Abstract
Sheep chromosome 3 (Oar3) has the largest number of QTLs reported to be significantly associated with resistance to gastro-intestinal nematodes. This study aimed to identify single nucleotide polymorphisms (SNPs) within candidate genes located in sheep chromosome 3 as well as genes involved in major immune pathways. A total of 41 SNPs were identified across 38 candidate genes in a panel of unrelated sheep and genotyped in 713 animals belonging to 22 breeds across Asia, Europe and South America. The variations and evolution of immune pathway genes were assessed in sheep populations across these macro-environmental regions that significantly differ in the diversity and load of pathogens. The mean minor allele frequency (MAF) did not vary between Asian and European sheep reflecting the absence of ascertainment bias. Phylogenetic analysis revealed two major clusters with most of South Asian, South East Asian and South West Asian breeds clustering together while European and South American sheep breeds clustered together distinctly. Analysis of molecular variance revealed strong phylogeographic structure at loci located in immune pathway genes, unlike microsatellite and genome wide SNP markers. To understand the influence of natural selection processes, SNP loci located in chromosome 3 were utilized to reconstruct haplotypes, the diversity of which showed significant deviations from selective neutrality. Reduced Median network of reconstructed haplotypes showed balancing selection in force at these loci. Preliminary association of SNP genotypes with phenotypes recorded 42 days post challenge revealed significant differences (P<0.05) in fecal egg count, body weight change and packed cell volume at two, four and six SNP loci respectively. In conclusion, the present study reports strong phylogeographic structure and balancing selection operating at SNP loci located within immune pathway genes. Further, SNP loci identified in the study were found to have potential for future large scale association studies in naturally exposed sheep populations.
Introduction
Assessment of livestock health conditions in developing countries for identification of priority diseases to be targeted for control, revealed helminth infections as one of the most important problems in sheep and goat [1], [2]. Gastro-intestinal parasitic infestations such as Haemonchus contortus, Teledorsagia circumcincta, Trichostrongyles, Nematodirus sp. impose severe constraints on sheep and goat production especially those reared by marginal farmers under low external input system. These parasites incur heavy losses to farmers in terms of body weight loss, direct cost of anthelminthic drugs, loss due to mortality, etc. For example, annual treatment cost for Haemonchus contortus alone had been estimated to be 26 million USD in Kenya, 46 million USD in South Africa and 103 million USD in India [3]. Emergence of strains resistant to anthelminthic drugs has further complicated the management of parasitic diseases in small ruminants [4], [5]. Breeding programs with the goal of enhancing host resistance to parasites may help to alleviate this problem in the long term. Genetic variation in host resistance exists for the major nematode species affecting sheep: Haemonchus contortus, Trichostrongylus colubriformis, Teledorsagia circumcincta and various Nematodirus species. Considerable variation has been reported among sheep breeds on their ability to resist gastro-intestinal nematodes (GIN). For example, indigenous sheep breeds like Red Maasai [6], Garole [7], Gulf Coast Native [8], Rhon [9] and Barbados Black Belly [10] were found to have relatively better resistance against GINs. Similarly, within-breed genetic variation has also been demonstrated in diverse sheep populations including Merino [11], Romney [12], Scottish Blackface [13], feral Soay sheep [14], etc. Estimation of genetic parameters revealed low to moderate heritability in different sheep populations (h2 = 0.149, Avikalin sheep [15] to h2 = 0.41, Armidale sheep [16]).
Exploration of genetic variation either within specific regions of genome or more specifically in candidate genes involved in innate and adaptive immune pathways may help to identify a set of DNA markers significantly associated with parasite resistance characteristics. The former approach in terms of quantitative trait locus (QTL) analysis is a powerful method to understand genotype-phenotype relationship. Several QTL studies on parasite resistance characteristics have been reported in sheep. A quick evaluation of Animal QTL database [17] revealed a total of 753 QTLs reported for various economic traits in sheep. Among these, 81 were found to be related to parasite resistance characteristics and distributed in all sheep chromosomes except chromosomes 5 and 19. However, such QTLs related to parasite resistance were found to be more concentrated in chromosome 3 (16 QTLs) followed by chromosome 14 (7 QTLs). Among different parasites, 44 of 81 QTLs have been reported on resistance to Haemonchus spp., 20 on Trichostrongyles spp., 11 on Nematodirus spp. and six on Strongyles spp (Figure S1a–e). Thus the complexity of this analysis is evident from the fact that multiple, significant QTL regions have been reported across the entire genome, but the identification of candidate causative genes has remained elusive. The lack of consensus overlap among reported QTLs has hindered the identification of candidate genes and genetic markers for selection in sheep [18]–[21].
One of the important objectives of QTL studies is to identify underlying causative gene polymorphisms associated with the trait. Different QTLs reported in chromosome 3 for parasite resistance characteristics were found to be distributed all over the chromosome with varying overlapping regions. Hence, different candidate genes within chromosome 3 along with genes involved in immune related KEGG pathways (KEGG-Kyoto Encyclopedia of Genes and Genomes) could be important targets for establishing underlying causative variations. It is expected that the potential causative polymorphisms within candidate genes are members of the same overarching KEGG pathway that lead to the phenotypic expression on parasite resistance characteristics in each population. Further, the extent of genetic diversity and population sub-structure at such polymorphic loci are critical for such a genotype-phenotype association study. Population stratification has been demonstrated to result in false positive associations in various species including humans [22], [23], dogs [24] and cattle [25], [26]. Considering the significance of genetic basis of parasite resistance in sheep, the Joint FAO/IAEA Division of International Atomic Energy Agency initiated a coordinated research project to document the phenotypic differences and underlying genetic variations in indigenous sheep and goat breeds of 12 countries from Asia, Africa and South America through on-farm artificial challenge and natural exposure under farmers' field conditions respectively. The ultimate goal of this project is to identify a common set of genetic markers that significantly influence parasite characteristics across different indigenous populations of sheep and goats. The present study thus aimed at exploring genetic variations within genomic regions containing significant QTLs and different candidate genes involved in immune pathways, identification of single nucleotide polymorphisms (SNPs) and genetic diversity analysis in sheep populations evolved under different environmental conditions. A preliminary study was performed on association of genotypes with host resistance characteristics against gastro-intestinal nematodes (i.e. fecal egg count, body weight change and packed cell volume measured in response to artificial challenge with infective L3 larvae of Haemonchus contortus parasite).
Materials and Methods
Animal Ethics Statement
All procedures for artificial challenge experiment at different locations in Argentina and Indonesia were approved respectively by the Institutional Committee for Care and Use of Experimental Animals of the National Institute of Agricultural Technology (CICUAE-INTA), Argentina (protocol number 35/2010) and Institutional Research Animal Facility, Bogor Agricultural University, Indonesia following the guidelines described in their institutional manuals. Experimental animals were challenged with infective L3 larvae of Haemonchus contortus and blood samples were collected from jugular vein under the supervision of qualified veterinarians for extraction of DNA and assessment of blood parameters. 42 days post challenge, animals were dewormed to clear parasites from the gut after the experiment. The experimental challenge in both locations did not involve animals from any endangered or protected species/breeds. Blood sample collection for DNA extraction and genotyping from remaining breeds were performed by local veterinarians in respective countries following good animal practice.
DNA samples for diversity analysis and association studies
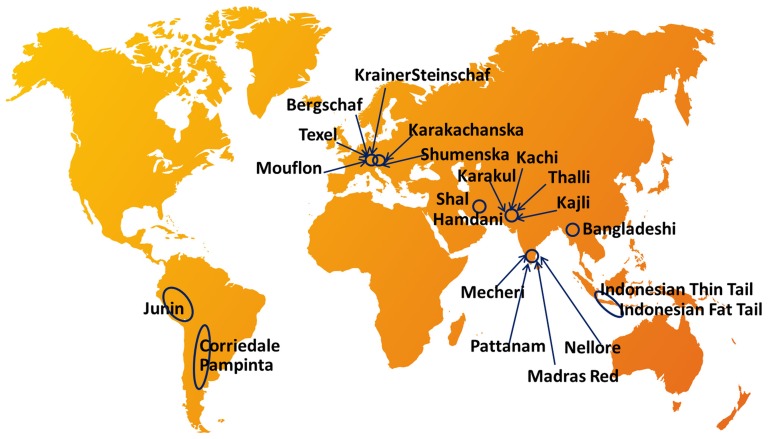
A total of 713 unrelated sheep from 22 different breeds/populations were utilized for diversity analysis in the present study. The sheep breeds/populations were distributed in three macro-environmental geographical locations including Asia (tropics), Europe (temperate) and South America (tropics) with the assumption that level of parasite load and infections vary significantly across these regions [27], [28]. The number of samples included for analysis from different breeds were as follows: Corriedale (102), Pampinta (34), Krainersteinschaf (42), Texel (21), Bergschaf (17), Mouflon (5), Karakachanska (20), Shumenska (14), Bangladeshi (17), Madras Red (60), Mecheri (64), Pattanam (54), Nellore (52), Indonesian Fat Tail (17), Indonesian Thin Tail (19), Shal (22), Hamdani (46), Thalli (17), Kachi (15), Karakul (17), Kajli (13) and Junin (45). The location of each of the sheep breeds/populations under study are presented in Figure 1. Artificial challenge experiments were carried out in two sheep breeds each from Argentina (Pampinta and Corriedale) and Indonesia (Indonesian Fat Tailed and Indonesian Thin Tailed) to generate phenotypes and DNA samples from these experimental animals were utilized for association study. Under artificial challenge trial, animals were dewormed before challenging with infective L3 larvae. Four to six weeks after deworming, blood samples were collected for DNA extraction and estimation of anemic parameters and experimental animals were challenged with a dose of 5000 infective L3 larvae cultured under in vitro conditions. All animals were maintained together during the entire trial in dry lot or under conditions of minimum additional parasite challenge. Body weight (BW), fecal egg count (FEC) and packed cell volume (PCV) were recorded at 0, 28, 35 and 42 days after artificial infection. 136 animals from Corriedale (66), Pampinta (34), Indonesian Fat Tail (17) and Indonesian Thin Tail breeds of sheep (19) were utilized for artificial challenge at experimental stations located in Argentina and Indonesia respectively.
Figure 1. Location of sheep breeds sampled for the present study.
Targeted re-sequencing, SNP identification and Genotyping
A total of 39 candidate genes were identified for targeted re-sequencing and to detect SNPs. The candidate genes were selected based on analysis of global list of sheep Entrez Gene IDs in bovine KEGG database using KEGGARRAY to identify candidate genes involved in pathways related to immune system. Oligonucleotide primers were designed for PCR amplification of partial regions of genes in a panel of eight or 16 unrelated animals from different breeds located in major geographical regions under study. Sequences generated from both ends were edited using Codon Code Aligner version 3.7.1 and secondary peaks were called to ascertain SNPs. The forward and reverse sequences were assembled to generate contigs using BioEdit version 7.1.3 (http://www.mbio. ncsu.edu/bioedit/bioedit.html). 44 novel SNPs were identified within the candidate genes under study for which competitive allele specific PCR (KASPar) assay based on FRET chemistry were developed for genotyping (KBiosciences, LGC Genomics, UK). Briefly, two forward primers one specific to each allele were designed with the respective proprietary tail sequence complementing the FAM or HEX fluorescence reporting system. A common reverse primer was designed for each genotyping assay. Thermal cycling parameters and recycling conditions were followed as per manufacturer's recommendations and are available on request. Endpoint allele discrimination module incorporated within the BioRad CFX96 (BioRad, USA) was utilized for calling the genotypes based on fluorescent intensity recorded for each of the two alleles. The emission data of all the samples in the plate were plotted in X and Y axis respectively for each allele and the genotypes were called based on distinct clustering. Quality of allele calling was confirmed by comparing the genotypes derived from KASPar assay with the available sequence data on individuals from the panel of unrelated animals. 38 out of 44 assays passed quality control and were subsequently utilized for genotyping large number of animals. Additionally, ten toll like receptor (TLR) genes were selected for in silico mining of SNP variations from sequences available at NCBI-GenBank database. A total of 14 non-synonymous SNPs within coding DNA regions of TLR genes were identified for development of genotyping assays (Details of SNPs identified and reference sequences used from NCBI-GenBank are given in Table S1). However, only three of these SNPs (within TLR5, TLR7 and TLR8 genes) were found to be polymorphic, while the remaining 11 were monomorphic in the populations under present study. A total of 41 SNPs were finally utilized for diversity analysis and association with parasite resistance characteristics.
Statistical Analysis
Basic diversity indices like allele frequency, genotype frequency, expected heterozygosity and test for Hardy Weinberg equilibrium were calculated using PEAS (Package for Elementary Analysis of SNP data) [29]. Allele sharing genetic distances (based on identical by state (IBS)) among pairs of individuals within and across different sheep breeds/populations were estimated using PEAS. Pair-wise allele sharing distance across different populations was utilized to construct the radial tree following UPGMA algorithm using PHYLIP version 3.5 [30]. Global F-statistics and pair-wise FST among different sheep breeds were computed using FSTAT software [31]. To investigate the sub-population structure of sheep breeds, pair-wise FST values among different sheep breeds were utilized to perform principal component analysis (PCA) using SPSS version 13.0. The first three principal components were used to draw the scattergram so as to understand underlying genetic structure and relationship of different breeds in three dimensional geometric space. The extent of population sub-structure was further explored using STRUCTURE with the assumption of different clusters, K = 1–15, 20, 25 and 30. Five replicate runs were performed for each K under admixture model without a priori population information. The number of burn in periods and MCMC repeats used for all the runs were 50000 and 100000 respectively. To identify the optimal ‘K’, the second order rate of change of L(K) with respect to ‘K’ was calculated by following the procedure reported elsewhere [32]. The results of STRUCTURE analysis were visualized using DISTRUCT [33].
Thirteen SNP loci within candidate genes involved in different immune related KEGG pathways and located in chromosome 3 were used to reconstruct haplotypes from unphased genotypic data. Reconstruction of haplotypes and estimation of haplotype frequencies were performed using PHASE for Windows, version 2.1 (www.stat.washington.edu/stephens) [34], [35]. The haplotype diversity and tests for departure from neutrality, Tajima's D, Fu and Li's D and Fu and Li's F were computed using DnaSP, version 4.10 [36]. The phased haplotypes were also utilized to perform analysis of molecular variance (AMOVA) and generate pair-wise FST values using ARLEQUIN version 3.1 [37]. Reduced Median network of haplotypes was constructed using NETWORK 4.5.1.2 with reduction threshold (r) of 10.0 [38].
A complete fixed effect model was employed for association of different genotypes at each SNP locus with fecal egg count (FEC), body weight change (BWC) and packed cell volume change (PCVC) measured 42 days post infective L3 larvae challenge. The data on FEC were subjected to log transformation before applying the program of least squares, LSMLMW [39]. The model included location of experimental stations and genotypes as fixed effects along with linear regression of breed effect on fecal egg count.
Results and Discussion
Immune pathway genes and SNP Discovery
A total of 243 sequences were generated by targeted re-sequencing of selected candidate genes (accession numbers are presented in Table 1) to identify 41 novel SNPs. Various details of SNPs including SNP ID, candidate gene name, chromosome location, genomic location, functional domain of the gene, alleles at each locus, SNP type, and strand genotyped are presented in Table 1. Among 41 SNPs identified, 27 were located in chromosome 3, the chromosome with maximum number of QTLs related to parasite resistance characteristics in sheep. Out of the remaining 14 SNPs, three were located in each of chromosomes 1 and 12, two in each of chromosomes 11, 16 and 27 and one in chromosome 8 and 13 respectively. The candidate genes selected for the study are involved in at least 18 KEGG pathways related to immune system (Table 2). The number of genes within each of these pathways varied from one to 14. JAK-STAT signaling pathway consisted a maximum of 14 genes followed by cytokine-cytokine receptor interaction and PIK3-AKT signaling pathways with eight genes each. The other major signaling pathways were Toll like receptor signaling pathway (5 genes) and chemokine signaling pathway (5 genes) followed by T cell receptor signaling pathway (4 genes). In a recent study, analysis of QTL and gene expression datasets following systems genetic approach revealed 14 KEGG pathways to be significant for parasite resistance in ruminants [21]. More than 50% of these immune related pathways have been included for candidate gene analysis in the present study. Altogether, 27 SNPs out of 41 SNP loci were within the candidate genes involved in immune pathways including 13 SNPs in chromosome 3. The location of SNPs in different functional domains of candidate genes varied considerably with 16 in 3′untranslated regions, 14 in exonic regions, 10 in intronic regions and one in 5′ flanking region upstream to start codon. Among the 14 SNPs located within exonic regions, eight were found to be non-synonymous mutations resulting in change of amino acid sequences while the remaining six were synonymous mutations.
Table 1. Details of candidate genes, chromosome location, genomic location and strand genotyped at 41 SNP loci under study.
| SNPID | Accessions Nos. | Gene Name | Alleles | Chr | Position | Strand | Domain | Synon/Non-Synon |
| PIK3R3_498 | KC734640-646 | Phosphoinositide-3-kinase, regulatory subunit 3 (gamma) | T/G | 1 | 20560912 | + | Intron | - |
| LEPR_260 | KC734602-617 | Leptin receptor | G/A | 1 | 42230500 | + | Intron | - |
| IL6R_227 | KC734807-813 | Interleukin 6 receptor | T/C | 1 | 110974347 | − | 3′UTR | - |
| ZDHHC17_190 | KC734734-740 | Zinc finger, DHHC-type containing 17 | G/A | 3 | 120038167 | + | 3′UTR | - |
| NAV3_591 | KC734618-624 | Neuron navigator 3 | G/C | 3 | 121343328 | − | Exon | Synonymous |
| ACVRL1_445 | KC734741 | Activin A receptor type II-like 1 | C/A | 3 | 134236531 | + | 3′UTR | - |
| FGD6_519 | KC734775-779 | FYVE, RhoGEF and PH domain containing 6 | G/A | 3 | 140073272 | − | Exon | G allele-D A allele-N |
| USP44_252 | KC734721-727 | Ubiquitin specific peptidase 44 | T/C | 3 | 140369758 | + | Exon | C allele-G T allele-S |
| ITGA5_111 | KC734594 | Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | G/C | 3 | 141110793 | + | Intron | - |
| GPR84_520 | KC734787 | G-protein coupled receptor 84 | G/A | 3 | 141144005 | + | 5′ flanking region | - |
| TARBP2_97 | KC734709-713 | TAR (HIV-1) RNA binding protein 2 | T/G | 3 | 142063154 | + | 3′UTR | - |
| ITGB7_538 | KC734595-601 | Integrin, beta 7 | G/A | 3 | 142354366 | + | Intron | - |
| CSRNP2_65 | KC734757-761 | Cysteine-serine-rich nuclear protein 2 (or) family with sequence similarity 130, member A1 | T/C | 3 | 144244455 | − | Exon | Synonymous |
| SLC11A2_174 | KC734676-682 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | T/C | 3 | 144430523 | + | 3′UTR | - |
| PTPRB_141 | KC734669-675 | Protein tyrosine phosphatase, receptor type, B | G/A | 3 | 158919598 | + | Intron | - |
| STAT2_486 | KC734684-688 | Signal transducer and Activator of Transcription 2 | G/C | 3 | 162936891 | + | 3′UTR | - |
| DDIT3_527 | KC734762-767 | DNA-damage-inducible transcript 3 | G/A | 3 | 173067524 | + | 3′UTR | - |
| GLI1_253 | KC734780-786 | GLI family zinc finger 1 | T/C | 3 | 173101195 | + | Exon | Synonymous |
| GLI1_576 | KC734780-786 | GLI family zinc finger 1 | T/G | 3 | 173101518 | + | Exon | T allele-N G allele-H |
| ZBTB39_51 | KC734728-733 | Zinc finger and BTB domain containing 39 | G/A | 3 | 173591120 | + | Exon | Synonymous |
| ANKRD52_113 | KC734742-746 | Ankyrin repeat domain 52 | G/A | 3 | 174303658 | + | Intron | - |
| ESYT1_157 | KC734769-774 | Extended synaptotagmin-like protein 1 (or) family with sequence similarity 62 (C2 domain containing), member A | T/C | 3 | 174409595 | + | Intron | - |
| TIMP3_716 | KC734714-720 | TIMP metallopeptidase inhibitor 3 | G/A | 3 | 189483881 | + | 3′UTR | - |
| CSF2RB_279 | KC734752-756 | Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | T/G | 3 | 193964280 | + | Exon | Synonymous |
| CSF2RB_557 | KC734752-756 | Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | T/C | 3 | 193964558 | + | Exon | C allele-P T allele-L |
| IL2RB_180 | KC734793-806 | Interleukin 2 receptor, beta | T/C | 3 | 194260690 | - | Exon | Synonymous |
| EM4b_574 | KC734768 | EM4b protein | C/T | 3 | 206971007 | + | Intron | - |
| CLEC1A_134 | KC734747-751 | C-type lectin domain family 1, member A | T/C | 3 | 220471614 | + | Intron | - |
| PTPN6_398 | KC734663-668 | Protein tyrosine phosphatase, non-receptor type 6 | G/A | 3 | 224998785 | + | Intron | - |
| SMCR7L_517 | KC734683 | Smith-Magenis syndrome chromosome region, candidate 7-like | G/A | 3 | 233018827 | − | 3′UTR | - |
| IL20RA_422 | KC734830-836 | Interleukin 20 receptor alpha | G/A | 8 | 66786499 | − | Exon | G allele-G A allele-E |
| STAT5B_385 | KC734694-708 | Signal transducer and activator of transcription 5B | G/A | 11 | 41843264 | + | 3′UTR | - |
| STAT3_138 | KC734689-693 | Signal transducer and activator of transcription 3 (acute-phase response factor) | G/A | 11 | 44531311 | + | 3′UTR | - |
| IL10_82 | KC734814-829 | Interleukin 10 | C/T | 12 | 1856307 | + | 3′UTR | - |
| TLR5_2276 | - | Toll like receptor 5 | C/G | 12 | 24704711 | + | Exon | G allele-S C allele-T |
| PIK3CD_443 | KC734625-639 | Phosphoinositide-3-kinase, catalytic, delta polypeptide | G/A | 12 | 46696545 | − | 3′UTR | - |
| IL2RA_388 | KC734788-792 | Interleukin 2 receptor, alpha | T/C | 13 | 18432828 | − | 3′UTR | - |
| PRLR_341 | KC734647-662 | Prolactin receptor | C/T | 16 | 42399902 | + | 3′UTR | - |
| PRLR_729 | KC734647-662 | Prolactin receptor | C/T | 16 | 42400290 | + | 3′UTR | - |
| TLR7_2491 | - | Toll like receptor 7 | G/A | 27 | 10360855 | + | Exon | A allele-K G allele-R |
| TLR8_1045 | - | Toll like receptor 8 | T/C | 27 | 10398449 | + | Exon | T allele-Y C allele-H |
Table 2. Details of candidate genes under study and involved in different KEGG immune pathways.
| KEGG Path ID | KEGG Pathway | No. Genes | Candidate Genes |
| bta04630 | Jak-STAT signaling pathway | 14 | CSF2RB,IL10,IL20RA,IL2RA,IL2RB,IL6R,LEPR,PIK3CD,PIK3R3,PRLR,PTPN6,STAT2,STAT3,STAT5B |
| bta04151 | PI3K-Akt signaling pathway | 8 | IL2RA,IL2RB,IL6R,ITGA5,ITGB7,PIK3CD,PIK3R3,PRLR |
| bta04060 | Cytokine-cytokine receptor interaction | 8 | CSF2RB,IL10,IL20RA,IL2RA,IL2RB,IL6R,LEPR, PRLR |
| bta04620 | Toll-like receptor signaling pathway | 5 | PIK3CD,PIK3R3,TLR5,TLR7,TLR8 |
| bta04062 | Chemokine signaling pathway | 5 | PIK3CD,PIK3R3,STAT2,STAT3,STAT5B |
| bta04660 | T cell receptor signaling pathway | 4 | IL10,PIK3CD,PIK3R3,PTPN6 |
| bta04640 | Hematopoietic cell lineage | 3 | IL2RA,IL6R,ITGA5 |
| bta04650 | Natural killer cell mediated cytotoxicity | 3 | PIK3CD,PIK3R3,PTPN6 |
| bta04662 | B cell receptor signaling pathway | 3 | PIK3CD,PIK3R3,PTPN6 |
| bta04145 | Phagosome | 2 | ITGA5 |
| bta04520 | Adherens junction | 2 | PTPN6,PTPRB |
| bta04664 | Fc epsilon RI signaling pathway | 2 | PIK3CD,PIK3R3 |
| bta04666 | Fc gamma R-mediated phagocytosis | 2 | PIK3CD,PIK3R3 |
| bta04670 | Leukocyte transendothelial migration | 2 | PIK3CD,PIK3R3 |
| bta04672 | Intestinal immune network for IgA production | 2 | IL10,ITGB7 |
| bta04010 | MAPK signaling pathway | 1 | DDIT3 |
| bta04142 | Lysosome | 1 | SLC11A2 |
| bta04340 | Hedgehog signaling pathway | 1 | GLI1 |
Minor allele frequency and genetic diversity within sheep breeds
The basic diversity measures estimated for different breeds under study: minor allele frequency, observed heterozygosity, expected heterozygosity and number of SNP loci deviating from Hardy-Weinberg equilibrium are presented in Table 3. The global minor allele frequency (MAF) across 41 SNP loci varied from 0.028 to 0.494 with a mean of 0.273. The power to detect genetic effect in a given study depends to a great extent on MAF of the tested alleles. Specifically, loci with a low MAF (<10%) have significantly lower power to detect weak genotype-phenotype associations than loci with a high MAF (>40%) [40], [41]. Further, previous studies have demonstrated that rare genotypes are more likely to result in spurious findings due to relatively higher standard error (within each test) and higher false discovery rate (under multiple testing procedures for many loci) [42]. In the present study, the global minor allele frequency was more than 0.10 in all but three SNP loci (FGD6_519, SMCR7L_517, TLR8_1045) thus indicating their suitability for association study. Examination of SNP loci within each breed revealed presence of both alleles in more than 90% of SNP loci, thus indicating high degree of polymorphism and possibility of these loci predating the radiation of sheep breeds under study. In addition, 49.8% of SNP loci showed MAF≥0.20 while 29.7% showed MAF≥0.30, suggesting the SNP set identified in the present study will likely have high utility for association analysis in different populations. The mean MAF within breeds varied from 0.167 (Pampinta) to 0.238 (Bangladeshi), although no significant difference in MAF was observed across different geographical regions: Asia, Europe and South America (Table 3). This is in contrast to earlier findings [43] which reported Asian and African breeds having excess of low MAF SNP (<0.10) compared to European populations. This reflects the absence of any ascertainment bias in the present study as the diversity panel was adequately represented with Asian sheep breeds for SNP discovery.
Table 3. Mean diversity indices and number of loci deviating from Hardy-Weinberg equilibrium in different sheep breeds at 41 SNP loci.
| Population | Code | Indices of Genetic Diversity | ||||
| MAF | Observed heterozygosity | Gene Diversity | No. Loci not in HWE | Mean IBS distance | ||
| South Asian populations | ||||||
| Bangladeshi | BAN | 0.238 | 0.282 | 0.314 | 8 | 0.272 |
| Mecheri | MEC | 0.194 | 0.266 | 0.262 | 6 | 0.206 |
| Madras Red | MRS | 0.202 | 0.295 | 0.282 | 4 | 0.229 |
| Nellore | NEL | 0.211 | 0.292 | 0.291 | 7 | 0.235 |
| Pattanam | PAT | 0.202 | 0.278 | 0.276 | 7 | 0.218 |
| Kachi | KAC | 0.204 | 0.234 | 0.279 | 12 | 0.253 |
| Kajli | KAJ | 0.193 | 0.278 | 0.262 | 10 | 0.221 |
| Karakul | KUL | 0.227 | 0.298 | 0.301 | 8 | 0.251 |
| Thalli | THA | 0.229 | 0.311 | 0.305 | 6 | 0.256 |
| Mean | 0.211 | 0.281 | 0.286 | 7.6 | 0.238 | |
| South East Asian Populations | ||||||
| Indonesian Fat Tailed | IFT | 0.209 | 0.262 | 0.282 | 4 | 0.250 |
| Indonesian Thin Tailed | ITT | 0.227 | 0.279 | 0.315 | 6 | 0.280 |
| Mean | 0.218 | 0.270 | 0.299 | 5.0 | 0.265 | |
| South West Asian Populations | ||||||
| Hamdani | HAM | 0.226 | 0.315 | 0.309 | 1 | 0.249 |
| Shal | SHA | 0.223 | 0.302 | 0.295 | 7 | 0.240 |
| Mean | 0.224 | 0.309 | 0.302 | 4.0 | 0.245 | |
| European Populations | ||||||
| Krainersteinschaf | KSF | 0.218 | 0.310 | 0.302 | 10 | 0.244 |
| KarakaChanska | KAR | 0.220 | 0.289 | 0.296 | 5 | 0.251 |
| Shumenska | SHU | 0.222 | 0.312 | 0.300 | 6 | 0.251 |
| Bergschaf | BER | 0.219 | 0.309 | 0.291 | 4 | 0.237 |
| Texel | TEX | 0.194 | 0.260 | 0.260 | 12 | 0.217 |
| Mean | 0.215 | 0.296 | 0.290 | 7.4 | 0.240 | |
| South American Populations | ||||||
| Junin | JUN | 0.218 | 0.299 | 0.304 | 4 | 0.250 |
| Pampinta | PAM | 0.167 | 0.230 | 0.237 | 7 | 0.197 |
| Corriedale | COR | 0.218 | 0.299 | 0.304 | 8 | 0.249 |
| Mean | 0.201 | 0.276 | 0.281 | 6.3 | 0.232 | |
The mean global observed and expected heterozygosities were 0.287 and 0.366 respectively. The mean observed heterozygosity within breeds varied from 0.230 (Pampinta) to 0.315 (Hamdani) while the mean expected heterozyosity varied from 0.237 (Pampinta) to 0.315 (Indonesian Thin Tail). Among different geographical regions, mean observed heterozygosity was highest in South West Asian sheep populations (0.309) followed by European populations [0.296] while South East Asian populations had the least mean observed heterozygosity (0.270). This is consistent with the fact that the diversity remains higher around the centre of domestication while decreasing with increasing geographic distance [44]. Among the European sheep breeds, Texel, the northern Europe originated sheep breed was having the lowest mean observed heterozygosity (0.260) as compared to other South or South Eastern European breeds [45]. Further, the overall mean observed heterozygosity of South West Asian and European sheep populations was found to be higher than gene diversity, although similar case was observed with respect to most South Asian sheep populations except Bangladeshi, Kachi and Karakul. The test for HWE showed significant deviations with a mean number of loci 7.6, 5, 4, 7.4 and 6.3 in South Asian, South East Asian, South West Asian, European and South American sheep populations. Among all the sheep populations, Hamdani was found to be in equilibrium at all the SNP loci except one further reiterating its high degree of genetic diversity.
Genetic distance within and between sheep breeds
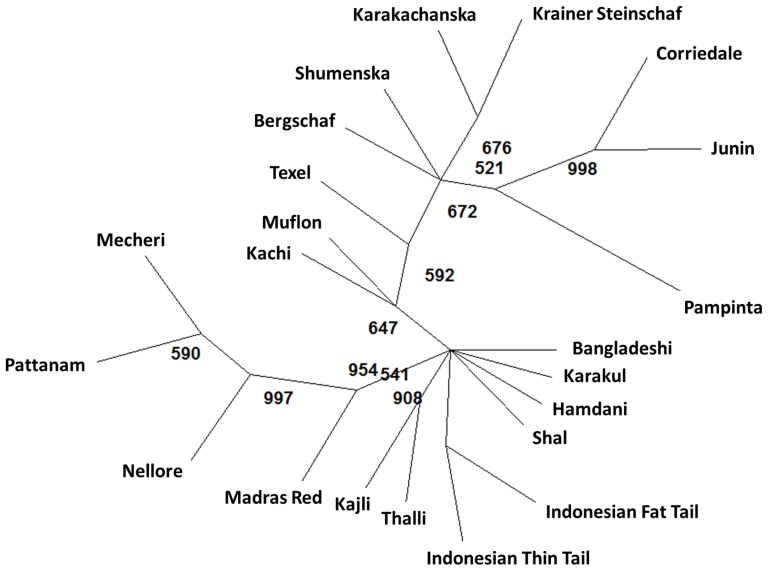
Allele sharing distance was calculated for all pair-wise combinations of individuals both within and across populations by subtracting average proportion of alleles shared from one [46]. The mean inter-individual allele sharing distance of all pair-wise combinations within breeds was 0.236 (SD = 0.06; n = 16907), while it ranged from 0.197 (Pampinta) to 0.280 (Indonesian Thin Tail). Across different geographical regions, the mean distance within breeds was lowest in South American populations (0.232) while it was highest in South East Asian populations (0.265). The mean distance between individuals derived from different breeds was 0.325 (SD = 0.07; n = 236921). Although the observed values were found to be higher than that reported for cattle, they were lower compared to the previous report in sheep [47]. The distribution of inter-individual distance values was found to be normal both within and across breeds (Figure S2). There was considerable overlap between inter-individual distances within and across breeds with almost equal proportion of pairwise combinations around the range of 0.28 to 0.30. This indicates that some individuals were found to be more closely related to individuals from other breed than from members of the same breed. To further investigate the genetic differentiation among different sheep breeds, pairwise allele sharing distance pair-wise FST (Table S2) and global F-statistics (Table S3) were estimated. The global FIT, FST and FIS were 0.227, 0.213 and 0.018 respectively while pairwise FST values ranged from 0.017 (Pattanam/Nellore) to 0.469 (Pampinta/Mouflon). 21.3% of total genetic variation was found to be due to between breed differences while 77.3% was due to within breed differences. The values are much higher than that reported for European sheep (13.1%) [45], Indian sheep (11.1%) [48] and European and South West Asian sheep (5.7%) [44] using microsatellite markers. The higher FST values observed could be understood from the fact that the samples were derived from wide geographic locations in the present study (Asia, Europe and South America). Further, phylogenetic analysis of pair-wise allele sharing distance revealed two major clusters with most of South Asian, South East Asian and South West Asian breeds clustering together while the European and South American sheep breeds clustered together separately (Figure 2). However, the three South American sheep breeds formed a sub-cluster together and interestingly found to be more closely related to Southern Europe sheep breeds than North European sheep. Similarly Karakul and Bangladeshi breeds were found to be clustering together with South West Asian sheep than with other South Asian breeds.
Figure 2. UPGMA radial tree constructed using pair-wise allele sharing distance among different sheep breeds.
(Numbers at nodes indicate bootstrap values out of 10000 resampled datasets).
Genetic structure of sheep breeds
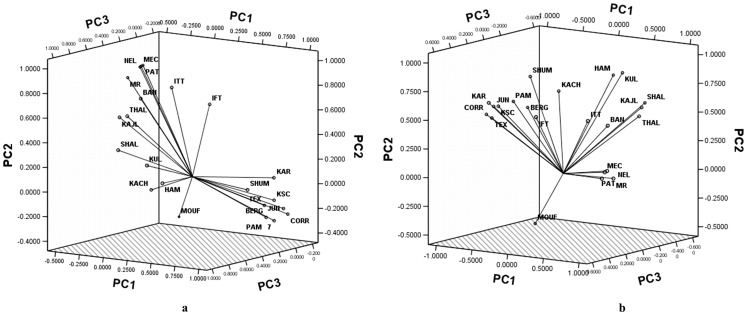
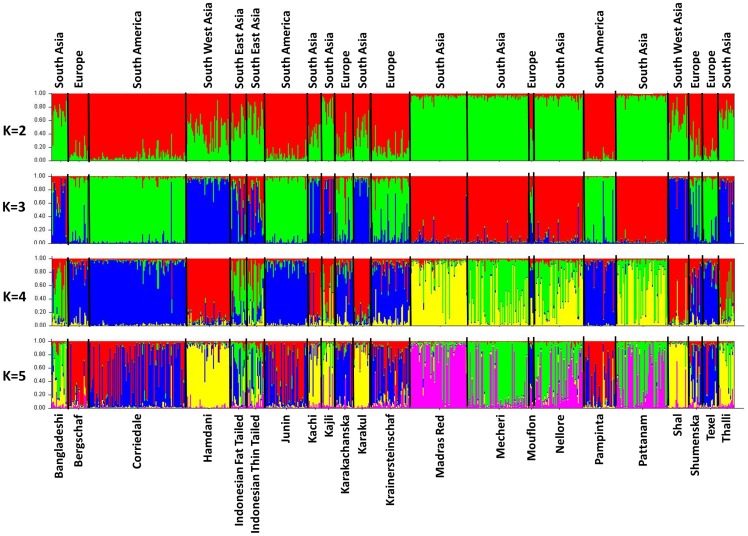
The pairwise FST were subjected to principal components analysis and the first three principal components (PCs) were plotted on a three dimensional scattergram to evaluate the genetic structure of sheep breeds (Figure 3a). The first, second and third principal components explained 42.01%, 38.12% and 7.15% of total genetic variation respectively. The clustering of sheep breeds followed their geographical origin and broadly differentiated into European and Asian groups. European Mouflon, which is more feral in nature was distinct from both these groups. However, analysis of a subset of SNP data at 27 loci located within immune pathway genes resulted in an additional distinct group with all the four South Indian sheep breeds clustering together closely (Figure 3b). In order to further understand the phylogeographic structure, analysis of molecular variance was performed to assess the SNP variation as a function of both breed membership and geographic origin (Table 4). Two types of groupings were assumed; the first grouping was with three major geographical groups Asia, Europe and South America; the second one with five geographical groups South Asia, South East Asia, South West Asia, Europe and South America. With grouping I, 14.16% of variation was due to differences in geographical groups and 11.54% due to between breed differences. In case of grouping II, among group variation marginally increased to 14.64% while between breed differences decreased to 9.51%. Further, with the analysis of subset of immune pathway SNP data at 27 loci, among group variation increased to 15.92% with grouping II showing a strong phylogeographic structure (Table 3). This is in contrast to earlier reports based on microsatellite genotypes [45], SNP genotypes [47] and mitochondrial DNA haplotypes [49], where much less variation was explained by grouping breeds into geographical regions. Similarly weak phylogeographic structure had been reported in domestic goats also [50]. All these studies concluded that the weak phylogeographic structure exhibited by domestic sheep and goat might be due to their small size and versatility enabling transportation and subsequent introgression in concert with human migration [47], [51]. The relatively strong phylogeographic structure observed in the present study is interesting from the fact that most of these SNP loci are within candidate genes involved in different immune pathways. The geographical locations of sampled individuals vary widely with respect to diversity and load of pathogens resulting in differences in the magnitude of natural selection pressure across these regions [27], [28]. Consequently, evolution of genes involved in immune system may either be highly optimized by natural selection process (purifying selection) or continue to evolve under low selection pressure (balancing selection) [52]. The genetic structure of sheep populations exhibited by a set of SNPs located in immune pathway genes could thus be different from those revealed by microsatellite or mitochondrial or genome wide SNP variations. In order to further clarify the breed demography and selection history, all the 41 SNP loci were subjected to Ewens-Watterson neutrality test to investigate whether the loci were influenced by selective forces within various sheep breeds/populations. 18 out of 41 SNP loci were found to deviate from selection neutrality in at least one of the breeds under study while the remaining 23 SNP loci were found to be selectively neutral (Table S4). The two subsets of data (23 neutral SNP loci and 18 non-neutral loci) were subjected to analysis of molecular variance. Among group variance at non-neutral SNP loci was found to be significant and higher (17.13%) than variance among populations within group as compared to selectively neutral SNP loci (11.01%) indicating the basis for strong phylogeographic structure observed in the present study (Table S5). Bayesian clustering was performed without prior population information using STRUCTURE program, and the second order rate of change of average likelihood at each K was calculated (K = 1–15, 20, 25, 30). ΔK reached its peak at K = 5, suggesting optimal K value appropriate for the dataset. When K = 2 was assumed, most of the individuals from Europe and South America were assigned to cluster 1, while all the four Indian breeds were assigned to cluster 2 (Figure 4). Individuals belonging to other breeds from South Asia (Kachi, Kajli, Karakul and Thalli), South West Asia (Hamdani, Shal) and South East Asia (Indonesian Fat Tail and Indonesian Thin Tail) were admixed and observed to be assigned in both the clusters. When K = 3 was assumed, European and South American sheep were mostly assigned to cluster 1, Indian sheep to cluster 2 and the South West Asian sheep along with Karakul to cluster 3. Indonesian sheep and other South Asian sheep were found to be admixed between cluster 2 and 3. With K = 4, the Indian sheep population got subdivided into two clusters while with K = 5, the subdivision of European cluster was evident. Further, Bayesian analysis was also performed with the subsets of genotypes at non-neutral and neutral SNP loci (Figure S3a and S3b respectively). The results revealed relatively better and more precise geographical clustering of animals with the non-neutral subset than the neutral subset of genotype data.
Figure 3. Three dimensional scattergram of first three principal components derived from pairwise FST (a) across 41 SNP loci and (b) across 27 SNP loci among different sheep populations.
(BAN-Bangladeshi; COR-Corriedale; PAM-Pampinta; JUN-Junin; BER-Bergschaf; TEX-Texel; KSF-Krainersteinschaf; MUF-Mouflon; KAR-Karakachanska; SHU-Shumenska; KUL-Karakul; THA-Thalli; KAC-Kachi; KAJ-Kajli; HAM-Hamdani; SHA-Shal; PAT-Pattanam; NEL-Nellore; MRS-Madras Red; MEC-Mecheri; IFT-Indonesian Fat Tail; ITT-Indonesian Thin Tail).
Table 4. Analysis of molecular variance among different sheep breeds based on (right) genotypes at 41SNP loci and (left) haplotypes reconstructed from genotypes at 13 immune SNP loci located in chromosome 3.
| Source of variation | Based on genotype data at 41 SNP loci | Based on haplotypic data from 13 SNP loci | ||||||||||
| d.f. | Sum of squares | Variance components | % of variation | P- value | d.f. | Sum of squares | Variance components | % of variation | P-value | |||
| No Groupings | ||||||||||||
| Among populations | 21 | 2205.51 | 1.57 | Va | 21.33 | 0.000 | 21 | 767.50 | 0.55 | Va | 22.22 | 0.000 |
| Within populations | 1404 | 8118.99 | 5.78 | Vb | 78.67 | 0.000 | 1404 | 2689.14 | 1.92 | Vb | 77.78 | 0.000 |
| Grouping-I (Asia, Europe, South America) * | ||||||||||||
| Among groups | 2 | 1075.13 | 1.10 | Va | 14.16 | 0.000 | 2 | 349.20 | 0.34 | Va | 13.27 | 0.000 |
| Among populations within groups | 19 | 1130.38 | 0.90 | Vb | 11.54 | 0.000 | 19 | 418.30 | 0.34 | Vb | 12.94 | 0.000 |
| Within populations | 1404 | 8118.99 | 5.78 | Vc | 74.30 | 0.000 | 1404 | 2689.14 | 1.92 | Vc | 73.80 | 0.000 |
| Grouping-II (South Asia, South East Asia, South West Asian, Europe, South America) ** | ||||||||||||
| Among groups | 4 | 1370.78 | 1.16 | Va | 14.64 | 0.000 | 4 | 493.56 | 0.41 | Va | 15.92 | 0.000 |
| Among populations within groups | 17 | 834.73 | 0.72 | Vb | 9.51 | 0.000 | 17 | 273.94 | 0.24 | Vb | 9.27 | 0.000 |
| Within populations | 1404 | 8118.99 | 5.78 | Vc | 75.86 | 0.000 | 1404 | 2689.14 | 1.92 | Vc | 74.81 | 0.000 |
Grouping-I: Asia – Bangladeshi, Madras Red, Mecheri, Pattanam, Nellore, Indonesian Fat Tailed, Indonesian Thin Tailed, Shal, Hamdani, Thalli, Kachi, Karakul, Kajli; Europe – Krainersteinschaf, Texel, Bergschaf, Mouflon, Karakachanska, Shumenska; South America – Junin, Pampinta, Corriedale.
Grouping-II: South Asia - Bangladeshi, Madras Red, Mecheri, Pattanam, Nellore, Thalli, Kachi, Karakul, Kajli; South East Asia - Indonesian Fat Tailed, Indonesian Thin Tailed; South West Asia – Hamdani, Shal; Europe – Krainersteinschaf, Texel, Bergschaf, Mouflon, Karakachanska, Shumenska; South America – Junin, Pampinta, Corriedale.
Figure 4. Bayesian clustering of 713 sheep under assumption of 2 to 6 clusters without a priori population information.
The breed names are given below the box plot and the geographical origin indicated above the box plot with the individuals of different breeds separated by vertical black lines.
Haplotype reconstruction and test for neutrality
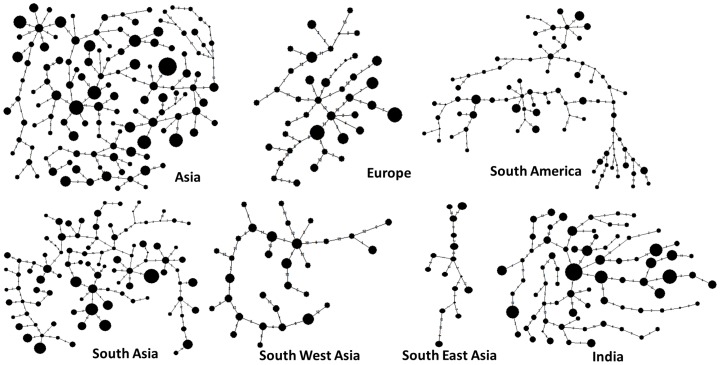
To investigate the influence of natural selection processes, unphased diploid genotypes at SNP loci located in chromosome 3 were utilized to reconstruct the haplotypes. Out of 27 SNP loci located in chromosome 3, 13 loci within immune pathway genes were used for haplotype phasing. Data on all 713 animals were utilized to reconstruct a total of 1426 haplotypes, of which 389 were found to be singletons. Predicted haplotype phases with best pair probabilities for each individual were retained for further analysis. Table 5 provides the results of tests for selective neutrality using three different statistics: Tajima's D, Fu and Li's D and Fu and Li's F. Significant deviations were found in Corriedale, Junin, Krainer Steinschaf, Texel and Nellore sheep breeds. All the three statistics showed significant deviation from neutrality in Junin breed, while two statistics in each of corriedale and Texel sheep breeds and one statistics in each of Krainersteinschaf and Nellore sheep breeds showed significant deviations from selective neutrality. When tested at regional level, Tajima's D and Fu and Li's F statistics revealed significant deviations in European, South American and South Asian populations. Similarly, the Indian sheep which clustered distinctly when analyzed at immune SNP loci, showed significant deviation from neutrality under Tajima's D and Fu and Li's F tests. However, Fu and Li's D statistic did not detect any significant deviation from neutrality in all these populations. All the test statistics that showed significant deviation both at breed and regional levels were found to be positive indicating balancing selection in force, probably with low selection pressure. In order to further examine this process, haplotype networks were constructed for each of the geographical regions under study. Star contraction is expected under strong purifying selection while few haplotypes with moderate frequencies and short branches are expected under a weak purifying selection. On the contrary, balancing selection is expected to retain multiple lineages with high and low frequency clusters and long branches [53]. Reduced Median (RM) networks were constructed for haplotypes derived from populations in different geographical regions (Figure 5). All the populations showed multiple lineages with several nodes of different sizes and long branches, thus confirming balancing selection in force as revealed by different tests for neutrality. This is further evident from the fact that many breeds under study were found to have either heterozygosity excess or near equal observed and expected heterozygosities. Although little information is available in sheep on immune gene polymorphisms across distinct geographical regions, few reports are available in human and cattle. In a study on innate immune genes including TLRs and defensins in Indian, European-American and African-American human populations, strong purifying selection was found to operate resulting in conservation of recognition motifs across a broad range of pathogens [52]. Similar observation was found with respect to TLR10 gene in a study on Bos taurus and Bos indicus cattle [54]. However, in case of adaptive immune genes like major histocompatibility complex, balancing selection with high genetic diversity and heterozygote advantage was found to be common [55], [56]. To the best of our knowledge, the present study is the first to report balancing selection forces operating in immune pathway genes of sheep. However, it needs to be noted that there may be local variations in the nature of selection as it could be modulated by local differences in pathogen diversity and load [52].
Table 5. Values of haplotype diversity and statistics for testing departures from neutrality (Haplotypes reconstructed from 13 SNPs in chromosome 3-R Method).
| Population/Breed | N | No. Haps | Haplotype diversity | Tajima's D | Fu and Li's D* | Fu and Li's F* | ||||
| Estimate | Variance | Value | P | Value | P | Value | P | |||
| Bangladeshi | 34 | 31 | 0.995 | 0.00007 | 1.82 | P>0.05 | 0.92 | P>0.10 | 1.41 | P>0.10 |
| Bergschaf | 42 | 26 | 0.954 | 0.00043 | 0.87 | P>0.10 | 1.02 | P>0.10 | 1.15 | P>0.10 |
| Corriedale | 204 | 131 | 0.992 | 0.00000 | 2.75 | P<0.05 | 1.48 | P>0.05 | 2.34 | P<0.02 |
| Hamdani | 92 | 45 | 0.971 | 0.00004 | 0.50 | P>0.10 | 0.93 | P>0.10 | 0.92 | P>0.10 |
| Indonesian Fat Tailed | 34 | 24 | 0.971 | 0.00022 | 0.73 | P>0.10 | 0.92 | P>0.10 | 1.01 | P>0.10 |
| Indoesian Thin Tailed | 38 | 31 | 0.990 | 0.00007 | 1.30 | P>0.10 | 0.97 | P>0.10 | 1.27 | P>0.10 |
| Junin | 90 | 68 | 0.991 | 0.00002 | 2.72 | P<0.01 | 1.51 | P<0.05 | 2.31 | P<0.02 |
| Kachi | 30 | 18 | 0.940 | 0.00072 | 0.66 | P>0.10 | 1.00 | P>0.10 | 1.05 | P>0.10 |
| Kajli | 26 | 22 | 0.985 | 0.00026 | 1.13 | P>0.10 | 0.48 | P>0.10 | 0.79 | P>0.10 |
| Karakachanska | 40 | 35 | 0.991 | 0.00007 | 1.52 | P>0.10 | 1.03 | P>0.10 | 1.40 | P>0.10 |
| Karakul | 34 | 25 | 0.979 | 0.00016 | 0.78 | P>0.10 | 1.04 | P>0.10 | 1.13 | P>0.10 |
| Krainer Steinschaf | 84 | 53 | 0.972 | 0.00010 | 1.27 | P>0.10 | 1.51 | P<0.05 | 1.70 | P>0.05 |
| Madras Red | 120 | 63 | 0.981 | 0.00001 | 1.56 | P>0.10 | 0.81 | P>0.10 | 1.29 | P>0.10 |
| Mecheri | 128 | 49 | 0.964 | 0.00005 | 0.92 | P>0.10 | 0.80 | P>0.10 | 1.01 | P>0.10 |
| Mouflon | 10 | 6 | 0.844 | 0.01060 | 0.17 | P>0.10 | 0.64 | P>0.10 | 0.59 | P>0.10 |
| Nellore | 104 | 64 | 0.987 | 0.00001 | 1.95 | P>0.05 | 1.42 | P>0.05 | 1.91 | P<0.05 |
| Pampinta | 68 | 48 | 0.986 | 0.00003 | 1.02 | P>0.10 | 0.41 | P>0.10 | 0.74 | P>0.10 |
| Pattanam | 108 | 51 | 0.973 | 0.00003 | 1.49 | P>0.10 | 0.75 | P>0.10 | 1.20 | P>0.10 |
| Shal | 44 | 24 | 0.967 | 0.00013 | 1.15 | P>0.10 | 0.33 | P>0.10 | 0.70 | P>0.10 |
| Shumenska | 28 | 24 | 0.989 | 0.00015 | 1.08 | P>0.10 | 1.01 | P>0.10 | 1.21 | P>0.10 |
| Texel | 34 | 26 | 0.964 | 0.00043 | 1.35 | P>0.10 | 1.44 | P<0.05 | 1.66 | P<0.05 |
| Thalli | 34 | 29 | 0.988 | 0.00013 | 1.32 | P>0.10 | 0.92 | P>0.10 | 1.23 | P>0.10 |
| Europe | 238 | 139 | 0.989 | 0.00000 | 2.66 | P<0.05 | 1.47 | P>0.05 | 2.31 | P<0.02 |
| South America | 362 | 193 | 0.993 | 0.00000 | 3.10 | P<0.01 | 1.44 | P>0.05 | 2.51 | P<0.02 |
| South East Asia | 72 | 51 | 0.987 | 0.00003 | 1.55 | P>0.10 | 1.48 | P>0.05 | 1.78 | P<0.05 |
| South Asia | 628 | 194 | 0.987 | 0.00000 | 2.57 | P<0.05 | 1.40 | P>0.05 | 2.25 | P<0.02 |
| South West Asia | 136 | 61 | 0.980 | 0.00001 | 1.13 | P>0.10 | 0.87 | P>0.10 | 1.15 | P>0.10 |
| India | 470 | 142 | 0.982 | 0.00000 | 2.15 | P<0.05 | 1.38 | P>0.05 | 2.02 | P<0.05 |
Figure 5. Reduced Median network of haplotypes reconstructed from 13 SNP loci related to immune genes in sheep chromosome 3.
(Asia – Bangladeshi, Madras Red, Mecheri, Pattanam, Nellore, Indonesian Fat Tailed, Indonesian Thin Tailed, Shal, Hamdani, Thalli, Kachi, Karakul, Kajli; Europe – Krainersteinschaf, Texel, Bergschaf, Mouflon, Karakachanska, Shumenska; South America – Junin, Pampinta, Corriedale; South Asia - Bangladeshi, Madras Red, Mecheri, Pattanam, Nellore, Thalli, Kachi, Karakul, Kajli; South East Asia - Indonesian Fat Tailed, Indonesian Thin Tailed; South West Asia – Hamdani, Shal; India – Madras Red, Maecheri, Nellore, Pattanam).
Association of immune pathway gene polymorphisms with fecal egg count
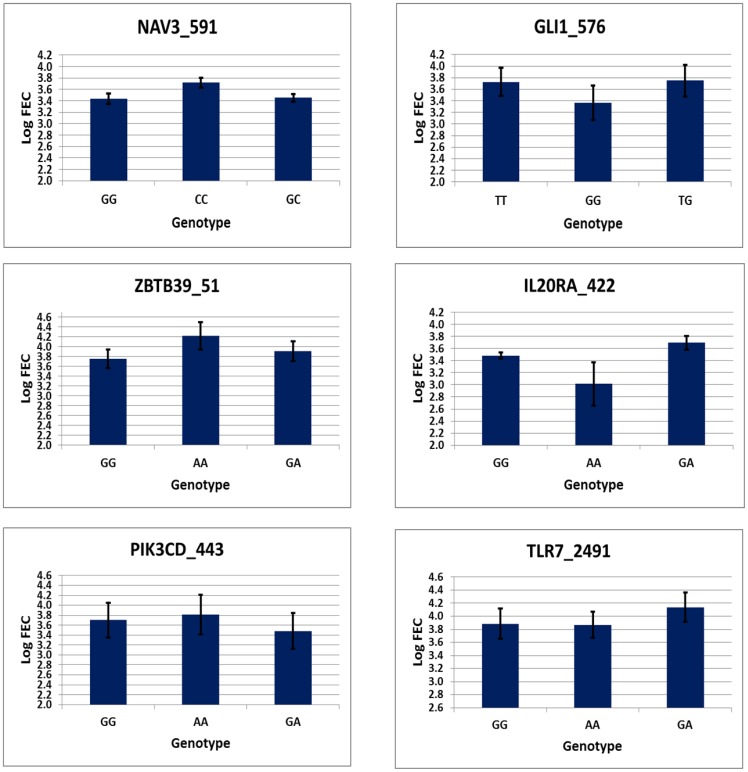
To evaluate the potential utility of the SNP loci for future association study on a large number of samples, a pilot analysis was performed with the phenotypes generated in four breeds after artificial challenge with infective L3 larvae of Haemonchus contortus. Phenotypic data (fecal egg count, body weight change and change in packed cell volume 42 days post challenge) on 136 animals was used for least squares analysis under a complete fixed effect model. The effect of location of experimental stations (Corriedale and Pampinta in South America; Indonesian Thin Tail and Indonesian Fat Tailed sheep in South East Asia) was not found to have significant influence on fecal egg count and packed cell volume while significant effect was observed with respect to body weight change (P<0.01). Although the locations are wide apart geographically, uniform protocol was followed across different experimental stations in terms of age of lambs selected for experiment, deworming and data recording schedule, however some differences did exist in terms of quality of pasture available for grazing, etc. Higher observed body weight change in animals challenged at Aguil Experimental Station (AES), Argentina was due to better growth performance of Pampinta lambs. Higher body weight achieved by Pampinta lambs were due to their genetic differences in growth rate (average pre-weaning weight gain of 295 g/day) and weight gain (average weaning weight of 33.4 kg) as compared to other breeds like Corriedale (218 g/day and 24.6 kg) [57] and Indonesian Fat Tailed sheep (47.7 g/day and 9.7 kg) [58]. The fixed effect on body weight change was adjusted before phenotype-genotype association analysis. Among different breeds, lowest mean fecal egg count (mean log FEC 3.23±0.16) and packed cell volume change (−1.57%) was observed in Indonesian fat tailed sheep while Corriedale showed highest mean values for these traits (mean logFEC 3.58±0.079 and PCV of −4.97%), although the differences were not statistically significant (P>0.05). Among the SNP loci examined, genotypes at two loci, NAV3_591 and GLI1_576, both located in chromosome 3 and within exonic regions of the respective genes (Neuron navigator and GLI family zinc finger 1) were found to have significant differences in their fecal egg count (Table 6). Among these, GLI1_576 locus was a non-synonymous change from Asparagine (T allele) to Histidine (G allele). The mean log transformed fecal egg count at NAV3 locus was 3.435, 3.717 and 3.453 for GG, CC and GC genotype groups respectively. Similarly, the mean log transformed fecal egg count at GLI1_576 locus was 3.724, 3.364 and 3.749 for TT, GG and TG respectively (Figure 6). Apart from these two loci, genotypes at ZBTB39_51, IL20RA_422, PIK3CD_433 and TLR7_2491 showed weak differences in their mean log transformed fecal egg count, although statistically not significant (P<0.10). However, it needs to be mentioned that none of these loci were found to have significant association when multiple testing correction factors were applied using Benjamini-Hochberg false discovery rate (FDR corrected P- value>0.10 for all the SNP loci). Similarly, with respect to body weight change, four loci (ACVRL1_445, GPR84_520, TARBP2_97 and SMCR7L_517) located in chromosome 3 were found to have significant association (P<0.05) but with higher FDR values (P>0.05). Association of genotypes with packed cell volume change 42 days post challenge revealed significance at six SNP loci (NAV3_591, CSRNP2_65, ANKRD52_113, ESYT1_157, TIMP3_716 and IL2RA_388), of which ESYT1_157 showed significant FDR corrected p value of 0.029. The mean change in packed cell volume of genotypes TT, CC and TC at this locus were −7.96%, −6.09% and −4.02% respectively. With respect to SNP loci showing significant or weak association with phenotypes (including fecal egg count, body weight change and packed cell volume change), some showed heterozygous advantage while few others had no heterozygous advantage. Unlike haplotype analysis conducted on many breeds within each region that showed balancing selection and heterozygous advantage, association analysis was performed in few selected breeds. Considering the results from neutrality tests, selection influence at a particular SNP locus vary across breeds and hence the balancing selection observed in haplotype networks within a particular region might be due to different alleles being favoured within and across loci in various breeds. However, it has to be noted that the preliminary association in the present study is based on relatively fewer number of animals and expected heterozygous advantage has to be tested in large populations.
Table 6. Least squares ANOVA of parasite resistance characteristics (log transformed fecal egg count, body weight change and packed cell volume change 42 days post challenge at different SNP loci in experimentally challenged sheep (Pampinta and Corriedale, Indonesian Thin Tailed and Indonesian fat tailed).
| SNPID | Chromosome | Fecal egg count | Body Weight | Packed cell volume | |||
| P-Value | FDR* | P-Value | FDR* | P-Value | FDR* | ||
| PIK3R3_498 | 1 | 0.385 | 0.727 | 0.584 | 0.940 | 0.797 | 0.846 |
| LEPR_260 | 1 | 0.937 | 0.998 | 0.161 | 0.516 | 0.741 | 0.844 |
| IL6R_227 | 1 | 0.776 | 0.998 | 0.716 | 0.955 | 0.377 | 0.736 |
| ZDHHC17_190 | 3 | 0.167 | 0.727 | 0.222 | 0.607 | 0.296 | 0.736 |
| NAV3_591 | 3 | 0.038 | 0.604 | 0.561 | 0.940 | 0.046 | 0.313 |
| ACVRL1_445 | 3 | 0.209 | 0.727 | 0.018 | 0.331 | 0.716 | 0.839 |
| FGD6_519 | 3 | 0.299 | 0.727 | 0.596 | 0.940 | 0.704 | 0.839 |
| USP44_252 | 3 | 0.110 | 0.643 | 0.892 | 0.955 | 0.538 | 0.809 |
| ITGA5_111 | 3 | 0.984 | 0.998 | 0.742 | 0.955 | 0.238 | 0.711 |
| GPR84_520 | 3 | 0.317 | 0.727 | 0.024 | 0.331 | 0.062 | 0.317 |
| TARBP2_97 | 3 | 0.506 | 0.830 | 0.045 | 0.463 | 0.055 | 0.317 |
| ITGB7_538 | 3 | 0.890 | 0.998 | 0.795 | 0.955 | 0.315 | 0.736 |
| CSRNP2_65 | 3 | 0.607 | 0.958 | 0.170 | 0.516 | 0.022 | 0.224 |
| SLC11A2_174 | 3 | 0.314 | 0.727 | 0.924 | 0.955 | 0.805 | 0.846 |
| PTPRB_141 | 3 | 0.998 | 0.998 | 0.923 | 0.955 | 0.260 | 0.711 |
| STAT2_486 | 3 | 0.292 | 0.727 | 0.751 | 0.955 | 0.341 | 0.736 |
| DDIT3_527 | 3 | 0.314 | 0.727 | 0.628 | 0.953 | 0.796 | 0.846 |
| GLI1_253 | 3 | 0.344 | 0.727 | 0.267 | 0.684 | 0.105 | 0.432 |
| GLI1_576 | 3 | 0.033 | 0.604 | 0.588 | 0.940 | 0.102 | 0.432 |
| ZBTB39_51 | 3 | 0.062 | 0.604 | 0.066 | 0.516 | 0.637 | 0.839 |
| ANKRD52_113 | 3 | 0.896 | 0.998 | 0.767 | 0.955 | 0.019 | 0.224 |
| ESYT1_157 | 3 | 0.658 | 0.998 | 0.092 | 0.516 | 0.001 | 0.029 |
| TIMP3_716 | 3 | 0.348 | 0.727 | 0.521 | 0.940 | 0.042 | 0.313 |
| CSF2RB_279 | 3 | 0.414 | 0.739 | 0.436 | 0.862 | 0.695 | 0.839 |
| CSF2RB_557 | 3 | 0.270 | 0.727 | 0.976 | 0.976 | 0.672 | 0.839 |
| IL2RB_180 | 3 | 0.390 | 0.727 | 0.151 | 0.516 | 0.699 | 0.839 |
| EM4b_574 | 3 | 0.138 | 0.707 | 0.403 | 0.862 | 0.451 | 0.803 |
| CLEC1A_134 | 3 | 0.924 | 0.998 | 0.441 | 0.862 | 0.325 | 0.736 |
| PTPN6_398 | 3 | 0.965 | 0.998 | 0.916 | 0.955 | 0.255 | 0.711 |
| SMCR7L_517 | 3 | 0.739 | 0.998 | 0.001 | 0.057 | 0.992 | 0.992 |
| IL20RA_422 | 8 | 0.085 | 0.604 | 0.932 | 0.955 | 0.410 | 0.764 |
| STAT5B_385 | 11 | 0.916 | 0.998 | 0.176 | 0.516 | 0.360 | 0.736 |
| STAT3_138 | 11 | 0.990 | 0.998 | 0.764 | 0.955 | 0.537 | 0.809 |
| IL10_82 | 12 | 0.862 | 0.998 | 0.437 | 0.862 | 0.236 | 0.711 |
| TLR5_2276 | 12 | 0.481 | 0.821 | 0.848 | 0.955 | 0.860 | 0.881 |
| PIK3CD_443 | 12 | 0.088 | 0.604 | 0.100 | 0.516 | 0.248 | 0.711 |
| IL2RA_388 | 13 | 0.823 | 0.998 | 0.166 | 0.516 | 0.006 | 0.113 |
| PRLR_341 | 16 | 0.374 | 0.727 | 0.142 | 0.516 | 0.486 | 0.809 |
| PRLR_729 | 16 | 0.955 | 0.998 | 0.822 | 0.955 | 0.509 | 0.809 |
| TLR7_2491 | 27 | 0.069 | 0.604 | 0.335 | 0.808 | 0.572 | 0.809 |
| TLR8_1045 | 27 | 0.313 | 0.727 | 0.106 | 0.516 | 0.572 | 0.809 |
FDR – Benjamini-Hochberg False Discovery Rate corrected P-value.
Figure 6. Least squares mean and standard error of log transformed fecal egg count 42 days post challenge at significant SNP loci in two Argentinian (Pampinta and Corriedale) and two Indonesian (Indonesian Thin Tailed and Indonesian fat tailed) sheep breeds.
In conclusion, the present study reports strong phylogeographic structure in sheep across Asia, Europe and South America and balancing selection operating at SNP loci located within immune pathway genes. Although the present association analysis is preliminary in nature, the SNP loci on chromosome 3 and those within immune pathway genes indicated their potential for future large scale association studies in naturally exposed populations.
Supporting Information
(a–c) QTLs related to gastro-intestinal nematode resistance in sheep (d) Chromosome-wise distribution of QTLs related to parasite resistance traits in sheep and number of SNP loci investigated in the present study (e) Quantitative trait loci (QTL) map of chromosome 3 related to parasite resistance traits in sheep (QTL Source data: Animal QTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/index).
(TIF)
Distribution of allele sharing distance (IBS) between pairs of individuals. Distance was plotted separately where pairs were drawn from within the same breed (blue bars) and from across the breeds (red bars).
(TIF)
Bayesian clustering of 713 sheep based on genotype data at (a) 18 non-neutral SNP loci (b) 23 neutral SNP loci under assumption of 2 to 6 clusters without a priori population information. The breed names are given below the box plot and the geographical origin indicated above the box plot with the individuals of different breeds separated by vertical black lines.
(TIF)
Details of SNPs identified in silico within TLR genes of sheep.
(DOCX)
Pairwise FST (lower triangle) and allele sharing distance (upper triangle) among different sheep breeds. (BAN-Bangladeshi; BER-Bergschaf; COR-Corriedale; HAM-Hamdani; IFT-Indonesian Fat Tailed; ITT-Indonesian Thin Tailed; JUN-Junin; KAC-Kachchi; KAJ-Kajli; KAR-Karakachanska; KUL-Karakul; KSF-Krainer Steinschaf; MRS-Madras Red; MEC-Mecheri; MUF-Mouflon; NEL-Nellore; PAM-Pampinta; PAT-Pattanam; SHA-Shal; SHU-Shumenska; TEX-Texel; THA-Thalli).
(DOCX)
Global F-Statistics among different sheep populations at 41 SNP loci.
(DOCX)
Results of Ewens Watterson Neutrality test at different SNP loci in various sheep breeds (1 – Significantly deviate from neutrality; 0 – No significant deviation from neutrality).
(DOCX)
Analysis of molecular variance among different sheep breeds based on (right) genotypes at 18 non-neutral SNP loci and (left) genotypes at 23 neutral SNP loci.
(DOCX)
Acknowledgments
The present study is part of the Coordinated Research Project D3.10.26 of Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, International Atomic Energy Agency, Vienna, Austria. The authors thank Gerrit J. Viljoen, Head Animal Production and Health Section, IAEA for his valuable comments and suggestions to improve the manuscript. The authors also express sincere thanks to anonymous reviewers for their critical suggestions to improve the quality of manuscript.
Funding Statement
The study is funded by International Atomic Energy Agency through grant of Research Contracts to member state counterparts under the Coordinated Research Project D3.10.26 “Genetic Variation on the control of resistance to infectious diseases to improve productivity in small ruminants”. The Research Contracts Administration Section of IAEA (funder) had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perry BD, Randolph TF, McDermott JJ, Sones KR, Thornton PK (2002) Investing in animal health research to alleviate poverty. International Livestock Research Institute, Nairobi, Kenya. [Google Scholar]
- 2. Nieuwhof GJ, Bishop SC (2005) Costs of the major endemic diseases of sheep in Great Britain and the potential benefits of reduction in disease impact. Anim Sci 81: 23–29. [Google Scholar]
- 3. Peter JW, Chandrawathani P (2005) Haemonchus contortus: Parasite problem No. 1 from Tropics - Polar Circle. Problems and prospects for control based on epidemiology. Trop Biomed 22: 131–137. [PubMed] [Google Scholar]
- 4. Molento MB, Fortes FS, Pondelek DA, Borges Fde A, Chagas AC, et al. (2011) Challenges of nematode control in ruminants: focus on Latin America. Vet Parasitol 180: 126–132. [DOI] [PubMed] [Google Scholar]
- 5. Kenyon F, Greer AW, Coles GC, Cringoli G, Papadopoulos E, et al. (2009) The role of targeted selective treatments in the development of refugia-based approaches to the control of gastrointestinal nematodes of small ruminants. Vet Parasitol 164: 3–11. [DOI] [PubMed] [Google Scholar]
- 6.Baker RL, Mugambi JM, Audho JO, Carles AB, Thorpe W (2002) Comparison of Red Maasai and Dorper sheep for resistance to gastro-intestinal nematode parasites, productivity and efficiency in a sub-humid and a semi-arid environment in Kenya. Proceedings of the seventh world congress on genetics applied to livestock production, Montpellier, communication 13–10.
- 7. Nimbkar C, Ghalsasi PM, Swan AA, Walkden-Brown SW, Kahn LP (2003) Evaluation of growth rates and resistance to nematodes of Deccani and Bannur lambs and their crosses with Garole,. Ani Sci 76: 503–515. [Google Scholar]
- 8. Miller JE, Bishop SC, Cockett NE, McGraw RA (2006) Segregation of natural and experimental gastrointestinal nematode infection in F2 progeny of susceptible Suffolk and resistant Gulf Coast Native sheep and its usefulness in assessment of genetic variation. Vet Parasitol 140: 83–89. [DOI] [PubMed] [Google Scholar]
- 9. Gauly M, Erhardt G (2001) Genetic resistance to gastrointestinal nematode parasites in Rhön sheep following natural infection. Vet Parasitol 102: 253–259. [DOI] [PubMed] [Google Scholar]
- 10. Gruner L, Aumont G, Getachew T, Brunel JC, Pery CY, et al. (2003) Experimental infection of Black Belly and INRA 401 straight and crossbred sheep with Trichostrongyle nematode parasites,. Vet Parasitol 116: 239–249. [DOI] [PubMed] [Google Scholar]
- 11. Woolaston RR, Windon RG (2001) Selection of sheep for response to Trichostrongylus colubriformis larvae:genetic parameters. Ani Sci 73: 41–48. [Google Scholar]
- 12. Morris CA, Vlassoff A, Bisset SA, Baker RL, Watson TG, et al. (2000) Continued selection of Romney sheep for resistance or susceptibility to nematode infection: estimates of direct and correlated responses. Ani Sci 70: 17–27. [Google Scholar]
- 13. Stear MJ, Bishop SC, Bairden K, Duncan JL, Gettinby G, et al. (1997) The heritability of worm burden and worm fecundity in lambs following natural nematode infection. Nature 389: 27.9288962 [Google Scholar]
- 14. Smith JA, Wilson K, Pilkington JG, Pemberton JM (1999) Heritable variation in resistance to gastrointestinal nematodes in an unmanaged mammal population. Proceedings of the Royal Society of London, Series B-Biological Sciences 266: 1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prince LL, Gowane GR, Swarnkar CP, Singh D, Arora AL (2010) Estimates of genetic parameters for faecal egg count of Haemonchus contortus infection and relationship with growth traits in Avikalin sheep. Trop Anim Health Prod 42: 785–791. [DOI] [PubMed] [Google Scholar]
- 16.Woolaston RR, Windon RG, Gray GD (1991) Genetic variation in resistance to internal parasites in Armidale experimental flocks, in: Gray G.D., Woolaston R.R. (Eds.), Breeding for disease resistance in sheep, Australian Wool Corporation, Melbourne, pp. 1–9. [Google Scholar]
- 17. Hu Z, Park CA, Wu X-L, Reecy JM (2013) Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nuc Acids Res 41 D1: D871–D879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davies G, Stear MJ, Benothman M, Abuagob O, Kerr A, et al. (2006) Quantitative trait loci associated with parasitic infection in Scottish Blackface sheep. Heredity 96: 252–258. [DOI] [PubMed] [Google Scholar]
- 19.Hadfield TS, Miller JE, Wu C, Bishop S, Davies G, et al.. (2007) Identification of putative QTL for parasite resistance in sheep. Plant and Animal Genome Conference XV Proceedings, Abstract P553.
- 20. Marshall K, Maddox JF, Lee SH, Zhang Y, Kahn L, et al. (2009) Genetic mapping of quantitative trait loci for resistance to Haemonchus contortus in sheep. Anim Genet 40: 262–72. [DOI] [PubMed] [Google Scholar]
- 21. Sayre BL, Harris GC (2012) Systems genetics approach reveals candidate genes for parasite resistance from quantitative trait loci studies in agricultural species. Anim Genet 43: 190–8. [DOI] [PubMed] [Google Scholar]
- 22. Helgason A, Yngvadóttir B, Hrafnkelsson B, Gulcher J, Stefánsson K (2005) An Icelandic example of the impact of population structure on association studies. Nat Genet 37: 90–95. [DOI] [PubMed] [Google Scholar]
- 23. Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, et al. (2008) Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet 4: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, et al. (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438: 803–819. [DOI] [PubMed] [Google Scholar]
- 25. Zenger KR, Khatkar MS, Cavanagh JA, Hawken RJ, Raadsma HW (2007) Genome-wide genetic diversity of Holstein Friesian cattle reveals new insights into Australian and global population variability, including impact of selection. Anim Genet 38: 7–14. [DOI] [PubMed] [Google Scholar]
- 26. McKay SD, Schnabel RD, Murdoch BM, Matukumalli LK, Aerts J, et al. (2008) An assessment of population structure in eight breeds of cattle using a whole genome SNP panel. BMC Genet 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guernier V, Hochberg ME, Guégan J (2004) Ecology drives the worldwide distribution of human diseases. PLoS Biol 2: 0740–0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones KE, Patel N, Levy M, Storeygard A, Balk D, et al. (2008) Global trends in emerging infectious diseases. Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu S, Gupta S, Li J (2010) PEAS V1.0: a package for elementary analysis of SNP data. Mol Ecol Res 10: 1085–1088. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein J (1993) PHYLIP : Phylogeny inference package, version 3.5. Department of Genetics, Washington University, Seattle, Washington. [Google Scholar]
- 31.Goudet J (2002) FSTAT version 2.9.3.2. Department of Ecology & Evolution, University of Lausanne, CH. [Google Scholar]
- 32. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 33. Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Molecular Ecol Notes 4: 137–138. [Google Scholar]
- 34. Stephens M, Smith N, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stephens M, Scheet P (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing data imputation. Am J Hum Genet 76: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rozas J (2009) DNA sequence polymorphism analysis using DSP. Methods Mol Biol 537: 337–350. [DOI] [PubMed] [Google Scholar]
- 37. Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 38. Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 39.Harvey WR (1987) Least squares analysis of data with unequal subclass numbers ARS H-4, USDA, Washington D.C.
- 40. Ardlie KG, Lunetta KL, Seielstad M (2002) Testing for population subdivision and association in four case-control studies. Am J Hum Genet 71: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tabangin ME, Woo JG, Martin LJ (2009) The effect of minor allele frequency on the likelihood of obtaining false positives. BMC Proc 3 Suppl 7: S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lam AC, Schouten M, Aulchenko YS, Haley CS, Koning D-J (2007) Rapid and robust association mapping of expression QTL. BMC Proc 1 Suppl 1: S144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kijas JW, Lenstra JA, Hayes B, Boitard S, Porto Neto LR, et al. (2012) Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol 10 2: e1001258 10.1371/journal.pbio.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peter C, Bruford M, Perez T, Dalamitra S, Hewitt G, et al. (2007) Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim Genet 38: 37–44. [DOI] [PubMed] [Google Scholar]
- 45. Lawson Handley LJ, Byrne K, Santucci F, Townsend S, Taylor M, et al. (2007) Genetic structure of European sheep breeds. Heredity 99: 620–631. [DOI] [PubMed] [Google Scholar]
- 46. Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, et al. (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368: 455–457. [DOI] [PubMed] [Google Scholar]
- 47. Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, et al. (2009) A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS One 4 3: e4668 10.1371/journal.pone.0004668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arora R, Bhatia S, Mishra BP, Joshi BK (2011) Population structure in Indian sheep ascertained using microsatellite information. Animal Genetics 42: 242–50. [DOI] [PubMed] [Google Scholar]
- 49. Meadows JR, Li K, Kantanen J, Tapio M, Sipos W, et al. (2005) Mitochondrial sequence reveals high levels of gene flow between breeds of domestic sheep from Asia and Europe. J Hered 96: 494–501. [DOI] [PubMed] [Google Scholar]
- 50. Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, et al. (2001) Multiple maternal origins and weak phylogeographic structure in domestic goats. Proc Nat Acad Sci, USA 98: 5927–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Naderi S, Rezaei HR, Taberlet P, Zundel S, Rafat SA, et al. (2007) Large-Scale Mitochondrial DNA Analysis of the Domestic Goat Reveals Six Haplogroups with High Diversity. PLoS ONE 2 10: e1012 10.1371/journal.pone.0001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mukherjee S, Sarkar-Roy N, Wagener DK, Majumder P (2009) Signatures of natural selection are not uniform across genes of innate immune system, but purifying selection is the dominant signature. Proc Nat Acad Sci, USA 106: 7073–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takahata N, Nei M (1990) Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genet 124: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seabury CM, Seabury PM, Decker JE, Schnabel RD, Taylor JF (2010) Diversity and evolution of 11 innate immune genes in Bos taurus and Bos taurus indicus cattle. Proc Nat Acad Sci, USA 107: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takahata N, Satta Y, Klein J (1992) Polymorphism and balancing selection at major histocompatibility complex loci. Genet 130: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cagliani R, Riva S, Pozzoli U, Fumagalli M, Comi GP, et al. (2011) Balancing selection is common in the extended MHC region but most alleles with opposite risk profile for autoimmune diseases are neutrally evolving. BMC Evol Biol 11: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Suarez VH, Busetti MR, Garriz CA, Gallinger MM, Babinec FJ (2000) Pre-weaning growth, carcass traits and sensory evaluation of Corriedale, Corriedale X Pampinta and Pampinta lambs. Small Rum Res 36: 85–89. [Google Scholar]
- 58. Sodiq A, Tawfiq ES (2004) Productivity and breeding strategies of sheep in Indonesia-A Review. J Agr. Rur Dev Trop Subtrop 105: 71–82. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a–c) QTLs related to gastro-intestinal nematode resistance in sheep (d) Chromosome-wise distribution of QTLs related to parasite resistance traits in sheep and number of SNP loci investigated in the present study (e) Quantitative trait loci (QTL) map of chromosome 3 related to parasite resistance traits in sheep (QTL Source data: Animal QTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/index).
(TIF)
Distribution of allele sharing distance (IBS) between pairs of individuals. Distance was plotted separately where pairs were drawn from within the same breed (blue bars) and from across the breeds (red bars).
(TIF)
Bayesian clustering of 713 sheep based on genotype data at (a) 18 non-neutral SNP loci (b) 23 neutral SNP loci under assumption of 2 to 6 clusters without a priori population information. The breed names are given below the box plot and the geographical origin indicated above the box plot with the individuals of different breeds separated by vertical black lines.
(TIF)
Details of SNPs identified in silico within TLR genes of sheep.
(DOCX)
Pairwise FST (lower triangle) and allele sharing distance (upper triangle) among different sheep breeds. (BAN-Bangladeshi; BER-Bergschaf; COR-Corriedale; HAM-Hamdani; IFT-Indonesian Fat Tailed; ITT-Indonesian Thin Tailed; JUN-Junin; KAC-Kachchi; KAJ-Kajli; KAR-Karakachanska; KUL-Karakul; KSF-Krainer Steinschaf; MRS-Madras Red; MEC-Mecheri; MUF-Mouflon; NEL-Nellore; PAM-Pampinta; PAT-Pattanam; SHA-Shal; SHU-Shumenska; TEX-Texel; THA-Thalli).
(DOCX)
Global F-Statistics among different sheep populations at 41 SNP loci.
(DOCX)
Results of Ewens Watterson Neutrality test at different SNP loci in various sheep breeds (1 – Significantly deviate from neutrality; 0 – No significant deviation from neutrality).
(DOCX)
Analysis of molecular variance among different sheep breeds based on (right) genotypes at 18 non-neutral SNP loci and (left) genotypes at 23 neutral SNP loci.
(DOCX)