Abstract
Sequence-specific control of gene expression on a genome-wide scale is an important approach for understanding gene functions and for engineering genetic regulatory systems. We have recently described an RNA-based method, CRISPR interference (CRISPRi), for targeted silencing of transcription in bacteria and human cells. The CRISPRi system is derived from the Streptococcus pyogenes CR ISPR (clustered regularly interspaced palindromic repeats) pathway, requiring only the coexpression of a catalytically inactive Cas9 protein and a customizable single guide RNA (sgRNA ). The Cas9-sgRNA complex binds to DNA elements complementary to the sgRNA and causes a steric block that halts transcript elongation by RNA polymerase, resulting in the repression of the target gene. Here we provide a protocol for the design, construction and expression of customized sgRNA s for transcriptional repression of any gene of interest. We also provide details for testing the repression activity of CRISPRi using quantitative fluorescence assays and native elongating transcript sequencing. CRISPRi provides a simplified approach for rapid gene repression within 1–2 weeks. The method can also be adapted for high-throughput interrogation of genome-wide gene functions and genetic interactions, thus providing a complementary approach to RNA interference, which can be used in a wider variety of organisms.
INTRODUCTION
Much of the information encoding the function and behavior of an organism is dictated by its transcriptome. As the first step in gene expression, transcription serves as a nexus of regulatory information. Understanding this fundamental cellular process requires experimental tools capable of systematically interrogating transcriptional regulation on a genome-wide scale. These tools should enable highly specific control of gene expression with programmable efficiency, and they should be able to regulate multiple genes in diverse organisms. Recently, we reported that the bacterial immune system–derived CRISPR pathway can be repurposed as a new RNA-guided DNA-binding platform to repress the transcription of any gene1. This CRISPR interfering system, which we refer to as CRISPRi, works as an orthogonal system in diverse organisms, including in bacterial and human cells, and it requires only a single protein and a customized sgRNA designed with a complementary region to any gene of interest. Here we provide a protocol for the design, construction and utilization of sgRNAs for sequence-specific silencing of genes at the transcriptional level.
The CRISPR system
About 40% of bacteria and 90% of archaea possess the endogenous CRISPR machinery, which uses small RNAs to recognize by base pairing and cleaves foreign DNA elements to confer genetic resistance to such elements in a sequence-specific manner2–5. Different types of CRISPR systems exist6–10. In the type II CRISPR system from Streptococcus pyogenes, a CRISPR-associated protein 9 (Cas9) and an RNA complex containing a CRISPR RNA (crRNA) and a trans-acting RNA (tracrRNA) have been shown as the factors responsible for targeted silencing of foreign DNAs11–14. It was previously shown that an engineered RNA chimera containing a designed hairpin could be used in place of the RNA complex, further minimizing the components required for CRISPR targeting12. The targeting specificity is determined both by base pairing between the RNA chimera and the target DNA and by the binding between the Cas9 protein and a short DNA motif commonly found at the 3′ end of the target DNA, called the protospacer adjacent motif (PAM)12,15,16. The PAM sequence, consisting of a 2- to 5-bp recognition site that varies depending on the CRISPR system and the host organism, is a key motif that is recognized by the CRISPR machinery for spacer acquisition and subsequent target interference17,18. Binding between the Cas9-sgRNA complex and the target DNA causes double-strand breaks within the target region because of the endonuclease activity of Cas9. Therefore, the CRISPR system provides a host-independent platform for site-selective, RNA-guided genome editing. Indeed, recent work has shown that this system can be used for efficient and multiplexed genome editing in a broad range of organisms including bacteria19, yeast20, fish21, mice22 and human cells23–26.
CRISPR interference
To repurpose the CRISPR system for transcription regulation, we have used a catalytically inactive version of Cas9 (dCas9) that lacks endonucleolytic activity. The dCas9 contains two point mutations in both its RuvC-like (D10A) and HNH nuclease (H840A) domains, and previous work has shown this mutant Cas9 to be deficient in nucleolytic activity in vitro12. We have shown in Escherichia coli1 that dCas9, when coexpressed with an sgRNA designed with a 20-bp complementary region to any gene of interest, can efficiently silence a target gene with up to 99.9% repression. The sgRNA is a 102-nt-long chimeric noncoding RNA12, consisting of a 20-nt target-specific complementary region, a 42-nt Cas9-binding RNA structure and a 40-nt transcription terminator derived from S. pyogenes. We have demonstrated that binding of the dCas9-sgRNA complex to the nontemplate DNA strand of the protein-coding region blocks transcription elongation, as confirmed by native elongating transcript sequencing (NET-seq) experiments. When the sgRNA targets the promoter region, it can sterically prevent the association between key cis-acting DNA motifs and their cognate trans-acting transcription factors, leading to repression of transcription initiation (Fig. 1). The silencing is inducible and fully reversible and is highly specific in bacterial cells as measured by RNA-seq. Repression efficiency can be tuned by introducing single or multiple mismatches into the sgRNA base-pairing region or by targeting different loci along the target gene. Multiple sgRNAs can be used simultaneously to regulate multiple genes, to synergistically control a single gene for enhanced repression or for tuning silencing to achieve a moderate level of gene repression. Thus, CRISPRi presents an efficient and specific genome-targeting platform for transcription control without altering the target DNA sequence, and it can potentially be adapted as a versatile genome regulation method in diverse organisms.
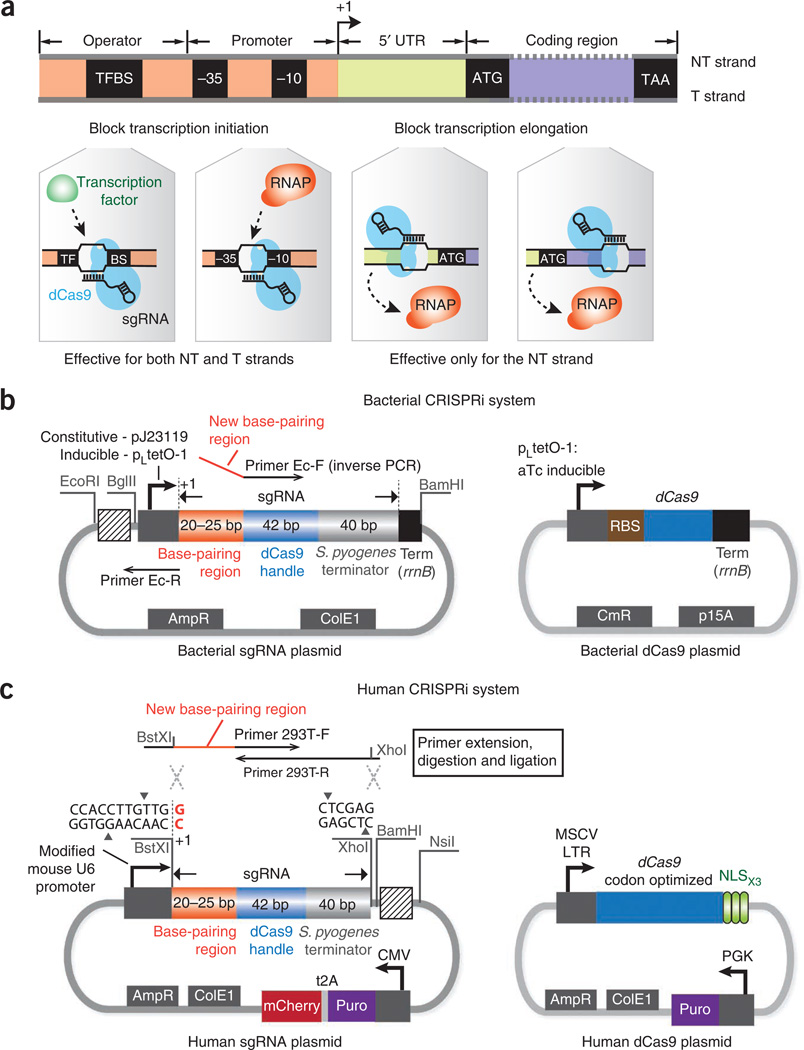
Figure 1.
The CRISPRi system for transcription repression in bacteria and human cells. (a) Depending on the target genomic locus, CRISPRi can block transcription elongation or initiation. When the dCas9-sgRNA complex binds to the nontemplate (NT) DNA strand of the UTR or the protein coding region, it can silence gene expression by blocking the elongating RNAPs. When the dCas9-sgRNA complex binds to the promoter sequence (e.g., the −35 or −10 boxes of the bacterial promoter) or the cis-acting transcription factor binding site (TFBS), it can block transcription initiation by sterically inhibiting the binding of RNAP or transcription factors to the same locus. Silencing of transcription initiation is independent of the targeted DNA strand. (b) The plasmid maps of the sgRNA and dCas9 expression vectors in E. coli. The sgRNA expression plasmid contains a promoter (constitutive—pJ23119 or inducible—pLtetO-1) with an annotated transcription start site (+1), an ampicillin-selectable marker (AmpR) and a ColE1 replication origin. The primer-binding sites for inverse PCR are highlighted. Three restriction sites EcoRI, BglII and BamHI are inserted to flank the sgRNA expression cassette to facilitate BioBrick cloning, so that new sgRNA cassettes can be repeatedly inserted into the striped box region. To ensure efficient transcription termination in E. coli, a strong terminator, rrnB, is added to the 3′ end of the sgRNA expression cassette. The dCas9 plasmid contains an aTc-inducible pLtetO-1 promoter, a strong ribosomal binding site (RBS), a chloramphenicol-resistance marker (CmR) and a p15A replication origin. (c) The plasmid maps used for sgRNA and dCas9 expression in human cells. The sgRNA expression plasmid is based on the pSico lentiviral vector that contains a mouse U6 promoter, an expression cassette consisting of a CMV promoter, a puromycin-resistance gene (Puro) and an mCherry gene for selection or screening of the plasmid, an ampicillin-selectable marker and a ColE1 replication origin for cloning in E. coli cells. Transcription of the U6 promoter starts at the last nucleotide G (red color) within the BstXI restriction site. New sgRNAs can be inserted between the BstXI and XhoI sites. The primer extension and insertion sites for sgRNA cloning are shown. The restriction sites BamHI and NsiI can be used to facilitate the BioBrick cloning: new sgRNA cassettes can be repeatedly inserted into the striped box region using BioBrick. The dCas9 plasmid contains a human codon-optimized dCas9 gene expressed from the murine stem cell virus (MSCV) long terminal repeat (LTR) promoter and is fused to three copies of the SV-40 NLS at the C terminus with a 3-aa linker. The plasmid also contains a puromycin-resistance gene controlled by the PGK promoter.
Comparison with other targeted genome regulation methods
Several targeted gene regulation techniques have been widely used in the past, such as RNA interference (RNAi)27,28 or engineered DNA-binding proteins, including zinc-finger29–31 or transcription activator–like effector (TALE)32–34 proteins. Although powerful, these methods have their own limitations. Although RNAi provides a convenient approach for perturbing genes on the mRNA level by using complementary RNAs, it is limited by off-target effects, low efficiency, toxicity and constrained use in particular organisms35. Compared with RNAi, it is likely that the performance of CRISPRi is more predictable and more specific owing to its simplicity and ease of design. Custom zinc-finger or TALE proteins, when coupled to effector domains, provide a versatile platform for achieving a wide variety of targeted regulatory functions. However, because of the repetitive nature of the TALE and zinc-finger proteins, construct development is time-consuming and expensive, making it difficult to build a comprehensive protein library large enough to perturb and interrogate genome-scale regulation or to simultaneously modulate multiple genes36. As the CRISPRi method is based on the use of sgRNAs with a gene-specific 20-nt-long complementary region, it presents a simple and inexpensive method for oligo-based gene regulation. As large-scale DNA oligonucleotide synthesis is becoming faster and cheaper, sgRNA libraries will allow targeting of large numbers of individual genes to infer gene function.
Limitations of the CRISPRi method
There are several potential limitations in using the CRISPRi method for targeted gene regulation. First, the requirement for an NGG PAM sequence for S. pyogenes Cas9 limits the availability of target sites in the genome. It has been shown that other Cas9 homologs use different PAM sequences14,24, and a given CRISPR system may tolerate various PAM sequences during target interference37,38. Indeed, recent studies have suggested that the S. pyogenes Cas9 protein could partially recognize an NAG PAM19, which might increase both the number of targetable genome sites and that of potential off-target sites. Therefore, exploiting different Cas9 homologs with different cognate PAMs, or exploring the PAM variability for a given CRISPR system, may either expand the targetable space if you are using more flexible PAMs or reduce potential off-target effects if you are using more stringent PAMs. Second, the targeting specificity is determined only by a 14-nt-long region (the 12 nt of the sgRNA and the 2 nt of the PAM), which might confer off-target effects in organisms with large genomes. The theoretical sequence length for unique targeting with a 14-nt recognition sequence is 268 Mb (414), which is only ~10% of the human genome. Genome-wide computational prediction of the redundant 14-nt recognition sites, with an eye toward the most up-to-date information regarding system-specific PAM variability, might be helpful to avoid off-target effects. Alternatively, choosing other Cas9 homologs with a longer PAM might reduce nonspecific targeting. Third, the level of transcriptional repression in mammalian cells varies between genes. Much work is needed to elucidate the rules for designing sgRNAs with higher efficiency, such as understanding the role of local DNA conformation and chromatin in binding and in regulatory efficiency39.
Experimental design
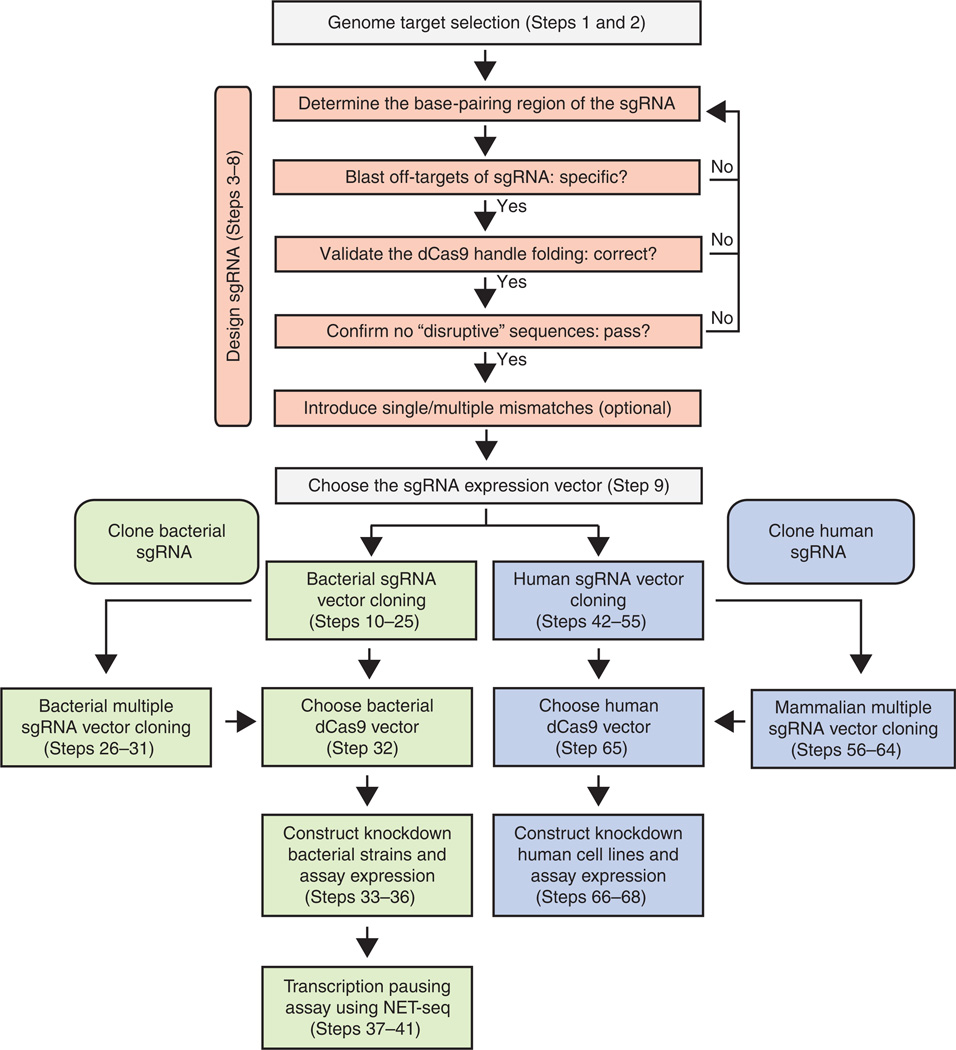
The general workflow for sgRNA design, cloning and expression for targeted gene regulation is summarized in Figure 2
Figure 2.
General workflow for the design, cloning and expression of sgRNAs. The orange boxes represent the sgRNA design steps. The green boxes show the cloning steps of sgRNAs for targeting genes in bacteria, and the blue boxes show the cloning of sgRNAs for human cells.
CRISPRi target site selection
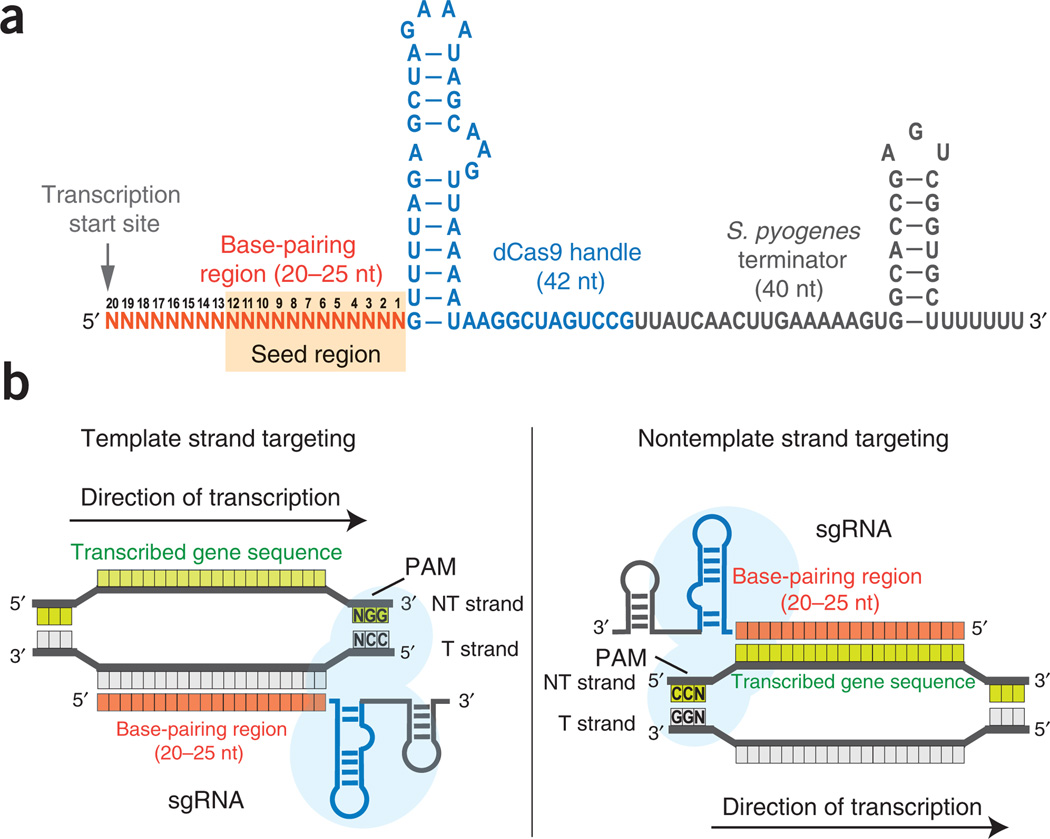
CRISPRi targeting is largely based on Watson-Crick base-pairing between the sgRNA and the target DNA sequence, enabling relative straightforward and flexible selection of targetable sites within a genome. As reported previously, the binding specificity of the dCas9-sgRNA complex to the target DNA is determined by sgRNA-DNA base pairing and an NGG PAM motif12. The PAM site is essential for dCas9 binding to the DNA, limiting the number of targetable sites within a genome. To avoid off-target effects, we recommend searching the genome for the 14-nt specificity region consisting of the 12-nt ‘seed’ region of the sgRNA and 2 of the 3-nt (NGG) PAM in the genome, in order to rule out additional potential binding sites (Fig. 3). Any sgRNA designed with more than one binding site should be discarded.
Figure 3.
Design of the sgRNAs. (a) The sgRNA is a chimera and consists of three regions: a 20–25-nt-long base-pairing region for specific DNA binding, a 42-nt-long dCas9 handle hairpin for Cas9 protein binding and a 40-nt-long transcription terminator hairpin derived from S. pyogenes. Transcription of sgRNAs should start precisely at its 5′ end. The 12-nt seed region is shaded in orange. (b) The schemes for designing sgRNAs to target the template (T) or nontemplate (NT) DNA strands. When targeting the template DNA strand, the base-pairing region of the sgRNA has the same sequence identity as the transcribed sequence. When targeting the nontemplate DNA strand, the base-pairing region of the sgRNA is the reverse-complement of the transcribed sequence.
Targeting different sites in a gene allows the dCas9-sgRNA complex to exhibit different regulatory functions. To block transcription elongation, target either the nontemplate DNA strand of the protein-coding region or the untranslated region (UTR). In this case, the bound dCas9-sgRNA complex will act as a roadblock to the elongating RNA polymerase (RNAP), leading to aborted transcription. To inhibit transcription initiation, the dCas9-sgRNA complex should target either the template or the nontemplate strand of RNAP-binding sites (e.g., the −35 or −10 boxes of the bacterial promoter) or the cis-acting motifs within the promoter (e.g., transcription factor binding sites, TFBS), thereby acting as a steric block to cognate protein factors (Fig. 1a).
The choice of the target sites also affects the level of transcription repression. By using fluorescent reporter genes in E. coli, we have observed that the repression is inversely correlated with the distance of the target site from the transcription start site1. Thus, to achieve better repression in bacteria, target sites within the 5′ end of the gene should be selected. In human cells, we recommend selecting multiple target sites within the promoter-proximal region (targeting either the template or nontemplate strand) or within the coding region (targeting the nontemplate strand), as epigenetic modifications and local chromatin structures might impede CRISPRi binding.
Chimeric sgRNA design
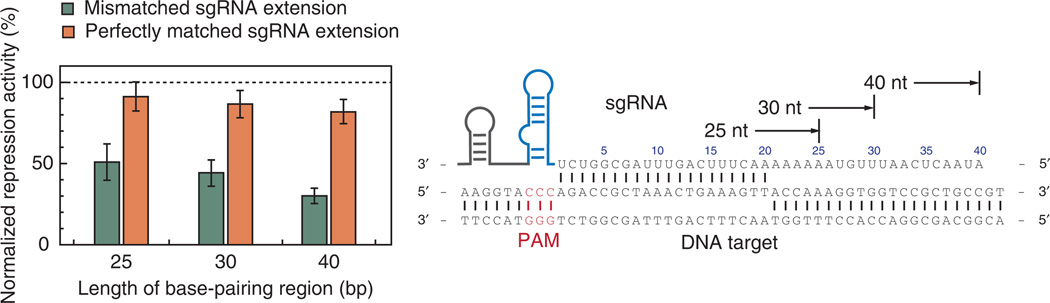
As shown in Figure 3, the chimeric sgRNA sequence is modularly composed of a base-pairing region (orange), a dCas9 handle hairpin (blue) and an S. pyogenes– derived terminator sequence (gray). We have shown that truncating the base-pairing region to <20 nt could substantially dampen the repression efficiency, whereas extending the 5′ sequence of the base-pairing region with perfectly matched nucleotides (up to 40 nt) did not markedly improve repression1. Therefore, we recommend using a length of 20–25 nt as the base-pairing region. We note that extending the 5′ sequence of the sgRNA with mismatched nucleotides also decreases the repression efficiency, implying that the 5′-nucleotide context is important for targeting and gene repression (Fig. 4). The interaction between dCas9 and the dCas9 handle hairpin is crucial for target binding. We suggest validating the secondary structure of the dCas9 handle for each sgRNA construct to ensure that this structure is maintained in different sequence contexts. The S. pyogenes terminator is introduced to mimic the natural RNA complex formation between crRNA and tracrRNA, which serves as a transcription terminator in bacterial cells and also probably stabilizes the sgRNA folding.
Figure 4.
Extensions to the base-pairing region with mismatched nucleotides decrease repression activity. Our results suggest that a precise 5′ end of the sgRNA is required for optimal targeted gene regulation. The sgRNA sequences used for testing the matched and mismatched extensions (additional 5, 10 or 20 nt) are shown on the right side, which are complementary to the mRFP gene. The data are normalized to the repression level achieved when using 20-nt-long sgRNAs (shown as the dotted line). Error bars show s.e.m. from three biological replicates.
Tuning the efficiency of sgRNA targeting
Single or multiple mutations introduced into the base-pairing region of the sgRNA can be used to tune the level of repression. We and others have shown that the 12-nt sequence that targets the region adjacent to the PAM site constitutes a seed region that is important for effective gene regulation12,19. Mismatches in this region generally reduce the repression by 70–90% (ref. 1). A single mismatch in the other 8-nt sequence, however, only causes a mild decrease in the repression activity (<50%). Although much work is needed to understand the design principles of using mismatches to fine-tune repression, mismatched sgRNAs could be used for achieving different levels of gene regulation.
Multiplexed gene targeting
The efficiency of gene repression can be enhanced by using multiple sgRNAs to simultaneously target the same gene. To achieve enhanced combinatorial repression, the target sites and PAMs of these sgRNAs should not overlap. Overlap between the target sites (either on the same strand or on the opposite strand) may result in competition and exclusive binding between multiple dCas9-sgRNA complexes, leading to a combinatorial effect that is lower than the repression achieved by each sgRNA individually. Similarly, multiple sgRNAs can be used to control multiple different genes simultaneously.
Cloning of sgRNAs
As the chimeric sgRNAs are short in length, differing only in the 20–25 nt base-pairing region, we have used inverse PCR (iPCR)40,41 to mutagenize the base-pairing region when the vector is small (<5 kb). The method only involves a single PCR step and a blunt-end ligation step and can be scaled up to clone many sgRNAs using 96-well plates. When the vector is big (>5 kb), oligo PCR and digestion or ligation methods should be used to avoid PCR errors that might be introduced by iPCR reactions.
Multiple sgRNA cloning
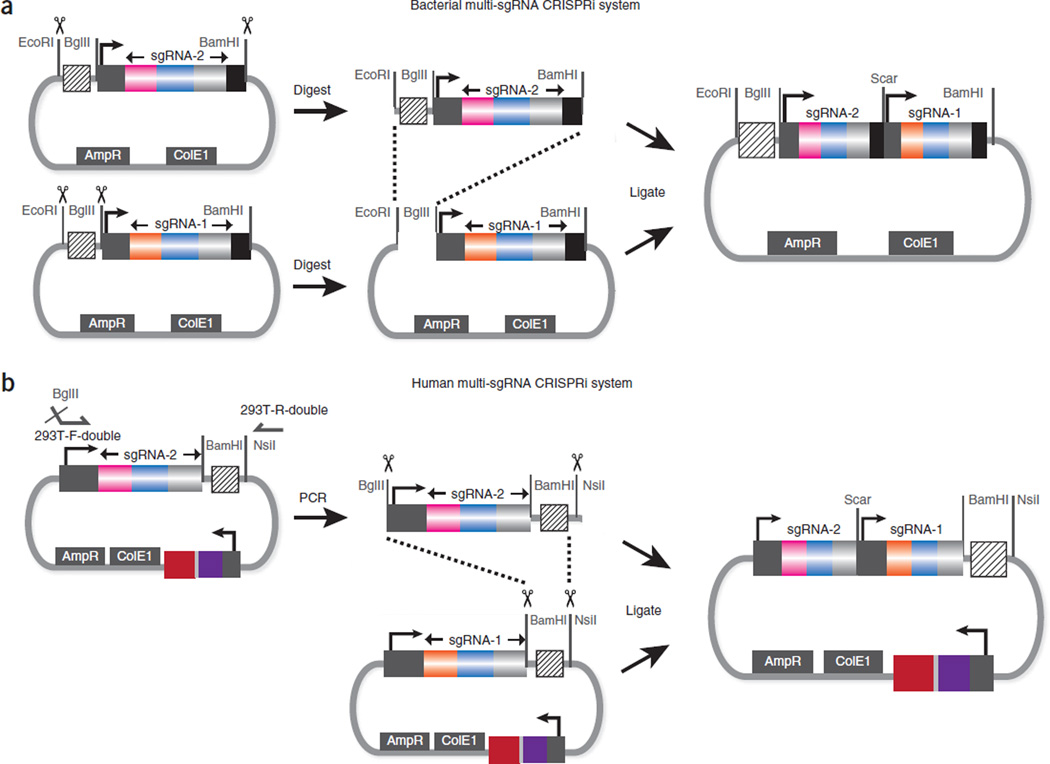
To clone multiple sgRNAs into a single expression vector, we inserted compatible restriction enzyme sites (BglII and BamHI) to flank individual sgRNA expression cassettes as BioBrick parts (Figs. 1b,c and 5)42. By using the BioBrick assembly42, a streamlined method for cloning of individual parts into multiple component genetic system, multiple sgRNA expression cassettes can be concatenated into a single expression vector.
Figure 5.
Cloning strategy for concatenating multiple sgRNA expression cassettes onto the same plasmid. (a) The cloning method for bacterial sgRNA expression. The donor sgRNA vector is digested using EcoRI and BamHI, and the backbone vector is digested using EcoRI and BglII. Ligation of the two fragments recreates the compatible restriction sites (BglII and BamHI), which can be used for the next round of sgRNA insertion. (b) The cloning method for human sgRNA expression. The main difference from a is that the insert is first PCR amplified to introduce a 5′ BglII site for subsequent BioBrick cloning.
Expression of sgRNAs
The CRISPRi system is highly efficient in bacterial cells, and leaky expression of either the dCas9 protein or the sgRNA from an inducible promoter can lead to a moderate (~80%) repression level, as compared with the dCas9 or sgRNA-only controls. Thus, although we previously used multiple- copy plasmids and strong promoters, expression systems based on low-copy plasmids, genome integration and/or weaker promoters can be used in bacteria for effective repression. The main constraint is that the sgRNA requires a well-defined 5′ end for effective target binding, as mismatches introduced into this region dampen silencing effects (Fig. 4). Therefore, promoters that start precisely at the 5′ end of the sgRNA, such as the BBa_J23119 synthetic E. coli promoter (http://parts.igem.org/), should be used for optimal repression43.
Design of functional validation assays
Functional assays using either flow cytometry or the measurement of β-galactosidase activity can be used to measure effects on protein expression if the target gene is fused to either a fluorescent protein or lacZ, respectively. Otherwise, quantitative methods such as qRT-PCR or genome-wide NET-seq could be used to measure the effects on transcription44,45. NET-seq can also be used to infer the exact genome loci that are being disrupted by the dCas9-sgRNA complex. All assays should be performed in the same cell type with similar growth conditions, as CRISPRi efficacy can vary between cell types and growth conditions. For simplicity, we have used the E. coli K12 MG1655 strain for functional assays, but other strains of E. coli and other bacterial species can also be used.
Controls
As the negative control for the sgRNA expression, we recommend using a truncated sgRNA that lacks the base-pairing region but which contains the dCas9-binding hairpin, allowing the sgRNA to form a complex with dCas9. The primers for constructing the negative control sgRNA are shown in Table 1. The negative control for dCas9 expression is the same vector, with the dCas9-coding sequence knocked out of it.
Table 1.
Primer sequences for sgRNA cloning.
| Name | Sequence | Purpose |
|---|---|---|
| Ec-F | 5′-N(20~25)GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC-3′ | Introduce new base-pairing sequences into E. coli sgRNAs |
| Ec-R | 5′-ACTAGTATTATACCTAGGACTGAGCTAGC-3′ | |
| Ec-F-control | 5′-GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC-3′ | Construct the negative control for the sgRNA vector = without the base-pairing region (PCR with Ec-R) |
| Ec-F-colony | 5′-GGGTTATTGTCTCATGAGCGGATACATATTTG-3′ | Colony PCR primers for E. coli sgRNA vector verification |
| Ec-R-colony | 5′-CGCGGCCTTTTTACGGTTC-3′ | |
| 293T-F | 5′-GGAGAACCACCTTGTTGGN(20~25)GTTTTA GAGCTA GAAATA GCAA GTTAAAATAAGGC-3′ | Construct sgRNAs on U6 vectors. The restriction sites are underlined, and the complementary sequence is in bold |
| 293T-R | 5′-CCTAGTACTCGAGAAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGA CTAGCCTTATTTTAACTT GCTATTTCTA GCTCTAAAAC-3′ |
|
| 293T-F-double | 5′-GCCGCGAGATCTGAGATCCGACGCGCCATC-3′ | Construct tandem sgRNAs on the U6 vector |
| 293T-R-double | 5′-AATAACTAATGCATGGCGGTAATACGGT-3′ | |
| 293T-F-seq | 5′-GAGATCCAGTTTGGTTAGTACCGGG-3′ | Sequencing primers for U6 vector |
| 293T-R-seq | 5′-ATGCATGGCGGTAATACGGTTAT-3′ |
MATERIALS
REAGENTS
sgRNA cloning
Plasmids: obtain plasmids from Addgene (http://www.addgene.org/crispr/qi/), including E. coli dCas9 plasmid (no. 44249), E. coli sgRNA plasmid (no. 44251), human dCas9 plasmid (nos. 44246 and 44247) and human sgRNA plasmid (no. 44248)
PCR primers for sgRNA cloning (Integrated DNA Technologies, custom DNA oligonucleotides)
Phusion hot start Flex 2× master mix (New England Biolabs, cat. no. M0536L) ▲ CRITICAL The high-fidelity DNA polymerase in this mix is important for iPCR, as a lower-fidelity enzyme might introduce errors during the PCR amplification step.
GoTaq Green master mix, 2× (Promega, cat. no. M7123)
QIAquick gel extraction kit (Qiagen, cat. no. 28706)
QIAquick 96 PCR purification kit (Qiagen, cat. no. 28181)
QIAquick PCR purification kit (Qiagen, cat. no. 28106)
QIAprep spin miniprep kit (Qiagen, cat. no. 27106)
UltraPure 10× TAE buffer (Invitrogen, cat. no. 15558-026)
UltraPure agarose (Invitrogen, cat. no. 16500500)
-
Ethidium bromide solution, 10 mg ml−1 (Sigma-Aldrich, cat. no. E8751)
! CAUTION Ethidium bromide is mutagenic; always wear nitrile gloves while working with it.
All-purpose HI-LO DNA mass ladder (Bionexus, cat. no. BN2050)
Restriction enzymes (all from New England Biolabs): BglII (cat. no. R0144S), BamHI-HF (cat. no. R3136S), EcoRI-HF (cat. no. R3101S), BstXI (cat. no. R0113L), XhoI (cat. no. R0146S), DpnI (cat. no. R0176S) and NsiI (cat. no. R0127S)
T4 DNA ligase (Roche, cat. no. 10481220001)
Quick blunt-end DNA ligation kit (New England Biolabs, cat. no. M2200S)
T4 polynucleotide kinase (New England Biolabs, cat. no. M0201S)
LB medium (Sigma-Aldrich, cat. no. L3022)
LB agar medium (Sigma-Aldrich, cat. no. L2897)
MOPS EZ rich defined medium kit (Teknova, cat. no. M2105)
Ampicillin, sterile filtered, 100 mg ml−1 (Sigma-Aldrich, cat. no. A5354)
Carbenicillin, sterile filtered, 100 mg ml−1 (Sigma-Aldrich, cat. no. C1613)
Chloramphenicol, sterile filtered, 34 mg ml−1 (Sigma-Aldrich, cat. no. C0378)
One Shot TOP10 chemically competent Escherichia coli (E. coli) (Invitrogen, cat. no. C4040-03)
Escherichia coli MG1655 strain (ATCC, cat. no. 700926)
Glycerol (Sigma-Aldrich, cat. no. G5516)
Exonuclease I and shrimp alkaline phosphatase for PCR samples cleanup (e.g., the USB ExoSAP-IT reagent by Affymetrix, cat. no. 78200 200 UL)
Metal spatula (Sigma-Aldrich, cat. no. Z283274)
ddH2O, sterile
NET-seq experiments
Nitrocellulose-mixed esters of cellulose membrane filters (Fisher Scientific, cat. no. E02WP09025)
Falcon tubes, 50 ml (Corning, cat. no. 14-432-22)
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
NP-40 (Sigma-Aldrich, cat. no. I8896)
Ammonium chloride (Sigma-Aldrich, cat. no. A9434)
Manganese(II) chloride (Sigma-Aldrich, cat. no. 244589)
EDTA, 0.5 M, pH 8 (Ambion, cat. no. AM9261)
SUPERase-In (Ambion, cat. no. AM2696)
Tagetin transcriptional inhibitor (Epicentre, cat. no. T9705H)
Protease inhibitor cocktail, complete, EDTA free (Roche, cat. no. 11873580001)
RQ1 RNase-free DNase (Promega, cat. no. M6101)
PD MiniTrap G-25 spin columns (GE Healthcare, cat. no. 28-9180-07)
Anti-FLAG M2 affinity gel (Sigma-Aldrich, cat. no. A2220)
FLAG peptide, 3× (Sigma-Aldrich, cat. no. F4799)
miRNeasy mini kit (Qiagen, cat. no. 217004)
Testing CRISPRi in HEK293 cells
HEK293 cells (ATCC, cat. no. CRL-1573)
Qiagen plasmid midi kit (Qiagen, cat. no. 12143)
Mirus TransIT-LT1 transfection reagent (Mirus, cat. no. 2300)
DMEM-high glucose medium (Invitrogen, cat. no. 11960-044)
l-Glutamine (Invitrogen, cat. no. 25031-081)
Penicillin-streptomycin (Invitrogen, cat. no. 15070-063)
Trypsin-EDTA solution, 0.05% (wt/vol) (Invitrogen, cat. no. 25300-062)
Lenti-X concentrator (Clontech, cat. no. 631232)
EQUIPMENT
MaxyClear microcentrifuge tubes, 1.7 ml (Axygen Scientific, cat. no. MCT-175-C-S)
Clear V-bottom polypropylene deep 96-well plates, 2 ml (Corning, cat. no. 3960)
Thermocycler with programmable temperature control, 96 wells (Bio-Rad)
PCR plates, 96 wells (Axygen Scientific, cat. no. PCR-96-FS-C)
Strip PCR tubes, 8 wells (Applied Biosystems, cat. no. N801-0580)
Multichannel pipettes (Rainin, cat. nos. L12-10XLS+, L12-200XLS+ and L12-1200XLS)
Gel electrophoresis system (PowerPac basic power supply, Bio-Rad, cat. no. 164-5050 and Sub-Cell GT system gel tray, Bio-Rad, cat. no. 170-4401)
Sterile 20-µl pipette tips for colony picking (Rainin, cat. no. SS-L10S)
Petri dishes, 60 mm × 15 mm (BD Biosciences, cat. no. 351007)
Incubator for bacteria plates (Thermo Scientific, cat. no. 50125590)
Microplate shaking incubator for bacteria suspension culture (Labnet, cat. no. LN-S2056A)
NanoDrop 8000 UV-visible spectrophotometer (Thermo Scientific)
Tissue culture plates, 100 mm (Cole-Parmer, cat. no. YO-01927-84)
Tissue culture plates, 24 wells (Cole-Parmer, cat. no. EW-01927-74)
Digital gel imaging system (GE)
BD-LSR II or alternate flow cytometer (BD Biosciences)
PROCEDURE
Genome target selection and sgRNA design ● TIMING 1–4 h
▲ CRITICAL Steps 1–8 describe the general sgRNA design procedure for both bacterial and mammalian cells. Steps 9–41 describe the sgRNA cloning and testing procedure for bacterial cells only and Steps 42–68 describe the sgRNA cloning and testing procedure for mammalian cells only.
-
1|
Determine the DNA sequence of genes to be targeted using available genome databases46–48.
-
2|
Select the DNA elements or loci to be targeted. For effective gene repression, the following elements or loci can be chosen1 (Fig. 1a): the nontemplate DNA strand of the 5′ portion of the gene-coding region; the nontemplate DNA strand of the 5′-UTR sequence; the −35 box or −10 box of the bacterial promoter (either the template or nontemplate strand); and cis-acting sites for trans-acting factor binding within the promoter (e.g., TFBS; either the template or nontemplate strand).
-
3|
Determine the sequence of the base-pairing region on the sgRNA (Fig. 3a). To target the template DNA strand, search in the transcribed gene sequence (which is the same as the nontemplate DNA strand) for 5′-N(20~25)-NGG-3′. The sequence of N(20~25) will be used directly as the base-pairing region of the sgRNA. NGG is the PAM sequence that is recognized by the S. pyogenes Cas9 protein. To target the nontemplate DNA strand, search in the transcribed gene sequence for 5′-CCN-N`(20~25)-3′. The reverse-complementary sequence of N`(20~25) will be used as the base-pairing region of the sgRNA.
▲ CRITICAL STEP Some promoters require a particular nucleotide as the transcription start site. For example, the human or mouse U6 promoters require a G at the 5′ end of the transcript for effective transcription. Therefore, to use the U6 promoters for sgRNA expression, the first nucleotide of the transcribed sgRNA should be a G to maximize U6 promoter activity. The search pattern should be modified accordingly to reflect this requirement: 5′-GN(19~24)-NGG-3′ or 5′-CCN-N`(19~24)C-3′ should be used for template or nontemplate strand targeting, respectively, and the sequence of GN(19~24) or the reverse-complement sequence of N`(19~24)C will be used as the base-pairing region of the sgRNA.
-
4|
Check the specificity of sgRNA binding in the genome using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov)49. Do a BLAST search for the 14-nt specificity sequence in the whole genome on both the sense and antisense strands to predict potential off-targets (full sequence: 5′-N12-NGG-3′; N12 is the seed region; Fig. 3a). Use only sgRNAs without predicted off-targets.
▲ CRITICAL STEP To compute the off-targets of large numbers of sgRNAs, computational algorithms for genome mapping of the short nucleotides should be used, such as Seqmap (http://www-personal.umich.edu/~jianghui/seqmap/)50. To use Seqmap, search the sequence 5′-N12-NGG-3′ in the genome sequence of the target organism using the parameter setting ‘1 /output_all_matches’, which allows the program to align the 5′-N12-NGG-3′ sequence to the genome sequence with one mismatch at the nucleotide N in NGG. Any sgRNA sequence that is mapped to multiple genomic loci should be discarded. Some Blast tools do not support mismatch functionality in their search algorithm. If this is the case, the NGG can be replaced by AGG/CGG/GGG/TGG and searched individually.
-
5|
Generate the full length of sgRNA by appending the dCas9 handle and S. pyogenes terminator sequence (Fig. 3a) to the 3′ end of the base-pairing region. The dCas9 handle sequence is 5′-GUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGC UAGUCCG-3′, and the S. pyogenes terminator sequence is 5′-UUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUUUU-3′.
-
6|
Predict the secondary structure folding of the dCas9 handle hairpin once it is appended to the sgRNA. The sgRNA requires a properly folded 42-nt hairpin for dCas9 protein-binding (the dot-bracket RNA format51 is (((((((.((((.)))).)))))))….). Predict the RNA secondary structure of the 62-nt region consisting of the base-pairing region and the 42-nt dCas9 handle using algorithms such as the online Quikfold algorithm from the UNAFold package52,53 (http://mfold.rna.albany.edu/?q=DINAMelt/Quickfold) or RNAfold from the ViennaRNA Package (http://www.tbi.univie.ac.at/~ronny/RNA/). Compute all suboptimal structures within 5% of the optimal structure. If any of the suboptimal folding results (for which the free energy is within 5% of the optimal fold) shows a properly folded dCas9 handle hairpin, keep the sgRNA and proceed.
-
7|
Confirm that the sgRNA sequence does not contain certain sequences, such as restriction enzyme sites for cloning (e.g., EcoRI, BglII and BamHI for bacterial sgRNAs; BstXI and XhoI for human sgRNAs) or transcription terminator sequences (e.g., more than four consecutive Us is a strong terminator for the U6 promoter54).
▲ CRITICAL STEP We have observed that the internal poly-U sequence can make the sgRNA non-functional, probably due to premature transcription termination at the poly-U sequence.
-
8|
If partial repression efficiency is desired, introduce single or multiple mismatches into the sgRNA base-pairing region by mutating the nucleotide into its complementary nucleotide (i.e., A to T, G to C and so on.). We have observed that mismatches at the 5′ end of the sgRNA generally cause a milder decrease of repression activity compared with the mismatches at the 3′ end1.
? TROUBLESHOOTING
-
9|
Choose sgRNA expression vectors. The bacterial sgRNA expression vector can be ordered from Addgene (ID no. 44251). Alternatively, other expression vectors can be used for sgRNA expression. These should contain a strong promoter that starts precisely at the 5′ end of sgRNA for optimal repression (Fig. 1b,c).
▲ CRITICAL STEP We have cloned an E. coli sgRNA expression vector (Addgene ID no. 44251) that contains a minimal synthetic σ70 promoter (pJ23119 (refs. 55,56); Fig. 1b) with an annotated transcription start site, an ampicillin-selectable marker and a ColE1 replication origin; and an inducible E. coli sgRNA expression vector that contains an anhydrotetracycline (aTc)-induced pLtetO-1 promoter57 with an annotated transcription start site, an ampicillin-selectable marker and a ColE1 replication origin. To express sgRNAs in mammalian cells, we have cloned a pSico-derived lentiviral mouse U6-based expression vector58 (Addgene ID no. 44248). The transcription start site of the U6 promoter is the last nucleotide G within the BstXI restriction enzyme site (Fig. 1c). The U6 vector also contains an expression cassette consisting of a CMV promoter, a puromycin-resistance gene and an mCherry gene for selection or screening purposes.
? TROUBLESHOOTING
High-throughput bacterial sgRNA cloning with iPCR ● TIMING 2–3 d (5 h hands-on time)
-
10|
Order primers for cloning sgRNAs (Ec-F and Ec-R; Table 1). The forward primers that contain different base-pairing regions should be ordered in the format of 96-well plates for a cheaper price and easy handling (e.g., from Integrated DNA Technologies).
-
11|In a 96-well PCR plate, set up the reactions for 5′ phosphorylation of both forward and reverse primers using T4 polynucleotide kinase as described below, and incubate the plate at 37 °C for 1 h. Multichannel pipettes should be used to minimize pipetting.
Component Amount per well (µl) Primer, 100 µM 0.5 T4 polynucleotide kinase buffer, 10× 0.5 T4 polynucleotide kinase 0.25 Nuclease-free water 3.75 Total 5 -
12|Combine the phosphorylation reactions of the pairs of forward and reverse primers, and use the primer mixes to prepare 25-µl PCR reactions:
Component Amount (µl) E. coli sgRNA plasmid (100 ng µl−1) 0.5 5′-phosphorylated forward and reverse primers 10 Phusion hot start Flex 2× master mix 12.5 Nuclease-free water 2 Total 25 -
13|Perform PCR on the samples with the following cycling conditions:
Cycle number Denature Anneal Extend 1 98 °C, 30 s 2–26 98 °C, 10 s 62 °C, 30 s 72 °C, 1 min 27 72 °C, 5 min -
14|
After the PCR reaction has completed, use gel electrophoresis to verify that the PCR was successful. Cast a 1% (wt/vol) agarose gel in TAE with ethidium bromide (0.5 µg ml−1). Run the gel at 20 V cm−1 for 15 min. It is not necessary to check all samples; checking a dozen random samples is usually sufficient. Successful PCR samples should show a ~2,500-bp product.
? TROUBLESHOOTING
-
15|
Purify the PCR reactions using a high-throughput 96-well PCR purification kit according to the manufacturer’s directions. This generates linearized PCR products that contain new sgRNA sequences.
-
16|
In a 96-well plate, digest 2 µl of the PCR product using DpnI to remove the template sgRNA vector in a 10-µl reaction according to the manufacturer’s instructions at 37 °C for 1 h.
-
17|
Without further purification or heat inactivation, self-ligate the DpnI digestion reactions using a Quick blunt-end DNA ligation kit.
-
18|
Transform the ligation reactions into One Shot TOP10 chemically competent E. coli cells according to the manufacturer’s instructions.
-
19|
Pick single colonies into a 2-ml-deep 96-well plate (usually two colonies for each sgRNA cloned), each well containing 300 µl of LB medium with proper selective antibiotics. Use a sterile 20-µl pipette tip to touch a single colony, and swirl the tip in the LB medium to dissolve the colony. Incubate the 96-well plate at 37 °C overnight.
-
20|Perform colony PCR on the colonies selected in Step 19. The purpose of the colony PCR is to generate linearized DNA fragments containing the sgRNA base-pairing region for sequencing reactions. Set up the following PCR reaction:
Component Amount (µl) Colony suspension from Step 19 1 Forward sequencing primer (Ec-F-colony), 10 µM 0.25 Reverse sequencing primer (Ec-R-colony), 10 µM 0.25 GoTaq Green master mix, 2× 12.5 Nuclease-free water 11 Total 25 -
21|Perform colony PCR with the following cycling conditions:
Cycle number Denature Anneal Extend 1 95 °C, 3 min 2–31 95 °C, 30 s 60 °C, 30 s 72 °C, 2 min 32 72 °C, 5 min -
22|
Clean up the PCR products using exonuclease I and shrimp alkaline phosphatase to remove residual primers and dNTPs, which interfere with the sequencing reactions. Follow the manufacturer’s instructions for the clean-up and heat-inactivation steps.
-
23|
Sequence the colony PCR products with primer Ec-F-colony to identify the clones with correct sgRNAs by using commercially available sequencing services.
? TROUBLESHOOT ING
-
24|
After sequencing verification, inoculate overnight culture of the correct clones (one clone per sgRNA) from the 96-well plate into 5 ml of LB culture with proper selection antibiotic. Incubate the culture at 37 °C for 4 h to overnight.
■ PAUSE POINT Correct clones from the 96-well overnight plate can be used to prepare glycerol stocks (500 µl of 40% (vol/vol) glycerol mixed with 500 µl of cell culture) and stored at −80 °C.
-
25|
Isolate plasmid DNA from the inoculated 5 ml of DNA cultures using the spin miniprep kit according to the manufacturer’s instructions.
Cloning of multiple bacterial sgRNAs into a single expression vector with BioBrick ● TIMING 2 d (6 h hands-on time)
-
26|
Digest the donor sgRNA vector using both EcoRI and BamHI, and digest the sgRNA recipient backbone vector using both EcoRI and BglII (Fig. 5a) according to the manufacturer’s instructions at 37 °C for 3 h.
-
27|
Separate the restriction enzyme-digested samples by electrophoresis in 1% (wt/vol) agarose gels; verify the fragment size by using a suitable DNA ladder. The correct size of the donor sgRNA fragment is ~500 bp, and the correct size of the backbone sgRNA fragment is 2,000 + 500 × N bp, where N is the number of sgRNA expression cassettes already cloned into the digested backbone vector.
? TROUBLESHOOTING
-
28|
Extract the donor and backbone DNA fragments using the gel extraction kit according to the manufacturer’s instructions.
-
29|
Ligate 150 ng of the donor DNA (sgRNA) with 50 ng of the backbone DNA using T4 DNA ligase according to the manufacturer’s instructions.
-
30|
Transform the reactions into One Shot TOP10 chemically competent E. coli cells following the manufacturer’s instructions.
-
31|
Identify correct clones by colony PCR and sequencing, as described in Step 23.
■ PAUSE POINT Correct clones can be stored as glycerol stocks at −80 °C.
Choice and cloning of bacterial dCas9 expression vector ● TIMING up to 3 d
-
32|
The bacterial dCas9 expression vector can be ordered from Addgene (ID no. 44249). Alternatively, the catalytically inactive dCas9 gene can be cloned onto a custom expression plasmid that is compatible with the sgRNA vector. The compatible vector should contain a different selection marker (e.g., chloramphenicol) than the one used in the sgRNA vector (e.g., ampicillin) but a compatible plasmid origin of replication similar to the one used in the sgRNA vector (e.g., the p15A and pSC101 origins are compatible with the ColE1 replication origin in the in the sgRNA vector57).
▲ CRITICAL STEP Weak promoters can be used to express dCas9 in E. coli, which is often sufficient for effective gene repression. In E. coli, we have cloned a bacterial dCas9 expression vector (Addgene ID no. 44249) that contains an aTc-inducible pLtetO-1 promoter, a chloramphenicol-resistance marker and a p15A replication origin.
E. coli gene repression strain construction ● TIMING 3 d (4 h hands-on time)
-
33|
Co-transform the sgRNA expression vector from Step 25 or 31 alongside the dCas9 expression plasmid from Step 32 into competent E. coli strains of interest (e.g., MG1655 cells) according to the standard protocols59. The negative control for sgRNA expression is an sgRNA expression vector without the 20-nt base-pairing region. The negative control for dCas9 expression is a blank dCas9 expression vector without the dCas9-coding sequence. Plate the transformed bacteria on LB agar plates supplemented with the appropriate antibiotics.
? TROUBLES HOOT ING
▲ PAUSE PO INT Correct clones can be kept as glycerol stocks at −80 °C, and aliquots can be used for further experiments.
-
34|
Pick single colonies and cultivate cells in 300 µl of MOPS EZ rich defined medium containing 100 µg ml−1 carbenicillin and 34 µg ml−1 chloramphenicol in 96-well 2-ml-deep well plates overnight at 37 °C with shaking at 1,200 r.p.m., as previously described60–62.
▲ CRITICAL STEP The choice of growth medium might affect CRISPRi repression efficiency due to varied growth conditions. For the purpose of performing fluorescence assays or other growth assays, we recommend using a defined growth medium that is colorless, such as MOPS EZ rich defined medium. This often improves experimental reproducibility and allows for more quantitative measurements.
-
35|
Add 1 µl of this overnight culture into 249 µl of fresh MOPS EZ rich defined medium with the same antibiotic concentrations and supplemented with 2 µM aTc. The aTc is added to induce the expression of the dCas9 protein and for the expression of sgRNAs from the pLtetO-1 promoter when used.
-
36|
Grow cells to early log phase (~3 h) before subjecting them to different experimental conditions (e.g., the presence of chemicals) to determine the cellular phenotype caused by the targeted gene repression. If the target genes are fluorescent reporters, fluorescence expression can be assayed using flow cytometry, according to the manufacturer’s instructions, and the absorbance and emission wavelength specifications for the particular fluorescent proteins used63. Each experiment should include triplicate cultures.
E. coli NET -seq for assaying CRISPRi-induced transcriptional pausing ● TIMING 1–2 weeks
-
37|
Cell culturing and collection. For each sample, inoculate an overnight culture of E. coli in EZ rich defined medium with the proper antibiotics at 37 °C. Dilute the overnight culture in 500 ml of EZ rich defined medium to an optical density at 600 nm (OD600) of 0.1 OD, and incubate the cells at 37 °C to early log phase: OD600 of 0.45 ± 0.05. Collect the cells by filtration with 0.22-µm nitrocellulose filters. As soon as the medium has passed through the filter, scrape the cells with a metal spatula and immediately freeze them in a 50-ml tube filled with liquid nitrogen to halt transcription.
■ PAUSE POINT Frozen cells can be kept at −80 °C for 3 months.
-
38|Cell mixer-milling. Prepare 25 ml of 10× cell lysis buffer:
Component Final concentration Amount (ml) Tris, 1 M, pH 8.0 200 mM 5 Triton X-100, 25% 4% (vol/vol) 4 NP-40, 20% 1% (vol/vol) 1.25 DEPC water 14.75 Total 25 Weigh 100 µg of frozen cells in a 50-ml tube prechilled with liquid nitrogen, and combine them with 500 µl of the following solution frozen in liquid nitrogen:Component Final concentration Amount (µl) Cell lysis buffer, 10× 1× 50 NH4Cl, 1 M 100 mM 50 MnCl2, 1 M 10 mM 5 Superase In, 20,000 U ml−1 50 U ml−1 1.25 Tagetin, 600 µM 15 µM 12.5 Proteinase inhibitor cocktail, 25× 1× 20 DEPC water 361.25 Total 500 µl Pulverize the mixture on a Qiagen TissueLyser II mixer mill six times at 15 Hz for 3 min. Resuspend the cell lysate on ice by pipetting, and add 110 µl of RQ1 DNase I; incubate the suspension for 20 min on ice. Add EDTA to lysate to a final concentration of 25 mM to quench the reaction, followed by centrifugation at 20,000g for 10 min at 4 °C. Load the lysate onto a PD MiniTrap G-25 column and elute with cell lysis buffer supplemented with 1 mM EDTA.
-
39|
Immunoprecipitate the transcription elongation complexes using an appropriate affinity tag on RNAP, such as the 3× FLAG epitope, as described in ref. 45.
-
40|
Purify the nascent RNA using the miRNeasy mini kit (Trizol) and convert it to a DNA library using a previously established library generation protocol64.
-
41|
Sequence the DNA library with deep coverage using a high-throughput next-generation sequencing platform, such as the Illumina Hiseq, and align to the appropriate reference genome. We recommend using Bowtie (http://bowtie-bio.sourceforge.net) for short read alignments.
Cloning of human sgRNA expression vector with oligo PCR ● TIMING 2–3 d (5 h hands-on time)
-
42|
Order primers (293T-F and 293T-R) for sgRNA cloning (Table 1).
▲ CRITICAL STEP For best results, long primers (longer than 90 bp) should be ordered as Ultramer oligos (Integrated DNA Technologies) purified with desalting to remove small molecule impurities, which improves the PCR reaction efficiency.
-
43|
Digest the human sgRNA expression vector (Addgene ID no. 44248) with restriction enzymes BstXI and XhoI at 37 °C, for 4–16 h according to the manufacturer’s instructions.
-
44|
Separate the restriction enzyme–digested samples by electrophoresis in a 1% (wt/vol) agarose gel using standard protocols. Verify the fragment size using a suitable DNA ladder; the correct band is at ~8 kb.
-
45|
Extract the ~8,000-bp-long vector DNA using the gel purification kit.
-
46|Prepare the oligo PCR reaction for generating the sgRNA cassette:
Component Amount (µl) Forward primer (293T-F), 10 µM 2.5 Reverse primer (293T-R), 10 µM 2.5 Phusion hot start Flex 2× master mix 12.5 Nuclease-free water 7.5 Total 25 -
47|Perform PCR with the following cycling conditions:
Cycle number Denature Anneal Extend 1 98 °C, 30 s 2–26 98 °C, 10 s 62 °C, 30 s 72 °C, 10 s 27 72 °C, 5 min -
48|
After the PCR has completed, use gel electrophoresis to verify that PCR reactions are successful. Successful PCR reactions should show a 133-bp-long product.
-
49|
Purify the PCR samples by using the PCR purification kit according to the manufacturer’s directions.
-
50|
Digest the PCR product using both BstXI and XhoI in a 50-µl reaction according to the manufacturer’s instructions, and then incubate the digestion reaction at 37 °C for 4 h.
-
51|
Clean up the digestion reaction using the PCR purification kit according to the manufacturer’s directions.
-
52|
Measure the concentration of the digestion product with the NanoDrop spectrophotometer.
-
53|
Ligate 300 ng of the digested sgRNA fragment (Step 51) with 50 ng of the digested backbone vector (Step 45), using T4 DNA ligase according to the manufacturer’s instructions.
-
54|
Transform the ligation reaction into One Shot TOP10 chemically competent E. coli cells according to the manufacturer’s instructions.
-
55|
Identify correct clones by sequencing as described in Step 23.
? TROUBLESHOOT ING
■ PAUSE POINT Correct clones can be stored as glycerol stocks at −80 °C.
Cloning of multiple human sgRNA expression cassettes into the same U6 vector with BioBrick ● TIMING 2 d (6 h hands-on time)
-
56|Set up the following PCR reaction to amplify the sgRNA expression cassette:
Component Amount (µl) Human sgRNA plasmid (donor vector, 100 ng µl−1) 0.5 Forward primer (293T-F-double), 100 µM 0.5 Reverse primer (293T-R-double), 100 µM 0.5 Phusion hot start Flex 2× master mix 25 Nuclease-free water 23.5 Total 50 -
57|Perform PCR with the following cycling conditions:
Cycle number Denature Anneal Extend 1 98 °C, 30 s 2–31 98 °C, 10 s 62 °C, 30 s 72 °C, 10 s 32 72 °C, 5 min -
58|
Purify the PCR samples by using the PCR purification kit according to the manufacturer’s directions.
-
59|
Digest the PCR product with both BglII and NsiI, and digest the human sgRNA plasmid (receptor vector) using both BamHI and NsiI according to the manufacturer’s instructions at 37 °C for 3 h (Fig. 5b).
-
60|
Separate the restriction enzyme–digested samples by electrophoresis in 1% (wt/vol) agarose gels using standard protocols. Verify the fragment size: the correct size of the donor sgRNA fragment is ~500 bp, and the correct size of the backbone sgRNA vector is ~8 kb.
-
61|
Extract the donor and backbone DNA fragments by using the gel purification kit according to the manufacturer’s instructions.
-
62|
Ligate the donor vector DNA fragment (150 ng) with the receptor vector DNA fragment (50 ng) by using a T4 DNA ligase according to the manufacturer’s instructions (e.g., Roche, T4 DNA ligase).
-
63|
Transform the ligation product into One Shot TOP10 chemically competent E. coli cells according to the manufacturer’s instructions.
-
64|
Identify correct clones using primers 293T-F-seq and 293T-R-seq by sequencing as described in Step 23.
■ PAUSE POINT Correct clones can be stored as glycerol stocks at −80 °C.
Choice and cloning of human dCas9 expression vector ● TIMING up to 3 d
-
65|
Order the human codon-optimized catalytically inactive dCas9 gene from Addgene (ID nos. 44246 or 44247). Alternatively, the codon-optimized dCas9 gene can be cloned onto different human expression vectors (e.g., Clontech MSCV-Puro plasmid) according to the manufacturer’s instructions.
▲ CRITICAL STEP The sequence encoding dCas9 should be fused with at least two copies of nuclear localization sequences (NLSs, e.g., SV40 NLS) for proper nuclear localization. We fused three copies of the SV-40 NLS to the 3′ end of dCas9 with a 3-aa linker (Addgene ID 44246). This protein can be expressed transiently, using standard expression vectors or stably using standard lentiviral or retroviral vectors. Importantly, because of the large size of dCas9, we have observed lower than expected viral titers for both standard lentiviral and retroviral systems. If higher titers are required, lentiviral supernatants can be concentrated using Lenti-X concentrator according to the manufacturer’s instructions.
Targeted gene repression in HEK293 cells ● TIMING 4 d (4 h hands-on time)
-
66|
Culture HEK293 cells in DMEM supplemented with 10% (vol/vol) FBS, 2 mM glutamine, 100 U ml−1 streptomycin and 100 µg ml−1 penicillin.
-
67|
Transfect the HEK293 cells using any DNA transfection reagent (e.g., Mirus TransIT-LT1) with the manufacturer’s recommended protocol. For transfection, seed a 24-well tissue culture plate with 50,000 HEK293 cells per well, 16–24 h before transfection. We transfect 0.5 µg of the dCas9 expression plasmid and 0.5 µg of the sgRNA expression plasmid per well.
? TROUBLESHOOTING
-
67|
As with all gene repression methods, the level of the overall effect depends on the level of suppression, the amount and half-life of pre-existing RNA and protein and the number of cell divisions before performing the measurement. We measured gene expression levels 72 h following transfection. To measure GFP expression by flow cytometry, trypsinize cells to a single-cell suspension and then analyze GFP fluorescence. The degree of repression is measured by comparing the median GFP fluorescence in cells expressing sgRNAs-targeting GFP with control cells that express the dCas9 vector alone or a nontargeting sgRNA.
▲ CRITICAL STEP To measure gene repression, we suggest using a puromycin selection step to ensure that all cells express dCas9 rather than relying on co-transfection of the sgRNA and Cas9-containing vectors. The dCas9 vector contains a puromycin-resistance marker, whereas the sgRNA vector contains a constitutive CMV promoter driving an mCherry gene. To select a pure population of Cas9-expressing cells, we treat HEK293 cells with 3 µg ml−1 puromycin for 24 h. We then analyze expression of our fluorescent protein reporter (GFP) in the mCherry-positive population of cells, defined as cells with greater than tenfold brighter mCherry median fluorescence over the negative control cells.
? TROUBLESHOOTING ?
? TROUBLESHOOTING ?
Troubleshooting advice can be found in Table 2.
● TIMING
Steps 1–9, genome target selection and sgRNA design: 1–4 h
Steps 10–25, high-throughput bacterial sgRNA cloning with iPCR: 2–3 d (5 h hands-on time)
Steps 26–31, cloning of multiple bacterial sgRNAs into a single expression vector with BioBrick: 2 d (6 h hands-on time)
Step 32, choice and cloning of bacterial dCas9 expression vector: up to 3 d
Steps 33–36, E. coli gene repression strain construction: 3 d (4 h hands-on time)
Steps 37–41, E. coli NET-seq for assaying CRISPRi-induced genome transcription pausing: 1–2 weeks (cDNA sample preparation takes 5–7 d, and sequencing/analysis takes 5–7 d)
Steps 42–55, cloning of human sgRNA expression vector with the oligo PCR method: 2–3 d (5 h hands-on time)
Steps 56–64, cloning of multiple human sgRNA expression cassettes into the same U6 vector with BioBrick: 2 d (6 h hands-on time)
Step 65, choice and cloning of human dCas9 expression vector: up to 3 d
Steps 66–68, targeted gene repression in HEK293 cells: 4 d (4 h hands-on time)
Table 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 8 | The mismatched sgRNAs do not function as expected | The mismatched sgRNAs have off-target binding sites in the genome; misfolded dCas9 handle hairpin | Perform Steps 4–7 again for the mismatched sgRNAs to check off-targets, dCas9 handle folding and poly-U sequence, etc. |
| 9 | The sgRNA activity is weak | Promoter is not strong enough; inaccurate transcription start site; alternative transcription start sites | Choose a stronger promoter with an accurately annotated transcription start site; remove alternative transcription start sites if necessary |
| 14 | No visible band | Degraded dNTPs | Use fresh stocks of dNTP mixtures |
| 23 | Clones contain too many mutations | Suboptimal PCR reaction conditions in Steps 12 and 13 | Decrease the amount of primers used for PCR (e.g., use 0.25 µl instead of 0.5 µl), and increase the annealing temperature (e.g., to 64 °C) |
| 27 | Digestion bands are smeared | Nonspecific digestion by the restriction enzymes | Use half the amount of restriction enzymes in Step 26 and reduce the digestion time (e.g., to 1 h) |
| 33 | Low transformation efficiency | Not enough plasmid DNA | Increase the amount of DNA used for transformation (e.g., double the amount of DNA used); use cells with higher transformation efficiency |
| 55 | No correct clone found | Incorrect amount of digested insert DNA used for ligation | Re-measure DNA concentration and adjust the molar ratio between insert and vector to ~5:1 |
| 67 | Low transfection efficiency | Low DNA quality Suboptimal amount of DNA used for transfection | Prepare DNA using high-quality plasmid preparation, such as the Qiagen plasmid midi kit Increase or decrease the amount of DNA to determine the optimal amount for transfection |
| 68 | No effect on gene expression | dCas9-sgRNA complex is unable to access the target site | Design new sgRNAs to target nearby sequence(s) |
ANTICIPATED RESULTS
The described protocol provides a number of sgRNA expression vectors that can be used in combination with the dCas9 expression vector to target any gene of interest in E. coli (e.g., strain MG1655) or in human cells (e.g., HEK293 cells) (Figs. 1b,c and 5). In our experience, the dCas9 and designed sgRNAs can robustly silence target genes with a 99–99.5% repression level in E. coli. There are several ways to tune the repression level, such as by using several, nonoverlapped sgRNAs per gene that could increase the repression level to 99.9% or by using mismatched sgRNAs that can decrease the repression level (Fig. 4). In human cells, we observed moderate but reproducible repression of gene expression using properly designed sgRNAs. For example, we observed ~50–60% gene repression using dCas9 and an sgRNA to repress a green fluorescent reporter gene that is stably inserted into the HEK293 genome. The use of sgRNAs to silence endogenous gene expression is dependent on the local chromatin state of the gene, as well as yet-to-be established mechanisms governing the binding affinity and dynamics of the dCas9-sgRNA complex to DNA. Therefore, we usually construct several sgRNAs for each genomic locus that we aim to control. The sgRNAs can be designed and cloned to bind to neighboring regions of a specific locus, all of which should contain an NGG PAM sequence. Because of differences in the epigenetic states among different types of cells, the silencing efficiency might vary in different cell types.
Acknowledgments
We thank the Jonathan Weissman lab and Wendell Lim lab for their support. L.S.Q. acknowledges support from the UCSF Center for Systems and Synthetic Biology. This work was supported by US National Institutes of Health (NIH) grant no. P50 GM081879 (L.S.Q., W.A.L.), the Howard Hughes Medical Institute (M.H.L., L.A.G., J.S.W., W.A.L.), a Howard Hughes Collaborative Initiative Award (J.S.W.) and a Ruth L. Kirschstein National Research Service Award (M.H.L.). X.W. is supported by A Foundation for the Author of National Excellent Doctoral Dissertation grant (grant number 201158).
Footnotes
AUTHOR CONTRIBUTIONS L.S.Q., M.H.L., L.A.G. and X.W. wrote the manuscript. J.S.W., W.A.L. and L.S.Q. supervised the research.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouns SJJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol. 2011;18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 8.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson TR, Saroj SD, Llewellyn AC, Tzeng Y-L, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 17.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 18.Shah SA, Erdmann S, Mojica FJM, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dicarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 26.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 28.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 29.Segal DJ, Barbas CF. Custom DNA-binding proteins come of age: polydactyl zinc-finger proteins. Curr. Opin. Biotechnol. 2001;12:632–637. doi: 10.1016/s0958-1669(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 30.Beerli RR, Barbas CF. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Segal DJ, Ghiara JB, Barbas CF. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc. Natl. Acad. Sci. USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Rossi J. RNAi mechanisms and applications. BioTechniques. 2008;44:613–616. doi: 10.2144/000112792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Sanchez M-J, et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 2012;85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 38.Fischer S, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012;287:33351–33363. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang SH. Inverse polymerase chain reaction. An efficient approach to cloning cDNA ends. Mol. Biotechnol. 1994;2:15–22. doi: 10.1007/BF02789286. [DOI] [PubMed] [Google Scholar]
- 41.Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 42.Shetty RP, Endy D, Knight TF. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat. Biotechnol. 2012;30:1002–1006. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 44.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Churchman LS, Weissman JS. Native elongating transcript sequencing (NET-seq) Curr. Protoc. Mol. Biol. 2012;4:14.1–14.17. doi: 10.1002/0471142727.mb0414s98. [DOI] [PubMed] [Google Scholar]
- 46.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keseler IM, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhagwat M, Young L, Robison RR. Using BLAT to find sequence similarity in closely related genomes. Curr. Protoc. Bioinformatics. 2012;37:10.8.1–10.8.24. doi: 10.1002/0471250953.bi1008s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 53.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 55.Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. USA. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP. Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic Acids Res. 2012;40:5775–5786. doi: 10.1093/nar/gks168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 60.Liu CC, et al. An adaptor from translational to transcriptional control enables predictable assembly of complex regulation. Nat. Methods. 2012;9:1088–1094. doi: 10.1038/nmeth.2184. [DOI] [PubMed] [Google Scholar]
- 61.Mutalik VK, Qi L, Guimaraes JC, Lucks JB, Arkin AP. Rationally designed families of orthogonal RNA regulators of translation. Nat. Chem. Biol. 2012;8:447–454. doi: 10.1038/nchembio.919. [DOI] [PubMed] [Google Scholar]
- 62.Liu CC, Qi L, Yanofsky C, Arkin AP. Regulation of transcription by unnatural amino acids. Nat. Biotechnol. 2011;29:164–168. doi: 10.1038/nbt.1741. [DOI] [PubMed] [Google Scholar]
- 63.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 64.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]