shown as gray bars. Based on crystal structures (eg 1qu7 corresponding to residues 292-518) and biochemical data, helices are represented as cylinders and nonhelical regions are shown as black lines. Residues enclosed in the rectangle are part of the N-terminal His tag appended to this CF construct. This representation was created using MSTools (http://ms.biomed.cas.cz/MSTools/DrawMap/DrawMap.php).
Discussion
By combining vesicle template assembly with HDX-MS measurements we have measured global exchange of chemoreceptor cytoplasmic fragments in functional signaling complexes to resolve a paradox regarding dynamics of this receptor domain. The HDX-MS results for exchange of CF alone in solution, 88% fast and 12% very slow, are consistent with previous experiments using NMR or tritium exchange, that observed 90% exchange in less than 15 min and a 10% core that does not exchange in 3 days.24,45 The dynamic nature of the cytoplasmic domain seemed incompatible with transmission of a subtle ~ 2 Å piston motion, so measuring the dynamics in a functional complex was critical to understanding the mechanistic details of the chemoreceptor signaling mechanism. Our results show that exchange rates of CF are significantly slowed in functional complexes, suggesting this domain is less dynamic, which is more compatible with signaling via tiny conformational changes. Furthermore, there are no large scale changes in exchange between signaling states, which is again consistent with there being only subtle conformational differences between these states.
CF in solution is alpha helical, elongated, typically monomeric, and highly dynamic:24 it most likely forms a fluctuating coiled-coil structure. The crystal structure of CF alone and in complexes shows that it dimerizes into a 4-helix bundle49,50, as independently predicted by extensive cysteine scanning studies of the cytoplasmic domain of the intact receptor.51 Table 5 compares the exchange properties of CF4E to two proteins of similar structure, a coiled-coil (GCN4)52 and a 4 helix bundle (acyl coenzyme binding protein, ACBP).53 CF4E in solution exhibits much faster exchange (~ 90% fast, 10% slow) than that of GCN4 and ACBP (~ 40% fast, 60% slow), consistent with our previous conclusion that CF has a fluctuating tertiary structure,24 and with the conclusion of disulfide scanning studies that this domain is a dynamic four-helix bundle.27 The hydrogen exchange rates of CF4E in functional complexes (~ 60% fast, 40% slow) are more consistent with those of GCN4 and ACBP. Thus CF in functional vesicle-bound complexes exhibits hydrogen exchange properties similar to stable proteins of similar structure. This behavior of CF is reminiscent of that of some intrinsically disordered proteins that only form stable structures upon binding to their protein partners.54
Table 5.
Hydrogen exchange rates for CF compared to similar protein structures.a
| Method | Fast: f0 + f1 | Slow: f2 + f3 | ||
|---|---|---|---|---|
| GCN4 (coiled coil)52 | NMR | ~ 43% k > 0.1 min−1 |
~ 57% k of 0.08 – 0.0005 min−1 |
|
| ACBP (four helix bundle)53 | NMR | ~ 36% k > 0.1 min−1 |
~ 64% k of 0.08 – 0.0002 min−1 |
|
| Asp receptor cytoplasmic fragment (CF) | MS | 88% k ≥ 0.5 min−1 |
12% k ≤ 0.0001 |
|
| CF in kinase-active complex | MS | 62% k ≥ 0.3 min−1 |
38% | |
| 24% k = 0.01 |
14% k ≤ 0.0001 |
|||
Published rates corrected for pH and temperature differences,55 to pH 7.1 and 25°C for comparison with CF data.
Large scale reductions in hydrogen exchange upon formation of protein-protein complexes have been reported in other systems. For example, binding of three different peptides to calmodulin reduces global hydrogen exchange of the protein, and this reduction is thought to be due to stabilization of calmodulin. The global exchange rates show a change similar to that observed in CF complexes: the number of protons which exchange at the slowest rates ~ 0.01 min−1 increases from zero in apo-calmodulin to ~ 20 (18%) in the peptide-bound complexes.56 Similarly, binding of the antitoxin IFS (immunity factor for SPN) to the bacterial toxin SPN (Streptococcus pyogenes NAD+ hydrolase), reduces exchange in both proteins. This reduced exchange is thought to be due to both decreased solvent accessibility at the SPN/IFS protein interface and to stabilization (increased hydrogen bonding) within each protein.57
Quantitative interpretation of the extent of protein stabilization corresponding to an observed change in hydrogen exchange rate depends on the type of exchange. Hydrogen exchange in stable proteins is typically in the EX2 limit, where the rate-determining step is the intrinsic exchange rate (kint).10 In this limit, the measured exchange rate (kHDX) depends on the fraction of time the amide site is in the open, exchange-competent conformation, given by the equilibrium constant for the opening event (Kopen): kHDX = kint Kopen. We can write the folding equilibrium constant as . CF complexes display EX2 exchange, but CF in solution exchanges too quickly to determine whether exchange is in the EX1 or EX2 limit. If we assume that both exhibit EX2 exchange, then the ratio of the exchange rates can be used to estimate the magnitude of the stabilization upon complex formation. For the 71 residues that show substantial slowing of exchange upon complex formation, , and a net estimated stabilization of the “closed” state by ΔΔG = 2.4 kcal/mol.
We have also shown that local hydrogen exchange measurements on template-assembled complexes are feasible, by incorporating a pepsin digest step, using tandem MS to identify peptides that cover 87% of the CF sequence, and demonstrating that only 20% amide backbone back exchange occurs for these peptides (on average) with our protocol. HDX-MS measurements of local exchange will be important in the chemoreceptor system to identify which CF regions are stabilized in functional complexes and to determine if there are differences in exchange between signaling states. Specific models have been proposed for changes in dynamics of the cytoplasmic domain between signaling states based on mutagenesis and cysteine crosslinking data.34,35 These models propose localized compensatory changes (increased dynamics in one region and decreased dynamics in another region) which would not have been detected by our global exchange measurements. Local hydrogen exchange measurements using the method we have demonstrated provide a promising approach for direct measurements of dynamics throughout the CF in different signaling states.
Here we have developed and demonstrated an HDX-MS method to monitor dynamics and conformational changes of proteins in functional membrane-bound complexes. Applications of HDX-MS to monitor dynamics have recently expanded to include membrane proteins in detergent micelles18–22 and in nanodiscs.58 Our method for HDX-MS of functional complexes assembled on vesicles provides a complementary approach that will be important for systems, such as chemoreceptor arrays, that are not stable in detergent and are too large for nanodiscs. Cytoplasmic domains of other membrane proteins such as receptor kinases can be assembled into functional complexes with vesicle template assembly36–39 for both global and local HDX-MS measurements. This tool is a promising approach for gaining insights into the structure and functional dynamics of membrane proteins.
Supplementary Material
Acknowledgments
The authors thank Dr. Daniel Fowler, Dr. Fe Sferdean, Dr. Michael Harris, and Dr. Sandy Parkinson for helpful discussions, and special thanks to Dr. Daniel Fowler and Yuzhou Tang for providing CheW and CheA proteins. This paper is dedicated to the memory of our colleague and friend, Bob Weis.
This research was supported by National Institutes of Health grant R01-GM085288. SSK was partially supported by a fellowship from the University of Massachusetts as part of the Chemistry-Biology Interface Training Program (National Research Service Award T32 GM08515).
ABBREVIATIONS
- CF
cytoplasmic fragment of E coli aspartate chemotaxis receptor
- DEAE
diethylaminoethanol
- DGS-NTA
1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid (Ni-chelating lipid)
- DMSO
dimethlysulfoxide
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- GuHCl
guanidine hydrochloride
- HDX-MS
hydrogen deuterium exchange mass spectrometry
- HPLC
high pressure liquid chromatography
- LC-ESI-MS
liquid chromatography electrospray ionization mass spectrometry
Footnotes
Activity measurements identifying conditions for maximal complex formation (Figure S1), gels demonstrating binding and release of components from vesicles (Figure S2), back exchange values for peptides identified by MS/MS (Table S1), and detailed methods for protein purification, assembly of functional complexes, and the strategy used for peptide identification. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Boehr DD, McElheny D, Dyson HJ, Wright PE. The Dynamic Energy Landscape of Dihydrofolate Reductase Catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 2.Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, Thian FS, Kobilka TS, Shaw DE, Mueller L, Prosser RS, Kobilka BK. The Dynamic Process of β(2)-Adrenergic Receptor Activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. Biased Signaling Pathways in β2-Adrenergic Receptor Characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, Maeda M, Tsujishita H, Shimada I. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat Commun. 2012;3:1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loo RR, Dales N, Andrews PC. The effect of detergents on proteins analyzed by electrospray ionization. Methods Mol Biol. 1996;61:141–160. doi: 10.1385/0-89603-345-7:141. [DOI] [PubMed] [Google Scholar]
- 6.Funk J, Li X, Franz T. Threshold values for detergents in protein and peptide samples for mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2986–2988. doi: 10.1002/rcm.2142. [DOI] [PubMed] [Google Scholar]
- 7.Rundlett KL, Armstrong DW. Mechanism of signal suppression by anionic surfactants in capillary electrophoresis-electrospray ionization mass spectrometry. Anal Chem. 1996;68:3493–3497. doi: 10.1021/ac960472p. [DOI] [PubMed] [Google Scholar]
- 8.Barrera NP, Robinson CV. Advances in the mass spectrometry of membrane proteins: from individual proteins to intact complexes. Annu Rev Biochem. 2011;80:247–271. doi: 10.1146/annurev-biochem-062309-093307. [DOI] [PubMed] [Google Scholar]
- 9.Morgner N, Montenegro F, Barrera NP, Robinson CV. Mass spectrometry--from peripheral proteins to membrane motors. Journal of Molecular Biology. 2012;423:1–13. doi: 10.1016/j.jmb.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyles SJ, Kaltashov IA. Methods to study protein dynamics and folding by mass spectrometry. Methods. 2004;34:88–99. doi: 10.1016/j.ymeth.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto T. Mass Spectrometry of Hydrogen/Deuterium Exchange of Escherichia coli Dihydrofolate Reductase: Effects of Loop Mutations. Journal of Biochemistry. 2004;135:487–494. doi: 10.1093/jb/mvh056. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Garcia-Ordonez RD, Pascal BD, Burris TP, Dodge JA, Griffin PR. Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer. Structure. 2010;18:1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HM, Yu X, Greig MJ, Gajiwala KS, Wu JC, Diehl W, Lunney EA, Emmett MR, Marshall AG. Drug binding and resistance mechanism of KIT tyrosine kinase revealed by hydrogen/deuterium exchange FTICR mass spectrometry. Protein Science. 2010;19:703–715. doi: 10.1002/pro.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B, Zhang ZY. Application of hydrogen/deuterium exchange mass spectrometry to study protein tyrosine phosphatase dynamics, ligand binding, and substrate specificity. Methods. 2007;42:227–233. doi: 10.1016/j.ymeth.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsutsui Y, Cruz, Dela R, Wintrode PL. Folding mechanism of the metastable serpin α1-antitrypsin. Proceedings of the National Academy of Sciences. 2012;109:4467–4472. doi: 10.1073/pnas.1109125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, Chance MR, Palczewski K. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20:826–840. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West GM, Chien EYT, Katritch V, Gatchalian J, Chalmers MJ, Stevens RC, Griffin PR. Ligand-dependent perturbation of the conformational ensemble for the GPCR β2 adrenergic receptor revealed by HDX. Structure. 2011;19:1424–1432. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung KY, Rasmussen SGF, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Sunahara RK. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rey M, Man P, Clémençon B, Trézéguet V, Brandolin G, Forest E, Pelosi L. Conformational dynamics of the bovine mitochondrial ADP/ATP carrier isoform 1 revealed by hydrogen/deuterium exchange coupled to mass spectrometry. Journal of Biological Chemistry. 2010;285:34981–34990. doi: 10.1074/jbc.M110.146209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehmood S, Domene C, Forest E, Jault JM. Dynamics of a bacterial multidrug ABC transporter in the inward- and outward-facing conformations. Proceedings of the National Academy of Sciences. 2012;109:10832–10836. doi: 10.1073/pnas.1204067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebling CM, Morgan CR, Stafford DW, Jorgenson JW, Rand KD, Engen JR. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82:5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeley SK, Weis RM, Thompson LK. The cytoplasmic fragment of the aspartate receptor displays globally dynamic behavior. Biochemistry. 1996;35:5199–5206. doi: 10.1021/bi9524979. [DOI] [PubMed] [Google Scholar]
- 25.Danielson MA, Bass RB, Falke JJ. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. Journal of Biological Chemistry. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winston SE, Mehan R, Falke JJ. Evidence that the Adaptation Region of the Aspartate Receptor Is a Dynamic Four-Helix Bundle: Cysteine and Disulfide Scanning Studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 29.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Müller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proceedings of the National Academy of Sciences. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrout AL, Montefusco DJ, Weis RM. Template-Directed Assembly of Receptor Signaling Complexes. Biochemistry. 2003;42:13379–13385. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
- 32.Montefusco DJ, Asinas AE, Weis RM. Liposome-mediated assembly of receptor signaling complexes. Meth Enzymol. 2007;423:267–298. doi: 10.1016/S0076-6879(07)23012-5. [DOI] [PubMed] [Google Scholar]
- 33.Besschetnova TY, Montefusco DJ, Asinas AE, Shrout AL, Antommattei FM, Weis RM. Receptor density balances signal stimulation and attenuation in membrane-assembled complexes of bacterial chemotaxis signaling proteins. Proc Natl Acad Sci USA. 2008;105:12289–12294. doi: 10.1073/pnas.0802868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swain KE, Gonzalez MA, Falke JJ. Engineered Socket Study of Signaling through a Four-Helix Bundle: Evidence for a Yin–Yang Mechanism in the Kinase Control Module of the Aspartate Receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Molecular Microbiology. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Shrout AL, Esposito EA, Weis RM. Template-directed Assembly of Signaling Proteins: A Novel Drug Screening and Research Tool. Chem Biol Drug Design. 2008;71:278–281. doi: 10.1111/j.1747-0285.2008.00627.x. [DOI] [PubMed] [Google Scholar]
- 38.Esposito EA, Shrout AL, Weis RM. Template-Directed Self-Assembly Enhances RTK Catalytic Domain Function. Journal of Biomolecular Screening. 2008;13:810–816. doi: 10.1177/1087057108322062. [DOI] [PubMed] [Google Scholar]
- 39.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proceedings of the National Academy of Sciences. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kott L, Braswell EH, Shrout AL, Weis RM. Distributed subunit interactions in CheA contribute to dimer stability: a sedimentation equilibrium study. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2004;1696:131–140. doi: 10.1016/j.bbapap.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Montefusco DJ, Shrout AL, Besschetnova TY, Weis RM. Formation and Activity of Template-Assembled Receptor Signaling Complexes. Langmuir. 2007;23:3280–3289. doi: 10.1021/la062717r. [DOI] [PubMed] [Google Scholar]
- 42.Fowler DJ, Weis RM, Thompson LK. Kinase-active Signaling Complexes of Bacterial Chemoreceptors Do Not Contain Proposed Receptor–Receptor Contacts Observed in Crystal Structures. Biochemistry. 2010;49:1425–1434. doi: 10.1021/bi901565k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asinas AE, Weis RM. Competitive and cooperative interactions in receptor signaling complexes. J Biol Chem. 2006;281:30512–30523. doi: 10.1074/jbc.M606267200. [DOI] [PubMed] [Google Scholar]
- 44.Wu J, Long DG, Weis RM. Reversible dissociation and unfolding of the Escherichia coli aspartate receptor cytoplasmic fragment. Biochemistry. 1995;34:3056–3065. doi: 10.1021/bi00009a037. [DOI] [PubMed] [Google Scholar]
- 45.Murphy OJ. Hydrogen Exchange Reveals a Stable and Expandable Core within the Aspartate Receptor Cytoplasmic Domain. Journal of Biological Chemistry. 2001;276:43262–43269. doi: 10.1074/jbc.M105585200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang J, Rand KD, Beuning PJ, Engen JR. False EX1 signatures caused by sample carryover during HX MS analyses. Int J Mass Spectrom. 2011;302:19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Molecular Microbiology. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 50.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proceedings of the National Academy of Sciences. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falke JJ, Kim SH. Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors. Curr Opin Struct Biol. 2000;10:462–469. doi: 10.1016/s0959-440x(00)00115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman EM, Kim PS. Periodicity of amide proton exchange rates in a coiled-coil leucine zipper peptide. Biochemistry. 1991;30:11615–11620. doi: 10.1021/bi00114a002. [DOI] [PubMed] [Google Scholar]
- 53.Kragelund BB, Knudsen J, Poulsen FM. Local perturbations by ligand binding of hydrogen deuterium exchange kinetics in a four-helix bundle protein, acyl coenzyme A binding protein (ACBP) Journal of Molecular Biology. 1995;250:695–706. doi: 10.1006/jmbi.1995.0409. [DOI] [PubMed] [Google Scholar]
- 54.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 55.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sperry JB, Huang RYC, Zhu MM, Rempel DL, Gross ML. Hydrophobic Peptides Affect Binding of Calmodulin and Ca as Explored by H/D Amide Exchange and Mass Spectrometry. Int J Mass Spectrom. 2011;302:85–92. doi: 10.1016/j.ijms.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sperry JB, Smith CL, Caparon MG, Ellenberger T, Gross ML. Mapping the protein-protein interface between a toxin and its cognate antitoxin from the bacterial pathogen Streptococcus pyogenes. Biochemistry. 2011;50:4038–4045. doi: 10.1021/bi200244k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morgan CR, Hebling CM, Rand KD, Stafford DW, Jorgenson JW, Engen JR. Conformational transitions in the membrane scaffold protein of phospholipid bilayer nanodiscs. Mol Cell Proteomics. 2011;10:M111 010876. doi: 10.1074/mcp.M111.010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.