Abstract
Human cytomegalovirus (HCMV) infection remains a significant problem in the setting of peripheral blood stem cell transplant (PBSCT), including primary infection resulting from transmission from a seropositive donor to a seronegative recipient (D+/R−). The lack of an animal model suitable for studying HCMV transmission after PBSCT is a major barrier in understanding this process and, consequently, the development of novel interventions to prevent HCMV infection. Our previous work demonstrated that human CD34+ progenitor cell engrafted NOD-scid IL2Rγcnull (NSG) mice support latent HCMV infection after direct inoculation, and reactivation after treatment with G-CSF. To more accurately recapitulate HCMV infection in the D+/R− PBSCT setting, granulocyte colony stimulating factor (G-CSF) mobilized peripheral blood stem cells (PBSCs) from seropositive donors were used to engraft NSG mice. All recipient mice demonstrated evidence of HCMV infection in liver, spleen, and bone marrow. These observations validate the NSG mouse model as a means to study HCMV transmission during PBSCT.
Introduction
Despite advances in diagnostics and therapeutics, human cytomegalovirus (HCMV) remains a significant cause of morbidity and mortality after peripheral blood stem cell transplant (PBSCT) and novel approaches to the prevention of HCMV infection are needed (1). HCMV seronegative patients who receive an allograft from an HCMV seropositive donor (D+/R−), while being at less risk for developing HCMV infection and disease than seropositive recipients (R+), will still develop post-transplant primary infection in up to 20% of cases (2–7). The donor graft is the most important source of virus early in the post-transplant period and retrospective analysis of D+/R− transplants described several factors associated with successful transmission of virus (5). However, the strict species specificity of cytomegaloviruses and the consequent lack of a suitable animal model system greatly hinder experimental validation of these findings as well as the development of preventative strategies in this population.
“Humanized” mice, transplanted with human cells and/or tissues, have recently been developed as tools to aid the in vivo study of pathogens with strict human tropism (8). We reported the first humanized mouse model of HCMV infection in which human CD34+ hematopoietic progenitor cell (HPC) -engrafted NOD-scid IL2Rγcnull (NSG) mice directly infected with HCMV supported latent viral infection, reactivation in human macrophages, and dissemination following granulocyte-colony stimulating factor (G-CSF)-induced mobilization of bone marrow hematopoietic cells (9). The experiments in this report were carried out to determine whether NSG mice would also demonstrate evidence of HCMV infection following transplantation of G-CSF mobilized PBSCs from HCMV seropositive donors, thereby recapitulating the D+/R− PBSCT and validating the NSG mouse model as a means of studying HCMV transmission and infection in this setting.
Methods
Mice
NOD-scid IL2Rγcnull (NSG) mice were maintained in an SPF facility according to procedures approved by the Institutional Animal Care and Use Committee at Oregon Health & Science University. Prior to transplant, mice were sublethally irradiated (100–250cGy) with a JL Shepherd Mark I 137Cs irradiator or a Rad Source RS 2000 X-ray irradiator. Following transplant, adult mice were provided with antibiotic water (1.1g/L neomycin sulfate and 167mg/ polymyxin B) until the time of tissue harvest.
Human donor cell transplantation
G-CSF mobilized peripheral blood stem cells (PBSC) were obtained from four donors after informed consent in accordance with Oregon Health & Science University Institutional Review Board policies. PBSC were transplanted intravenously into the retroorbital plexus of 8–12 week old NSG mice. Mice were sacrificed at 6–8 weeks following transplant or sooner if they appeared sick. Peripheral blood, bone marrow, liver, and spleen were collected at the time of euthanasia. Peripheral blood, bone marrow, and a portion of the spleen were processed immediately following euthanasia to obtain leukocytes for flow cytometry. Portions of each liver and spleen were placed in RNAlater (Invitrogen) for preservation until processed for DNA. Human cell engraftment was assessed by flow cytometry analysis of adult mouse peripheral blood as previously described (9).
Quantification of HCMV genomic DNA
Total DNA was extracted from tissue or isolated cells with DNAzol (Invitrogen) utilizing the manufacturer’s suggested protocol with the addition of a second DNA precipitation to remove any residual contamination. Briefly, 0.025g of tissue was placed in an eppendorf tube containing 1ml of DNAzol reagent with 1mm glass beads. The samples were homogenized for 3 minutes in a Beadbeater tissue homogenizer. Total DNA was precipitated by the addition of 100% ethanol (EtOH) followed by centrifugation (16,000xg for 10 minutes). The DNA pellet was washed twice with 75% EtOH, resuspended in dH2O, and precipitated a second time with the addition of sodium acetate and EtOH. DNA was analyzed by quantitative real time (RT)-PCR using a primer and probe set directed against the HCMV US28 gene sequence: probe=TGA TCC CGC TCA GTG T; forward primer=GAA CTC ATG CTC GGT GCT TTC; and reverse primer=CTT TGT GGC GCG ACT GAG A). This primer/probe set was identified using Primer Express software (Applied Biosystems). PCR reactions were performed using Fast Advanced Master Mix (Applied Biosystems) and the following reaction conditions: thermal activation of AmpliTaq Gold (20 sec. at 95°C) followed by a total of 40 cycles of PCR (1 sec. at 95°C and 20 sec. at 60°C) using the ABI Prism Step One Plus Sequence Detection System (Applied Biosystems). PCR results were analyzed using ABI Prism Step One Sequence Detection Software. HCMV bacterial artificial chromosomal DNA was used as quantification standards. The sensitivity of detection of this assay was ~50 HCMV genomic copies.
Results and Discussion
The objective of this study was to determine whether transplantation of G-CSF mobilized PBSCs from HCMV seropositive donors would result in successful infection of recipient NSG mice, similar to what occurs during transplantation of allografts from HCMV seropositive donors to seronegative recipients (5). Archived G-CSF-mobilized PBSCs from three different HCMV-seropositive donors (donors 1–3) were used to transplant a total of 12 sublethally-irradiated NSG mice (four mice per donor). As a control, three NSG mice were transplanted with G-CSF mobilized PBSCs from an HCMV seronegative donor (donor 4). All mice received approximately 2x106 unfractionated cells intravenously and were evaluated for overall health daily after transplant. The cohort of mice that received PBSCs from donor 1 were sacrificed one week after transplant as they began to appear sickly, with weight loss and loss of normal fur, consistent with xenogenic graft-versus host disease (GVHD). Mice that received PBSCs from donors 2, 3, and 4 appeared well until the planned time of harvest at 6, 8, and 6 weeks after transplant, respectively. Peripheral blood, spleen, and bone marrow from each mouse (except donor 1) were analyzed for engraftment by flow cytometry and for HCMV DNA by real time quantitative PCR.
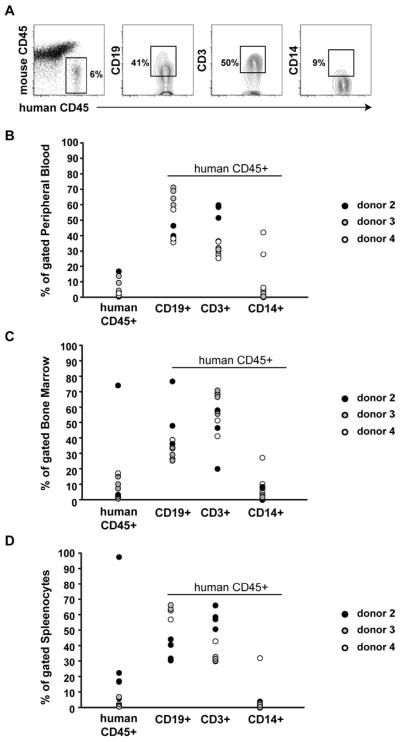
At the time of harvest 6–8 weeks after transplant, mice that received PBSCs from donors 2–4 displayed significant levels of human CD45+ cells in the peripheral blood (mean 5.5% +/− 5.5), bone marrow (mean 12.6% +/− 21.2), spleen (mean 17.3% +/− 29.3) (Figure 1). Although the majority of engrafted human CD45+ cells were either CD3+ T-cells or CD19 + B-cells, a significant number of CD14 expressing myelomonocytic cells were also detected (mean % +/− standard deviation for peripheral blood, bone marrow, and spleen = 8 +/− 14, 6.3 +/− 7.8, and 5.1 +/− 10.2, respectively).
Figure 1. Engraftment of NSG mice after transplantation of G-CSF mobilized PBSCs.
(A) Representative donor lineage analysis in xenografts. Flow cytometry of splenocytes from a mouse transplanted with PBSCs from donor 2 is shown. (B-D) Peripheral blood (B), bone marrow (C), and splenocytes (D) from mice transplanted with PBSCs from donors 2, 3, and 4 were analyzed at the time of tissue harvest for engraftment of human cell populations. Donor 1 human CD45+ engraftment was <0.5% in all mice.
Quantitative PCR for HCMV DNA performed on samples obtained from mice transplanted with PBSCs from HCMV seronegative donor 4 yielded low-level positive results, with a mean range of 33 – 127 copies per cell in the bone marrow, liver and the spleen (Table 1). These results likely reflect some degree of non-specific amplification of either human or, less likely, mouse DNA. Using the maximum quantity of HCMV detected in any sample in the cohort of mice transplanted from HCMV seronegative donor 4 (185 copies/γg DNA) as a baseline, 11 of 12 NSG mice that were transplanted with PBSCs from the three seropositive donors had virus detected at > 10-fold above baseline in bone marrow, liver, and spleen. Since HCMV will not replicate in these native mouse tissues, this finding most likely reflects detection of disseminated HCMV present in cells derived from transplanted human PBSC product. PCR assays without template were performed with each PCR run and were consistently negative (0 copies), indicating that the results obtained were not due to cross-contamination with HCMV DNA. Quantitative PCR analysis of HCMV DNA present in total stem cell product from each donor prior to transplant was unsuccessful, likely due to the extremely low amount of virus present (10).
Table 1.
NSG mice demonstrate evidence of HCMV infection after PBSCT from HCMV-seropositive donors
| viral load¶
|
|||
|---|---|---|---|
| bone marrow | liver | spleen | |
|
| |||
| Donor 1 | |||
| 1 | 13130 | 2060 | 1455 |
| 2 | 110991 | 9350 | 8322 |
| 3 | 24243 | 696 | 2355 |
| 4 | 11162 | 16809 | 612 |
|
| |||
| Average | 39881 | 7229 | 3186 |
|
| |||
| Donor 2 | |||
| 1 | 18443 | 419 | 214 |
| 2 | 951 | 1013 | 2574 |
| 3 | 25252 | 2100 | 9029 |
| 4 | 1170 | 2627 | 544 |
|
| |||
| Average | 11454 | 1540 | 3090 |
|
| |||
| Donor 3 | |||
| 1 | 513 | 1240 | 167 |
| 2 | 3039 | 352 | 92 |
| 3 | 2741 | 539 | 349 |
| 4 | 863 | 2351 | 305 |
|
| |||
| Average | 1789 | 1121 | 228 |
|
| |||
| Donor 4⌘ | |||
| 1 | 141 | 121 | 0 |
| 2 | 106 | 185 | 0 |
| 3 | 134 | 46 | 100 |
|
| |||
| Average | 127 | 118 | 33 |
HCMV genomic copies per μg DNA
HCMV seronegative donor
Based on these results, we conclude that G-CSF mobilized PBSCs obtained from HCMV seropositive donors represent a source of infectious virus that is capable of transmission and dissemination in the NSG mice model. This work complements and expands upon previous studies utilizing NSG mice as an in vivo model of HCMV latency and reactivation (9). This model can be used to determine the cellular reservoir(s) of latent virus in PBSCs that is necessary and sufficient for transmitting HCMV from donor to recipient, as standard cell separation techniques can be used to the enrich or deplete the stem cell product of specific cell types prior to transplant into NSG hosts. Of particular interest are myeloid lineage cells such as CD34+ progenitor cells and CD14+ monocytes, which have been previously described as harboring latent HCMV (11). Defining the cell populations that harbor latent HCMV may lead to novel strategies to prevent transmission during PBSCT.
Why only 15–20% of seropositive donors successfully transmit HCMV to seronegative recipients is not clear. The previous lack of an experimental model limited the analysis of transmission to retrospective clinical studies. The NSG mouse model now permits the experimental evaluation of factors inherent to the allograft that have been found to correlate with transmission in such studies (5). In addition, the NSG mouse provides a method for testing novel hypotheses, such as whether the risk of transmission is related to the quantitative viral load in the allograft. Thus, the NSG mouse model represents a unique opportunity to gain novel insight into the fundamental mechanisms of HCMV transmission and latency after PBSCT.
Acknowledgments
This work was supported by research grants from the National Institutes of Health to JAN (AI21640) and WHF (HL069133). MH was supported by a faculty development award from the Sunlin and Priscilla Chou Foundation.
Footnotes
Contribution: MH, DCG, DNS, KLH, and CNK performed experiments. MH, DCG, DNS, WHF, and JAN designed the research and analyzed results. MH and DCG made the tables and figures. MH wrote the paper.
Conflict-of-interest disclosure: the authors declare no competing financial interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boeckh M, Geballe AP. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest. 2011;121:1673–1680. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckh M, Nichols WG. The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood. 2004;103:2003–2008. doi: 10.1182/blood-2003-10-3616. [DOI] [PubMed] [Google Scholar]
- 3.George B, Pati N, Gilroy N, et al. Pre-transplant cytomegalovirus (CMV) serostatus remains the most important determinant of CMV reactivation after allogeneic hematopoietic stem cell transplantation in the era of surveillance and preemptive therapy. Transpl Infect Dis. 2010;12:322–329. doi: 10.1111/j.1399-3062.2010.00504.x. [DOI] [PubMed] [Google Scholar]
- 4.Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. High risk of death due to bacterial and fungal infection among cytomegalovirus (CMV)-seronegative recipients of stem cell transplants from seropositive donors: evidence for indirect effects of primary CMV infection. The Journal of infectious diseases. 2002;185:273–282. doi: 10.1086/338624. [DOI] [PubMed] [Google Scholar]
- 5.Pergam SA, Xie H, Sandhu R, et al. Efficiency and risk factors for CMV transmission in seronegative hematopoietic stem cell recipients. Biol Blood Marrow Transplant. 2012;18:1391–1400. doi: 10.1016/j.bbmt.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker CM, van Burik JA, De For TE, Weisdorf DJ. Cytomegalovirus infection after allogeneic transplantation: comparison of cord blood with peripheral blood and marrow graft sources. Biol Blood Marrow Transplant. 2007;13:1106–1115. doi: 10.1016/j.bbmt.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Matthes-Martin S, Lion T, Aberle SW, et al. Pre-emptive treatment of CMV DNAemia in paediatric stem cell transplantation: the impact of recipient and donor CMV serostatus on the incidence of CMV disease and CMV-related mortality. Bone marrow transplantation. 2003;31:803–808. doi: 10.1038/sj.bmt.1703927. [DOI] [PubMed] [Google Scholar]
- 8.Legrand N, Ploss A, Balling R, et al. Humanized mice for modeling human infectious disease: challenges, progress, and outlook. Cell Host Microbe. 2009;6:5–9. doi: 10.1016/j.chom.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith MS, Goldman DC, Bailey AS, et al. Granulocyte-colony stimulating factor reactivates human cytomegalovirus in a latently infected humanized mouse model. Cell Host Microbe. 2010;8:284–291. doi: 10.1016/j.chom.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slobedman B, Mocarski ES. Quantitative analysis of latent human cytomegalovirus. Journal of virology. 1999;73:4806–4812. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair J. Human cytomegalovirus: Latency and reactivation in the myeloid lineage. J Clin Virol. 2008;41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]