Abstract
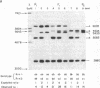
Spacer-length (sl) variation in ribosomal RNA gene clusters (rDNA) was surveyed in 502 individual barley plants, including samples from 50 accessions of cultivated barley, 25 accessions of its wild ancestor, and five generations of composite cross II (CCII), an experimental population of barley. In total, 17 rDNA sl phenotypes, made up of 15 different rDNA sl variants, were observed. The 15 rDNA sl variants comprise a complete ladder in which each variant differs in length from adjacent variants by approximately equal to 115 nucleotide pairs. Studies of four rDNA sl variants in an F2 population showed that these variants are located at two unlinked loci, Rrn1 and Rrn2, each with two codominant alleles. Using wheat-barley addition lines, we determined that Rrn1 and Rrn2 are located on chromosomes 6 and 7, respectively. The nonrandom distribution of sl variants between loci suggests that genetic exchange occurs much less frequently between than within the two loci, which demonstrates that Rrn1 and Rrn2 are useful as new genetic markers. Frequencies of rDNA sl phenotypes and variants were monitored over 54 generations in CCII. A phenotype that was originally infrequent in CCII ultimately became predominant, whereas the originally most frequent phenotype decreased drastically in frequency, and all other phenotypes originally present disappeared from the population. We conclude that the sl variants and/or associated loci are under selection in CCII.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allard R. W., Kahler A. L., Weir B. S. The effect of selection on esterase allozymes in a barley population. Genetics. 1972 Nov;72(3):489–503. doi: 10.1093/genetics/72.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N., Treco D., Taylor B., Eicher E. M. Distribution of ribosomal gene length variants among mouse chromosomes. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4677–4680. doi: 10.1073/pnas.79.15.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg M. T., Kahler A. L., Allard R. W. Estimation of life cycle components of selection in an experimental plant population. Genetics. 1978 Aug;89(4):765–792. doi: 10.1093/genetics/89.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Thoday J. M., Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982 Feb 18;295(5850):564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- Gerlach W. L., Bedbrook J. R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979 Dec 11;7(7):1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim-Elahi A. Computer program for calculating recombination values from electrophoretic data. J Hered. 1984 Mar-Apr;75(2):150–151. doi: 10.1093/oxfordjournals.jhered.a109895. [DOI] [PubMed] [Google Scholar]
- Krystal M., D'Eustachio P., Ruddle F. H., Arnheim N. Human nucleolus organizers on nonhomologous chromosomes can share the same ribosomal gene variants. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5744–5748. doi: 10.1073/pnas.78.9.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]