Abstract
Cell–cell communication, facilitating the exchange of small metabolites, ions and second messengers, takes place via aqueous proteinaceous channels called gap junctions. Connexins (cx) are the subunits of a gap junction channel. Mutations in zebrafish cx43 produces the short fin (sof b123) phenotype and is characterized by short fins due to reduced segment length of the bony fin rays and reduced cell proliferation. Previously established results from our lab demonstrate that Cx43 plays a dual role regulating both cell proliferation (growth) and joint formation (patterning) during the process of skeletal morphogenesis. In this study, we show that Hapln1a (Hyaluronan and Proteoglycan Link Protein 1a) functions downstream of cx43. Hapln1a belongs to the family of link proteins that play an important role in stabilizing the ECM by linking the aggregates of hyaluronan and proteoglycans. We validated that hapln1a is expressed downstream of cx43 by in situ hybridization and quantitative RT-PCR methods. Moreover, in situ hybridization at different time points revealed that hapln1a expression peaks at 3 days post amputation. Expression of hapln1a is located in the medial mesenchyme and the in the lateral skeletal precursor cells. Furthermore, morpholino mediated knock-down of hapln1a resulted in reduced fin regenerate length, reduced bony segment length and reduced cell proliferation, recapitulating all the phenotypes of cx43 knock-down. Moreover, Hyaluronic Acid (HA) levels are dramatically reduced in hapln1a knock-down fins, attesting the importance of Hapln1a in stabilizing the ECM. Attempts to place hapln1a in our previously defined cx43–sema3d pathway suggest that hapln1a functions in a parallel genetic pathway. Collectively, our data suggest that Cx43 mediates independent Sema3d and Hapln1a pathways in order to coordinate skeletal growth and patterning.
Introduction
Gap junctions play a critical role in coupling tissue function and they have long been hypothesized to play a role in the maintenance of homeostasis, morphogenesis, cell differentiation, growth control and the process of skeletogenesis in multicellular organisms [1]. Gap junctions are proteinaceous channels formed by the docking of two connexons between neighboring cells, and they mediate the exchange of low molecular weight metabolites (<1000 Da), ions and second messengers between the contacting cells [2]. Each connexon or hemichannel is made up of six connexins, each protein containing a four pass transmembrane domain. The syndrome oculodentodigital dysplasia (ODDD), characterized by abnormalities in craniofacial elements, limbs and dentition, has been linked to missense mutations in the GJA1 gene locus in humans [3]. At least 24 separate point mutations in GJA1, which codes for Connexin43 (Cx43), have been identified in patients with ODDD [3]–[5]. The CX43 knock out (CX43−/−) mouse dies perinatally because of cardiac malformations [6], [7]. Similarly, targeted gene knock-down of cx43 results in embryonic heart defects in zebrafish, signifying the essential role of cx43 during development [8]. The skeletal defects seen in the CX43−/− KO mouse model exhibited hypomineralization of craniofacial bones and severely delayed ossification of the appendicular skeleton [9]. Moreover, the ODDD phenotype is similar to a set of craniofacial abnormalities observed in the targeted CX43 knock-down chick model [10], [11]. In zebrafish, a homozygous mutation in cx43 causes the short fin phenotype, characterized by shorter tail fins due to defects in the fin skeleton. The mechanism by which CX43 based mutations cause skeletal defect phenotypes is largely unknown. However, it is apparent that the function of Cx43 in the vertebrate skeleton is conserved.
We utilize the zebrafish short fin mutant (sof b123) to address the role of Cx43 during skeletal development. Our lab has found that the mutation in zebrafish cx43 gene causes the short fin (sof b123) phenotypes characterized by short bony fin ray segments, short fins, and reduced cell proliferation. The sof b123 mutant exhibits reduced levels of cx43 mRNA and Cx43 protein, without a lesion in the coding sequence [8]. Moreover, three additional alleles that cause missense mutations also resulted in reduced gap junctional intercellular communication (GJIC), short segments, and reduced cell proliferation [12]. Furthermore, morpholino mediated knock-down of Cx43 in wild type zebrafish completely recapitulate all the phenotypes produced by sof alleles [13]. In contrast to sof b123, the another long fin (alf dty86) mutant exhibits fin overgrowth and overlong segments due to stochastic joint failure [14]. These phenotypes are opposite those of sof, and we have shown that alf dty86 exhibits increased levels of cx43 mRNA [15]. Indeed, cx43 knock-down in alf dty86 rescues the segment length phenotype, suggesting that Cx43 over expression contributes to the alf dty86 phenotypes. (note that the mutation causing alf dty86 phenotypes is not located in the cx43 gene [15]). We interpret the cx43-dependent loss of joints in alf dty86 and the premature joint formation (i.e. short segment) phenotype in sof to indicate that Cx43 suppresses joint formation. At the same time, Cx43 is positively associated with cell proliferation. Thus, Cx43 functions in more than one way, both positively influencing cell proliferation and negatively influencing joint formation, thereby concomitantly regulating bone growth and skeletal patterning during the process of fin regeneration.
An important, yet poorly understood question with respect to mutations in connexin genes in general is, how does GJIC impact tangible cellular events like cell division and differentiation? One hypothesis is that Cx43 based GJIC can influence gene expression patterns [16], [17]. Our lab exploited the availability of the two mutants, sof b123 and alf dty86, in order to identify genes whose expression depends on Cx43. Thus, we utilized a novel microarray strategy to identify a set of candidate genes, which are both downregulated in sof b123 and upregulated in alf dty86. The first gene validated from this microarray is semaphorin3d (sema3d) [18]. Here, we provide molecular and functional validation of another gene identified from the microarray analysis, hapln1a (hyaluronan and proteoglycan link protein 1a). In mouse and human, the orthologous protein Hapln1 has also been referred to as either cartilage link protein (Crtl1) or link protein (LP). The function of Hapln1 is to “link” hyaluronic acid (HA) with proteins termed proteoglycans (PG) in the extracellular matrix (ECM). Remarkably, the mouse knockout for CRTL1 (aka HAPLN1) causes dwarfism, craniofacial abnormalities, and perinatal lethality in the mouse [19].
The ECM is a complex mixture of proteins and carbohydrates that forms a dense network surrounding cells. Little is known about the functional role of the ECM during zebrafish fin regeneration. Pharmacological treatments that alter the localization of PGs and collagen, as well as the expression levels of matrix metalloproteases, have been found to restrict outgrowth during fin regeneration [20], [21]. Even less is known about the particular role of the Hapln1a-based ECM, including HA, in the regenerating fin. HA is a large molecular weight carbohydrate polymer with a molecular mass up to 106–107 Da [22], [23]. The structure of the ECM is stabilized by Haplns by virtue of forming stable associations between HA and PGs [24]–[26]. Unsulfated glycosaminoglycans (such as HA) are prominent in the blastemas of late stage regenerating cichlid fins, although their function has not been elucidated [27]. Apart from contributing to the physicochemical properties, ECM turnover and remodeling are critical events during tissue injury and wound repair. HA is also known to regulate cell migration, proliferation, and differentiation through activation of HA-specific cell surface receptors [28]–[30]. This is the first study to evaluate the effects of destabilizing the Hapln1a-based ECM during zebrafish fin regeneration. We find that reduced Hapln1a causes skeletal growth and patterning defects during fin regeneration, perhaps via destabilization of HA. Furthermore, we find that hapln1a functions downstream of cx43, providing novel insights into how skeletal morphogenesis could be influenced by Cx43.
Materials and Methods
Statement on the ethical treatment of animals
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols used for this manuscript were approved by Lehigh's Institutional Animal Care and Use Committee (IACUC) (protocol identification #128, approved 11/14/2012). Lehigh University's Animal Welfare Assurance Number is A-3877-01. All experiments were performed to minimize pain and discomfort.
Animal procedures
The following zebrafish (Danio rerio) strains were used in this study: wild-type C32, sof b123 [31] and alf dty86 [14]. Fish were maintained at a constant temperature of 25°C with 14 light: 10 dark photoperiod [32]. Fish were anaesthetized in 0.1% tricaine and caudal-fin amputations were performed at the 50% level using razor blades. Fin regeneration was then allowed to proceed until the desired time period and the regenerated fins were harvested from anaesthetized fish. Fins were fixed overnight in 4% paraformaldehyde (PFA) in PBS and dehydrated in 100% methanol at −20°C. Knockdown and whole mount in situ hybridization experiments were performed in triplicate with 5 fins per trial. For histochemistry on sectioned tissue, a minimum of 10 sections were evaluated from each of 3 different fins. For quantitative analyses, student's t-tests were completed to determine statistical significance (p<0.05).
In situ hybridization on whole mount and cryosectioned fins
RNA probes were generated from PCR amplified linear DNA in which the reverse primer contained the T7 RNA polymerase binding site. The primers used in this study for ISH are summarized in Table 1. Digoxygenin (DIG) labeled RNA probes were synthesized using DIG labeling mix (Roche) and in situ hybridization on regenerated whole fins was carried out as described [18]. Briefly, for the whole mount method, fins stored in methanol were sequentially rehydrated with methanol/PBST and then treated with 5 µg/ml proteinase K for 45 min and re-fixed in 4%PFA in PBS for 20 min followed by extensive washes with PBST and finally the pre-hybridization process was carried out with HYB (50% formamide, 5X SSC, 10 mM citric acid, 0.1% Tween20, heparin and tRNA) at 65°C for 0.5–1 h. Hybridization with DIG labeled probes suspended in HYB was carried out overnight at 65°C followed by sequential washes with HYB/PBST and finally with PBST. Anti-DIG Fab (Roche) fragments were used at 1∶5000 dilution, followed by extensive PBST washes and short washes in staining buffer. Subsequently fins were transferred to a staining solution containing NBT and BCIP and allowed to develop under dark conditions until purple color was observed. The reaction was stopped by washing in PBST and fixing overnight with 4% PFA. Finally the fins were mounted on a glass slide and analyzed using a Nikon Eclipse 80i microscope. Pictures were taken using a digital Nikon camera.
Table 1. Primers and Morpholinos.
| Gene | Primers for ISH | Primers for qRT-PCR | Morpholinos |
| hapln1a | F = GGTTCTCCGCTTGGCAGCG RT7 = TAATACGACTCACTATAGGGCCCATCCCTGCCTAAGACC | F = AACGACTATGGCACATACCGG R = AAGGTTGTACCGCCCCAAA | Splice MO = ATCAAAAGTGATTCTTACCTTGTGC ATG MO = GCCACAGAAAACAGAGCAATCATCT 5MM MO = GCCAGACAAAAGAGACCAATGATCT |
| sema3d | F = CGAAGTGTAGTACCATTTACG RT7 = TAATACGACTCACTATAGGGTATGAGGATCATATGTCC | F = TGGATGAGGAGAGAAGCCGAT R = GCAGGCCAGCTCAACTTTTT | MO = TGTCCGGCTCCCCTGCAGTCTTCAT 5MM = TGTGCCGCTGCCCTCCACTCTTCAT |
| actin | F = TTTTGCTGGAGATGATGCCC R = CCAACCATCACTCCCTGATGT |
The T7 RNA polymerase binding site is in bold in the reverse primers. F = Forward primer; RT7 = Reverse primer; MO = targeting morpholino; ATG MO = targets translation initiation; Splice MO = targets splicing event; 5MM = control morpholino with 5 mismatch pairs to target sequence. All primers and MOs are shown in the 5′-3′orientation.
For in situ on sections, fins were fixed overnight with 4% PFA in PBS after harvest. After a brief methanol wash, fins were dehydrated in 100% methanol and stored at −20°C until use. Before sectioning, fins were sequentially rehydrated in a methanol-PBS series of washes and then were embedded in 1.5% agarose/5% sucrose dissolved in PBS and equilibrated overnight in 30% sucrose. Fins were mounted in OCT and cryosectioned (15 µm sections) using a Reichertâ Jung 2800 Frigocut cryostat. Sections were collected on Superfrost Plus slides (Fisher) and allowed to air dry overnight at room temperature. Sections can be stored at −20°C for up to a year. Before use, the slides were brought to room temperature for at least an hour. Using a marking pen (ImmEdge Pen H-4000; PAP pen, VWR Laboratories), sections were circled. An appropriate amount of probe was pre-hybridized with a mixture of 1X salt solution (NaCl, Tris HCl, Tris Base, Na2HPO4.7H20, NaH2PO4, and 0.5 M EDTA) containing 50% deionized formamide (Sigma), 10% dextran sulfate, 1 mg/mL tRNA, and 1X Denhart's (Fisher) at 70°C for 5 mins. Hybridization with DIG–labeled antisense probes was carried out at 65°C overnight. The following day, slides were taken through a series of washes in a solution of 1X SSC, 50% formamide and 0.1% Tween-20 at 65°C. Slides were brought to room temperature and washed extensively in MABT (100 mM Maleic acid, 150 mM NaCl, and 0.1% Tween-20) and incubated in a blocking solution (MABT, Goat serum and 10% milk) for at least 2 hours. Anti-DIG Fab fragments (pre-absorbed against zebrafish fin tissue) were used at 1∶5000 dilution and incubated overnight at 4°C. On day 3, slides were washed 4X in MABT (30 min each) followed by 2X short washes (5–10 min) with staining buffer (100 mM Tris, 9.5, 50 mM MgCl2, 100 mM NaCl, and 0.1% Tween20). Slides were then transferred to 10% polyvinyl alcohol (PVA; MW: 86,000) staining solution with NBT/BCIP stock solution (Roche) and allowed to develop overnight at 37°C. When a purple color started to appear on the sections, the reaction was stopped by washing the slides with PBST for at least 3 h and then stored in PBST at 4°C until imaging. Sections were mounted in 100% glycerol and examined on a Nikon Eclipse 80i microscope.
Morpholino mediated gene knock-down in regenerating fins
All morpholinos (MOs) used in this study were fluorescein tagged, obtained from Gene Tools, LLC and used at a 1 mM concentration for injection. The sequences for MOs used in this study can be found in Table 1. Injection and electroporation were carried out as described previously [18]. Briefly, 3 dpa fish were anesthetized and approximately 50 nl of MO (targeting or mismatch [MM] control) was injected into either the dorsal or the ventral half of the tail fin. The un-injected half of the fin served as the internal control. Following injection, the entire fin was electroporated using a CUY21 Square Wave electroporator (Protech International Inc). MO-positive fish were selected 24 hpe (hours post electroporation) by examination under a fluorescence microscope. For H3P staining and qRT-PCR analysis, the fins were harvested 1 dpe, and for the analysis of regenerate length and segment length, the fins were harvested 4 dpe. For each MO (i.e., targeting or mismatch) 5–6 fish were injected on one half of the fin with the un-injected side serving as an internal control. Reproducibility was confirmed by testing the MO in three independent experiments. Statistical significance was determined using the student's t-test (P<0.05).
Immunochemistry and detection of HA
The following primary antibodies were used: Rabbit anti-histone-3-phosphate (anti-H3P, Millipore, 1∶200); Mouse anti-Hapln1a antibody (MD Bioproducts, 1∶500). The following secondary antibodies were used: anti-mouse Alexa 488 or 546 (1∶200); anti-rabbit Alex 546 (1∶200). For H3P staining, whole fins were harvested 1 dpe, after MO mediated knock-down and the experiment was carried out as described [18]. HA was detected by biotinylated-HABP (US Biological, 1∶300) followed by fluorescently labeled streptavidin-Alexa-546 conjugate, (Invitrogen, 1∶200). For detection of Hapln1a or HA after MO injection (targeting or MM control), the fins were harvested 1 dpe, fixed in 4% PFA overnight and then stored in Methanol. Cryosectioning was done as described [18] and the sections were allowed to dry overnight. The sections were then rehydrated in PBS, twice for 10 min followed by two washes with Immunostaining block (2% BSA, 0.1% Tween in PBS). Then, sections were blocked for another 1 h at room temperature with the immunostaining block and then incubated in primary antibody or biotin-HABP overnight at 4°C. The sections were washed with block (3x, 15 min each), incubated at room temperature for 1 hr with secondary antibody or streptavidin-Alexa-546 (pre-absorbed for 1 hr. at room temperature with fixed zebrafish fins to reduce background staining), washed (3x, 15 min each), incubated with HOECHST stain for 10 min at room temperature, followed by a quick wash with distilled water. They were blotted dry and then mounted for imaging. To test for specificity of HABP, WT fin sections were treated with bovine hyaluronidase (Sigma-H3506, final concentration 200 U/ml) in 10 mM phosphate buffer containing 100 mM NaCl (pH 6.0) for 2 hours at 37°C [33], [34]. Untreated sections were incubated in buffer without enzyme. Following treatment the sections were washed two times in Immunostaining block before staining for HABP as described above.
Lysate preparation and Immunoblotting
For WT and sof b123, 5 dpa regenerating fins were harvested from 10–15 fish. For MO injected fins, the injections were performed at 3 dpa fish (20–25 fish) and harvested the next day. Regenerating fins were harvested into 300–500 µl of RIPA lysis buffer, supplemented with protease inhibitor (Thermo scientific, Halt™ Protease and Phosphatase Inhibitor Cocktail) and homogenized using a tissue homogenizer (Bio-Gen, PRO 200) at high speed (5X) for 5 seconds with 10 second cooling intervals. Homogenized samples were centrifuged at 200 g for 10 min at 4°C and supernatant was used for further analysis. The protein samples were concentrated using a lyophilizer (Edwards-Freeze dryer super modulyo) and the pellet was resuspended in a minimal volume of PBS containing (Thermo scientific, Halt™ Protease and Phosphatase Inhibitor Cocktail, 100X) and the protein levels estimated by Bradford's assay.
To release Hapln1a from HA for detection by immunoblotting, equal concentrations of Hapln1a MO and MM lysates were treated with 100 U/ml of bovine hyaluronidase (Sigma) for 2 hours at 37°C as described [35]. Immunoblotting was performed as described [13] using mouse anti-Hapln1a (MD Bioproducts, 1∶500) and anti-α-tubulin (Sigma, 1∶3000) in combination with peroxidase-conjugated goat anti-mouse IgG (1∶10,000). Signal detection was performed using ECL Prime western blotting detection reagent (Amersham™ - GE Healthcare).
Measurements
The regenerate length, segment length, and the number of dividing cells were estimated/calculated as described [18]. For each experiment, at least 5 fish were evaluated in triplicate and a student's t-test was performed to evaluate statistical significance. Image Pro software was used to measure the regenerate length from the plane of amputation to the tip of the fin and segment length between two joints under bright field. Cell number was calculated by counting the number of histone-3-phosphate (H3P) positive cells from the distal-most 250 µm2 of the 3rd fin ray.
Quantitative real-time PCR
For qRT-PCR analysis, TRIZOL RNA extraction was made from the 5 dpa regenerating fins of wild-type, sof b123, and alf dty86 and 1 dpe for MO injected fins (targeting or MM). A minimum of 10 fins was used for total RNA extraction. For each sample, 1 µg of total RNA were reverse transcribed with SuperScript III reverse transcriptase (Invitrogen) using oligo-dT primers. Primers for qPCR analysis of hapln1a, sema3d and actin were designed using Primer express software (Table 1). Three independent RNA samples were used for the experimental comparison and qPCR for each gene was done in duplicates. The samples were analyzed using Rotor-Gene 6000 series software (Corbette Research) and the average cycle number (CT) was determined for each amplicon. Delta CT (ΔCT) values represent normalized expression levels of the test with respect to actin, the internal control. The relative level of gene expression, which is the fold difference, was determined using the delta delta CT (ΔΔCT) method (i.e., 2−ΔΔCT).
Results
Hapln1a is expressed downstream of cx43
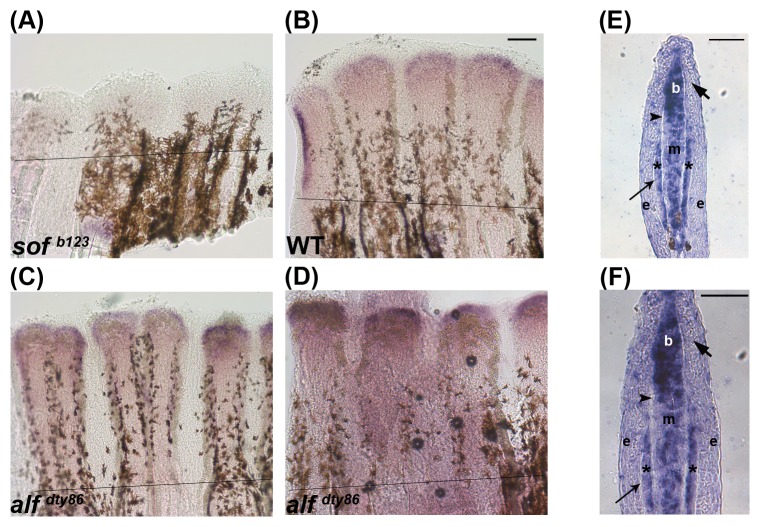
Recently, we described a microarray strategy to identify genes that function downstream of Cx43. We identified ∼50 genes that were both downregulated in sof b123 and upregulated in alf dty86 [18]. One of the candidate genes identified was hapln1a. To validate that hapln1a is expressed downstream of cx43, we compared the expression levels of hapln1a in WT, sof b123, and alf dty86 regenerating fins by whole mount in situ hybridization (Figure 1). As expected, hapln1a mRNA expression appeared to be downregulated in sof b123. We noticed some variability among alf dty86 regenerating fins, where some fins exhibited upregulation and some appeared more similar to WT regenerating fins (two representative images are shown in Figure 1). Therefore, we performed quantitative RT-PCR (qPCR) to evaluate the level of hapln1a expression in sof b123 and alf dty86 regenerating fins compared with WT (Table 2). As expected, hapln1a is downregulated in sof b123. However, we did not observe an upregulation of hapln1a in alf dty86, suggesting that among a population of alf dty86 regenerating fins, there is little difference in the expression level of hapln1a between WT and alf dty86.To determine the tissue-specific expression of hapln1a, we performed in situ hybridization on cryosections. The longitudinal section of a regenerating fin ray reveals several outer epidermal layers, including the basal layer of epidermis, which is adjacent to the mesenchyme. The mesenchyme is located medially. Within the mesenchymal compartment, a blastema is established at the distal, or growing end, of each ray. Blastemas are comprised largely of dividing cells that contribute to new tissue growth (reviewed in [36], [37]). The hapln1a mRNA is expressed throughout the mesenchyme with slightly higher expression in the blastema, and to a lesser extent in the skeletal precursor cells (Figure 1).
Figure 1. Validation of microarray results by in situ hybridization using whole mount in situ hybridization on 5 dpa regenerating fins.
(A) hapln1a expression is reduced in sof b123 compared with WT (B). alf dty86 expression is variable (C,D). (E,F) In situ hybridization on a WT 5 dpa cryosection reveals compartment specific expression of hapln1a, mainly in the blastema (b), in the mesenchyme (m), and skeletal precursor cells (*). The thick arrow identifies the basal layer of the epidermis, which underlies the epidermis (e). The thin arrow identifies lepidotrichia and the arrowhead identifies the actinotrichia. The amputation plane is indicated in panels A-D. Scale bar in E and F is 50 µm.
Table 2. Quantitative RT-PCR confirms changes in gene expression.
| Samples compared | Gene | ΔCT(Mut/MO) Gene-Actina | ΔCT (WT/MM) Gene-Actina | ΔΔCT = ΔCT((Mut/MO) – (WT/MM)) b | Fold difference relative to WT/MM c |
| sof b123 with WT | hapln1a | 6.51±0.13 | 4.99±0.10 | 1.52±0.13 | 0.35 (0.32–0.38) |
| alf dty86 with WT | hapln1a | 4.97±0.25 | 4.99±0.10 | −0.02±0.25 | 1.01 (0.85–1.20) |
| cx43 KD with MM | hapln1a | 6.57±0.23 | 5.67±0.08 | 0.90±0.23 | 0.53 (0.46–0.63) |
| sema3d KD with MM | hapln1a | 4.89±0.20 | 5.13±0.19 | −0.24±0.20 | 1.18 (1.03–1.36) |
| hapln1a KD with MM | cx43 | 2.78±0.10 | 2.84±0.19 | −0.06±0.10 | 1.04 (0.97–1.12) |
| hapln1a KD with MM | sema3d | 6.69±0.15 | 6.90±0.15 | −0.21±0.15 | 1.15 (1.04–1.28) |
a. The ΔCT value is determined by subtracting the average Actin CT value from the average Gene CT value. The standard deviation of the difference is calculated from the standard deviations of the gene and actin values using the Comparative Method.
b. The calculation of ΔΔCT involves subtraction by the ΔCT calibrator value. This is subtraction of an arbitrary constant, so the standard deviation of ΔΔCT is the same as the standard deviation of the ΔCT value.
c. The range given for gene relative to WT/MM is determined by evaluating the expression: 2∧ –ΔΔCT with ΔΔCT + s and ΔΔCT – s, where s = the standard deviation of the ΔΔCT value.
In order to provide a secondary test to demonstrate cx43-dependence of hapln1a, we examined its expression in fins treated for cx43 knock-down [13]. Indeed, the expression of hapln1a is reduced in WT fins treated for cx43 knock-down compared to cx43 mismatch-treated fins by both in situ hybridization (Figure S1) and by qPCR (Table 2). In contrast, cx43 expression is not reduced in WT fins treated for hapln1a knock-down (Table 2), indicating that cx43 expression does not depend on hapln1a. In summary, we find that hapln1a is reduced both in sof b123 and in cx43 knock-down fins, providing independent confirmation that hapln1a is expressed in a cx43-dependent manner. Together, these data support the conclusion that hapln1a is molecularly downstream of cx43.
To evaluate when during fin growth hapln1a may be required in WT regenerating fins, we examined the expression pattern of hapln1a in non-regenerating fins, and at 2 dpa, 3 dpa, 5 dpa and 8 dpa (Figure 2). In non-regenerating fins hapln1a expression was not detectable. By 2 dpa, the expression of hapln1a was slightly up-regulated, and by 3 dpa hapln1a was expressed strongly within each fin ray. At 5 dpa, there was a clear decrease in expression, and its expression was decreased even more by 8 dpa. Thus, hapln1a expression is expressed at 2 dpa and is maximally expressed at 3 dpa. The maximum expression at 3 dpa corresponds with the time point at which maximum rate of regeneration has been observed [38].
Figure 2. hapln1a expression pattern at different time points in regenerating fin.
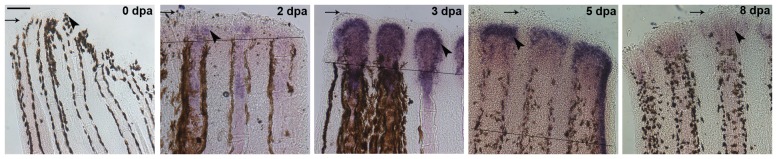
Whole mount in situ hybridization for hapln1a on regenerating fins at different time points. The hapln1a gene is not expressed during normal fin growth (uncut). Expression of hapln1a initiates around 2 dpa and is maximally expressed at 3 dpa, followed by gradual reduction at 5 dpa and 8 dpa. Arrow identifies the distal end of the fin; the arrowhead identifies region of staining (or where staining would be observed in uncut fins). The amputation plane is indicated for 2 dpa, 3 dpa, and 5 dpa, and is out of the field of view for the 8 dpa image. Scale bar is 50 µm.
Hapln1a knock-down is functionally downstream of Cx43
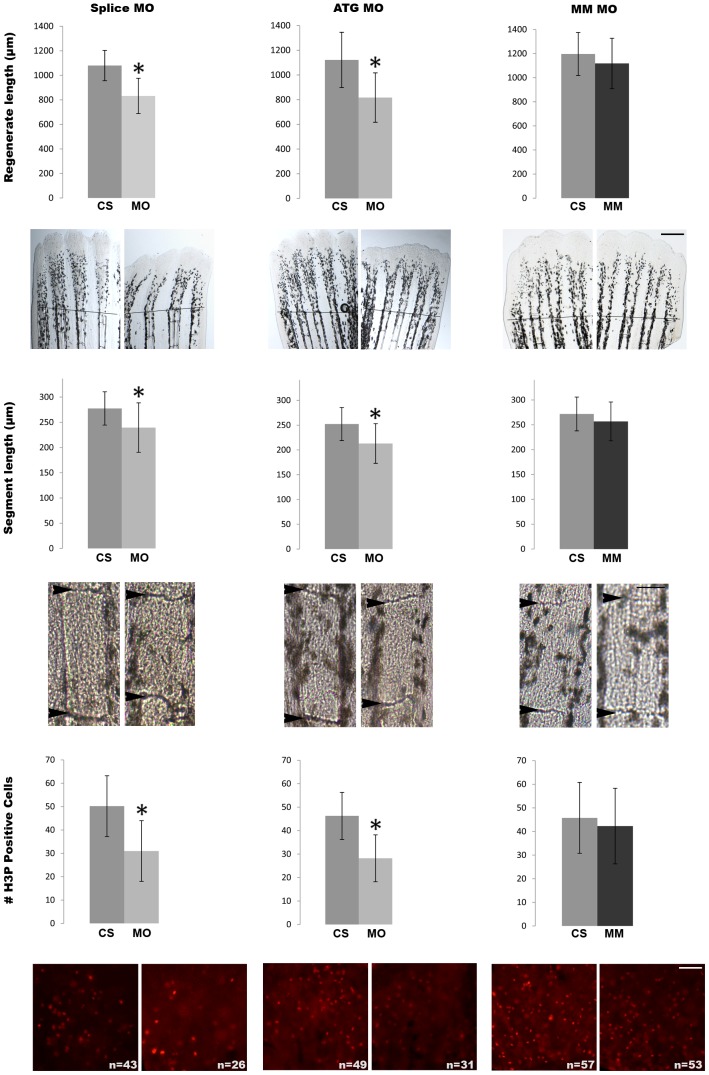
In order to determine if hapln1a is functionally downstream of cx43, we completed morpholino-mediated gene knock-down of hapln1a in WT regenerating fins [13], [15]. Two targeting morpholinos (MOs) were generated, an ATG blocker (ATG MO) that inhibits protein translation and a splice blocker (Splice MO) that was designed to inhibit the splicing of intron 1. As a control, we used a mismatch MO (MM MO), which includes five mismatches to the target sequence for the ATG-blocking MO (Table 1). All MOs are conjugated to fluorescein, permitting validation of cellular uptake. Following microinjection and electroporation, MO-positive fish were selected for fluorescein-positive cells at 24 hours post electroporation (hpe). Positive fins were either harvested for analysis of cell proliferation or permitted to regenerate for 4 additional days for evaluation of segment length and regenerate length. The effect of hapln1a knock-down on cell proliferation was evaluated by counting the number of mitotic cells detected by H3P immunostaining. Regenerate length was measured as the distance between the amputation plane and the distal end of the fin. Segment length was measured as the distance between the first two joints in the regenerate. Interestingly, we found that hapln1a knock-down with both the ATG MO and Splice MO exhibited the same phenotypes as cx43 knock-down. Thus, knock-down fins exhibited reduced fin length (Figure 3A), reduced segment length (Figure 3B), and reduced cell proliferation (Figure 3C). The specificity of these knock-downs is demonstrated by the use of two independent gene-targeting MOs and also by the mismatch control. Collectively, these data reveal that cx43 and hapln1a act in a common pathway to promote cell proliferation and to inhibit joint formation.
Figure 3. Morpholino mediated knock-down of hapln1a recapitulates all cx43-dependent phenotypes.
(Top row) Total regenerate length was measured in MO and MM treated fins. Hapln1a knock-down resulted in reduced fin length (*). The dotted line represents the amputation plane. (Middle row) Segment length was measured in MO and MM treated fins. Hapln1a knock-down resulted in reduction of segment length (*). Arrowheads identify joints. (Bottom row) Total number of H3P positive cells (red) was counted. Images of representative whole fins stained for H3P are shown for each condition. Knock-down of Hapln1a resulted in reduced cell proliferation (*) compared to the mismatch control. MO represents targeting morpholino; ATG MO targets translation initiation; Splice MO targets splicing event; MM represents control morpholino with 5 mismatches to target sequence; CS represents uninjected/untreated side. All experiments were completed in triplicate with a minimum of five fins treated with MO per trial. Statistical significance was determined by student's t-test where p<0.05. By student's t-test, there is no statistical difference between MM and CS. Error bars represent the standard deviation. Scale bar is 50 µm.
Hapln1a knock-down destabilizes HA
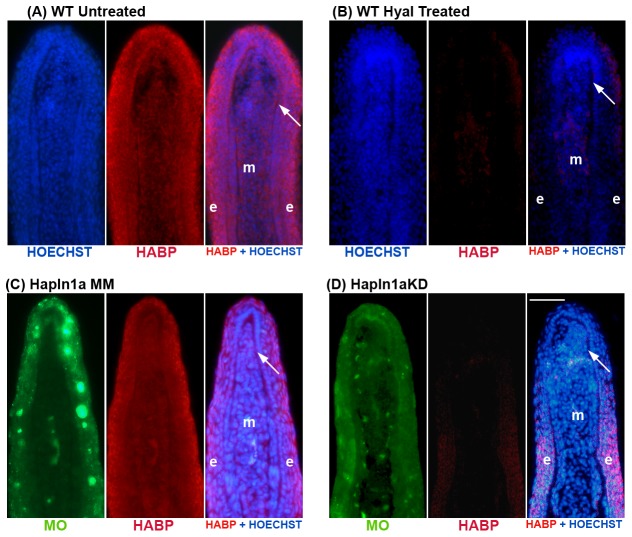
To evaluate the effect of hapln1a knock-down on HA in WT regenerating fins, we completed a MO-mediated hapln1a knock-down (i.e. using the ATG MO) and evaluated both Hapln1a staining and HA staining on the treated fins. The success of hapln1a MO-mediated protein knock-down was further confirmed by immunostaining. Thus, comparison of the MM MO treated fins with the ATG MO treated fins revealed that Hapln1a protein is reduced following hapln1a knock-down (Figure 4A-B). Moreover, reduced Hapln1a protein levels in ATG MO treated fins was confirmed through immunoblotting (Figure 4C). Hapln1a protein is glycosylated and typically detected as several bands above its predicted molecular weight of 38 kD [35]. All bands are reduced following Hapln1a knock-down. Since Hapln1a is crucial for the stabilization of the HA-PG network in the ECM, we also evaluated the level of HA in hapln1a knock-down fins. We utilized biotinylated HA-binding protein (HABP-biotin) in combination with streptavidin-Alexa546 to detect HA. First, we demonstrate that HABP is specific for the detection of HA by treating fin sections with the HA-degrading enzyme hyaluronidase. Endogenous levels of HA may be observed in WT 5 dpa fins (untreated). In contrast, treatment of fin sections with hyaluronidase greatly reduced the staining by HABP (Figure 5A-B), suggesting that HA is required for detection. Importantly, we also observed a clear reduction in the amount of detectable HA following hapln1a knock-down (Figure 5C-D). These findings strongly support the hypothesis that HA is destabilized in the absence of sufficient Hapln1a.
Figure 4. Morpholino mediated knock-down of hapln1a in WT regenerating fins results in reduced Hapln1a.
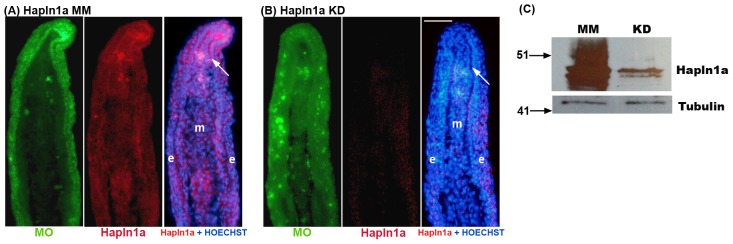
Immunostaining for Hapln1a and HOECHST staining for DNA (blue). The green reveals the location of the targeting and control MOs, which are fluorescein tagged. (A) Longitudinal section of a fin ray treated with Hapln1a control morpholino (MM). (B) Longitudinal section of a fin ray knocked down for Hapln1a with a targeting morpholino (KD). Compared to the control MM fins, Hapln1a knock-down (KD) fins exhibit reduced staining for Hapln1a. (C) Immunoblots confirming reduced levels of Hapln1a protein following Hapln1a knockdown. Hapln1a-MO treated fins (KD) were compared to control morpholino (MM). Tubulin was used as a loading control. Arrow identifies the basal layer of the epidermis; m, mesenchyme; e, epithelium. Scale bar is 50 µm.
Figure 5. Morpholino mediated knock-down of hapln1a in WT regenerating fins results in reduced HA levels.
HA is detected by biotin-HABP with streptavidin-Alexa-546. HOECHST detects DNA (blue). The green reveals the location of the targeting and control MOs, which are fluorescein tagged. (A,B) Fin sections from 5 dpa WT regenerating fins were either untreated or treated with hyaluronidase. The reduced signal in the hyaluronidase-treated sections demonstrates that HABP detects HA. (C) Longitudinal section of a fin ray treated with Hapln1a control morpholino (MM). (D) Longitudinal section of a fin ray knocked down for Hapln1a with a targeting morpholino (KD). Compared to the control MM fins, Hapln1a knock-down (KD) fins exhibit reduced staining for HA. Arrow identifies the basal layer of the epidermis; m, mesenchyme; e, epithelium. Scale bar is 50 µm.
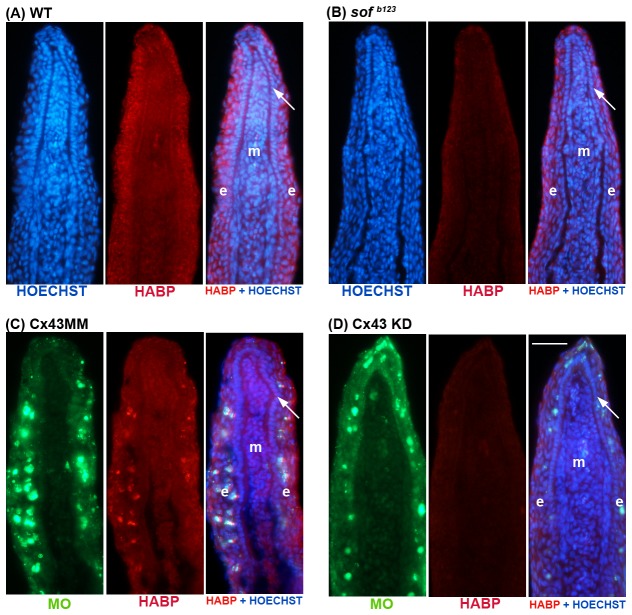
To further demonstrate that hapln1a functions in a cx43-dependent manner, we next evaluated HA levels in both sof b123 regenerating fins and in fins treated for cx43-knockdown [13]. Since hapln1a levels depend upon Cx43 activity, we predicted that reduced Cx43 would similarly lead to reduced levels of HA. Indeed, this is what we found. In sof b123 regenerating fins compared with WT regenerating fins, we observed reduced levels of HA (Figure 6A-B). Similarly, in WT fins treated for MO mediated cx43 knockdown, HA levels are reduced compared with the MM control-treated fins (Figure 6C-D). Together with our previous findings, these data strongly support the conclusion that hapln1a functions in a common pathway with cx43, and that this pathway mediates cell proliferation and joint formation at least in part through influencing the Hapln1a-based ECM.
Figure 6. HA levels are reduced in sof b123 and in cx43-KD regenerating fins.
Immunostaining for HA (Red) and HOECHST staining for DNA (blue). In the panels treated for cx43-MO or MM, the green reveals the location of the MOs, which are fluroescein tagged. HA levels were detected as described in Figure 4. (A,C) HA levels in WT 5 dpa regenerating fins and in WT fins treated for cx43-MM. (B,D) HA levels are reduced in sof b123 regenerating fins and in WT fins treated for cx43-MO. Arrow identifies the basal layer of the epidermis; m, mesenchyme; e, epithelium. Scale bar is 50 µm.
Hapln1a functions in a cx43-dependent and sema3d-independent genetic pathway
In our recent study, we found that Cx43 and Sema3d function in a common molecular pathway to promote cell proliferation and joint formation [18]. We next wished to determine how hapln1a contributes to this previously defined pathway. Our prior findings suggest that Sema3d utilizes the Nrp2a receptor to mediate cell proliferation and the PlxnA3 receptor to mediate joint formation. Indeed, in addition to Cx43 knock-down, both Sema3d- and PlxnA3 knock-down rescues joint formation in alf dty86. Therefore, we first tested if hapln1a knock-down functioned similarly. Interestingly, segment length was not rescued in alf dty86 fins by hapln1a knock-down (Figure 7). Failure of hapln1a knock-down to rescue segment length in alf dty86 was not due to a failure of the knock-down since both Hapln1a and HA were similarly reduced (i.e. evaluated by immunostaining for Hapln1a protein or for HA levels as completed above, see Figure S2). The finding that hapln1a knock-down does not rescue joint formation in alf dty86 is consistent with our finding that hapln1a is not up-regulated in alf dty86. Moreover, this suggests that hapln1a functions independently of sema3d-plxna3-dependent joint formation. We next evaluated changes in hapln1a and sema3d gene expression following reciprocal knock-down experiments. Thus, in sema3d knock-down fins hapln1a is not affected (Table 2), suggesting that hapln1a is not downstream of sema3d. Similarly, in hapln1a knock-down fins sema3d expression is not affected (Table 2), suggesting that sema3d is not downstream of hapln1a. Collectively, our findings provide evidence that hapln1a and sema3d both function downstream of cx43, but independently of each other. Therefore, these data suggest that at least two pathways contribute to cx43-dependent cell proliferation and cx43-dependent joint formation (Figure 7A). Future studies will be completed to determine if the Sema3d and Hapln1a gene products instead functionally interact in a common pathway.
Figure 7. Hapln1a knock-down does not rescue segment length in alf dty86.
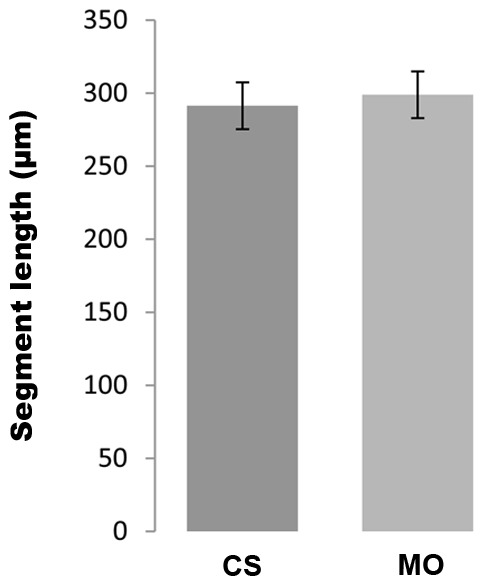
Compared to the uninjected control side (CS) the Hapln1a morpholino treated side (MO) does not show significant difference in the segment length. Statistical significance was determined by Student's t-test where p<0.05. Error bars represent the standard deviation.
Discussion
This is the first report to provide a functional analysis of hapln1a in zebrafish. Our finding that Hapln1a functions downstream of Cx43 is supported by multiple independent lines of evidence. The hapln1a gene exhibited reduced expression levels in sof b123 and cx43 knock-down fins compared to WT regenerating fins by in situ hybridization. Additionally, qPCR analysis confirmed that hapln1a expression is dependent upon the level of cx43 expression. Knock-down of Hapln1a using two independent MOs recapitulated all of the Cx43-dependent phenotypes, namely reduced regenerate length, reduced segment length, and reduced cell proliferation. Thus, hapln1a functions downstream of cx43. Moreover, we found that HA levels were strongly reduced in Hapln1a knock-down fins, suggesting that the loss of Hapln1a protein influences the stability of HA. The finding that HA levels are similarly reduced in sof b123 and in cx43 knock-down fins provides additional evidence that cx43 and hapln1a function in a common pathway. Attempts to include hapln1a in the established cx43-sema3d pathway suggest that hapln1a may function independently of sema3d (Figure 8A). Therefore, Cx43 promotes cell proliferation and suppresses joint formation via the coordination of (at least) two downstream pathways. We find it interesting that Hapln1a is located in the ECM that physically connects the medial cx43-positive compartment of dividing cells [8] and the lateral sema3d-positive compartment of skeletal precursor cells [18]. The functional significance of this observation is unclear.
Figure 8. Model depicting Cx43-Hapln1a mediated effect on skeletal patterning during fin regeneration.
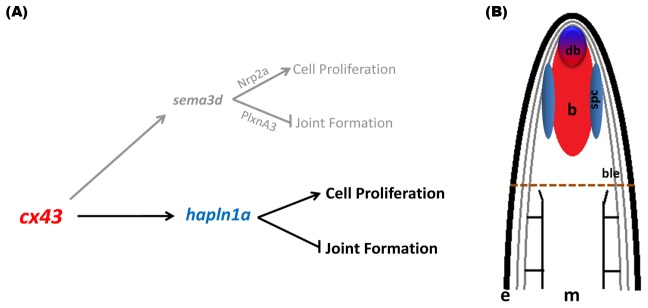
(A) Proposed pathway for Hapln1a mediated effects of Cx43. Hapln1a functions in a cx43-dependent, but sema3d-independent pathway, positively influencing cell proliferation and inhibiting joint formation. (B) Cartoon illustrating the compartments of gene in the regenerating fin: The cx43 mRNA is expressed throughout the mesenchyme (red) accompanied by cx43-dependent hapln1a upregulation in the same compartment (red), but primarily in the distal blastema (blue-red) and to a lesser extent in the proximal skeletal precursor cells (blue). e, epithelium; m, mesenchyme; b, blastema; db, distal blastema; spc, skeletal precursor cells; ble, basal layer of epidermis.
Hapln1a belongs to the hyaluronectin superfamily, which includes four members in mammals (HAPLNs1-4) and five in zebrafish, where the hapln1 gene appears to have duplicated into hapln1a and hapln1b. These genes share high degrees of sequence homology, especially in the amino acid sequence coding for the link module [25], [39]. Hapln1 was first identified in cartilage, however it can also be found in noncartilaginous tissue like sclera [40], aorta [41], brain [42], dermis of the embryonic skin [43], and in chicken embryonic mesonephros [44]. During zebrafish embryogenesis, expression of hapln1a has been observed in multiple tissues, including somites, floor plate, hypochord, and rhombomeres [45]. During jaw and skull formation, hapln1a expression co-localized with the PG aggrecan in the pharyngeal arches and with the PG dermacan in the pectoral fins [45]. Functional studies were not performed as part of these studies. However, reduced Hapln1 function is correlated with skeletal defects in other animals, including human. For example, single-nucleotide polymorphisms identified in the HAPLN1 gene have recently been associated with spinal osteoarthritis in aging female populations [46]. Moreover, targeted gene knockout of CRTL1 (aka HAPLN1) in the mouse reveals that Crtl1/Hapln1 is indispensible during skeletal development. For example, the CRTL1−/− mouse showed defects in cartilage and bone development with short limbs and craniofacial abnormalities, suggesting a prominent function for Crtl1 during chondrocyte differentiation [19]. In addition to these skeletal defects, CRTL1−/− mice die perinatally and exhibit a spectrum of myocardial defects. These defects have been attributed to the reduction in the PG versican [47], which may promote cell proliferation [48]–[50]. Interestingly, cartilage-specific expression of transgenic Crtl1/Hapln1a inhibits perinatal lethality and rescues skeletal abnormalities in CRTL1−/− mice [51], reinforcing the importance of the HA-PG network during heart development and skeletogenesis. These studies provide evidence that the “link” function of Hapln1 is critical for stability of components of the ECM and for regulation of cell differentiation and cell proliferation. Our findings on the function of Hapln1a in the regenerating fin are consistent with the role of Hapln1 in the developing mouse skeleton. Future studies will be focused on defining how Hapln1a mediates skeletal growth and patterning.
We propose that Cx43 activity in the blastema activates gene expression of hapln1a in the medial mesenchyme (Figure 8B). Secretion of Hapln1a in the ECM establishes the HA-PG network, stabilizes HA, and contributes to the regulation of cell proliferation and joint formation via unknown mechanisms. The Hapln1a-dependent ECM might be involved in providing the necessary microenvironment for the surrounding cells. Alternatively, Hapln1a may be required to maintain a stable population of HA, which in turn mediates signaling pathways through interaction with HA-specific cell surface receptors. Indeed, it is known that HA has diverse functions in skeletal biology including bone remodeling [22], bone resorption [52], and osteogenesis [53], [54]. Further, there is growing evidence that the ECM can influence interactions between locally secreted growth factors and their receptors [55]–[59]. Continued studies are required to determine if Hapln1a plays direct or indirect roles in influencing cellular behaviors required during fin skeletal morphogenesis.
Conclusions
The identification of the Cx43-dependent Hapln1a pathway is novel and reveals tangible roles for the ECM during bone growth and skeletal patterning. We find that reduced Hapln1a levels are correlated with reduced HA levels, which may provide insights into the underlying mechanism of Hapln1a function. Combined with known skeletal defects associated with the loss of Hapln1 in the mouse, and with skeletal diseases associated with Hapln1 polymorphisms in human, these findings demonstrate that the role of Hapln1a is conserved in zebrafish. Therefore, continued studies designed to elucidate the mechanism of Hapln1a-dependent cell proliferation and joint formation will provide new and relevant insights into skeletal development in all vertebrates.
Supporting Information
Cx43 knock down results in reduced expression levels of hapln1a . Whole mount in situ hybridization shows reduced expression of hapln1a in WT fins treated for cx43 knock-down (Cx43KD) fins compared to WT fins treated for cx43 mismatch control (Cx43MM) control fins.
(TIF)
The hapln1a -KD was effective in alf dty86 . Immunostaining for Hapln1a or HA (Red) and HOECHST staining for DNA (blue). The green reveals the location of the targeting and control MOs, which are fluroescein tagged (A) Longitudinal section of an alf dty86 fin ray treated with Hapln1a control morpholino (MM). (B) Longitudinal section of an alf dty86 fin ray knocked down for Hapln1a with a targeting morpholino (KD). Compared to the control MM fins, Hapln1a knock-down (KD) fins exhibit reduced staining for Hapln1a. HA was detected by biotinylated-HABP followed by streptavidin-Alexa 546 conjugate. (C) Longitudinal section of an alf dty86 fin ray treated with Hapln1a control morpholino (MM). (D) Longitudinal section of an alf dty86 fin ray knocked down for Hapln1a with a targeting morpholino (KD). Compared to the control MM fins, Hapln1a knock-down (KD) fins exhibit reduced staining for HA. Arrow identifies the basal layer of the epidermis; m, mesenchyme; e, epithelium. Scale bar is 50 µm.
(TIF)
Acknowledgments
The authors would like to thank Rebecca Jefferis for maintenance and care of the fish colony, and members of the Iovine lab for critical discussions regarding this manuscript.
Funding Statement
The research was funded by the National Institutes of Health (HD047737) and National Science Foundation (IOS 1145582). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Oyamada M, Oyamada Y, Takamatsu T (2005) Regulation of connexin expression. Biochim Biophys Acta 1719: 6–23. [DOI] [PubMed] [Google Scholar]
- 2. Goodenough DA, Goliger JA, Paul DL (1996) Connexins, connexons, and intercellular communication. Annu Rev Biochem 65: 475–502. [DOI] [PubMed] [Google Scholar]
- 3. Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, et al. (2003) Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet 72: 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kjaer KW, Hansen L, Eiberg H, Leicht P, Opitz JM, et al. (2004) Novel Connexin 43 (GJA1) mutation causes oculo-dento-digital dysplasia with curly hair. Am J Med Genet A 127A: 152–157. [DOI] [PubMed] [Google Scholar]
- 5. Richardson R, Donnai D, Meire F, Dixon MJ (2004) Expression of Gja1 correlates with the phenotype observed in oculodentodigital syndrome/type III syndactyly. J Med Genet 41: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reaume AG, de Sousa PA, Kulkarni S, Langille BL, Zhu D, et al. (1995) Cardiac malformation in neonatal mice lacking connexin43. Science 267: 1831–1834. [DOI] [PubMed] [Google Scholar]
- 7. Ya J, Erdtsieck-Ernste EB, de Boer PA, van Kempen MJ, Jongsma H, et al. (1998) Heart defects in connexin43-deficient mice. Circ Res 82: 360–366. [DOI] [PubMed] [Google Scholar]
- 8. Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL (2005) Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol 278: 208–219. [DOI] [PubMed] [Google Scholar]
- 9. Lecanda F, Warlow PM, Sheikh S, Furlan F, Steinberg TH, et al. (2000) Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol 151: 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Becker DL, McGonnell I, Makarenkova HP, Patel K, Tickle C, et al. (1999) Roles for alpha 1 connexin in morphogenesis of chick embryos revealed using a novel antisense approach. Dev Genet 24: 33–42. [DOI] [PubMed] [Google Scholar]
- 11. Law LY, Lin JS, Becker DL, Green CR (2002) Knockdown of connexin43-mediated regulation of the zone of polarizing activity in the developing chick limb leads to digit truncation. Dev Growth Differ 44: 537–547. [DOI] [PubMed] [Google Scholar]
- 12. Hoptak-Solga AD, Klein KA, DeRosa AM, White TW, Iovine MK (2007) Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett 581: 3297–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, et al. (2008) Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol 317: 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg, C P, Jiang, Y J, Kane, D A, Kelsh, R N, Mullins, M C, Odenthal J, Warga, R M, Nusslein-Volhard C (1996) Genetic analysis of fin formation in the zebrafish, Danio rerio. Development 123: 255–262. [DOI] [PubMed] [Google Scholar]
- 15. Sims K Jr, Eble DM, Iovine MK (2009) Connexin43 regulates joint location in zebrafish fins. Dev Biol 327: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stains JP, Civitelli R (2005) Cell-cell interactions in regulating osteogenesis and osteoblast function. Birth Defects Res C Embryo Today 75: 72–80. [DOI] [PubMed] [Google Scholar]
- 17. Stains JP, Lecanda F, Screen J, Towler DA, Civitelli R (2003) Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J Biol Chem 278: 24377–24387. [DOI] [PubMed] [Google Scholar]
- 18. Ton QV, Iovine M (2012) Semaphorin3d mediates Cx43-dependent phenotypes during fin regeneration. Dev Biol 366: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watanabe H, Yamada Y (1999) Mice lacking link protein develop dwarfism and craniofacial abnormalities. Nat Genet 21: 225–229. [DOI] [PubMed] [Google Scholar]
- 20. Andreasen EA, Mathew LK, Lohr CV, Hasson R, Tanguay RL (2007) Aryl hydrocarbon receptor activation impairs extracellular matrix remodeling during zebra fish fin regeneration. Toxicol Sci 95: 215–226. [DOI] [PubMed] [Google Scholar]
- 21. Bai SX, Wang YL, Qin L, Xiao ZJ, Herva R, et al. (2005) Dynamic expression of matrix metalloproteinases (MMP-2, -9 and -14) and the tissue inhibitors of MMPs (TIMP-1, -2 and -3) at the implantation site during tubal pregnancy. Reproduction 129: 103–113. [DOI] [PubMed] [Google Scholar]
- 22. Bastow ER, Byers S, Golub SB, Clarkin CE, Pitsillides AA, et al. (2008) Hyaluronan synthesis and degradation in cartilage and bone. Cell Mol Life Sci 65: 395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Contreras EG, Gaete M, Sanchez N, Carrasco H, Larrain J (2009) Early requirement of Hyaluronan for tail regeneration in Xenopus tadpoles. Development 136: 2987–2996. [DOI] [PubMed] [Google Scholar]
- 24. Knudson CB, Knudson W (1993) Hyaluronan-binding proteins in development, tissue homeostasis, and disease. FASEB J 7: 1233–1241. [PubMed] [Google Scholar]
- 25. Neame PJ, Barry FP (1993) The link proteins. Experientia 49: 393–402. [DOI] [PubMed] [Google Scholar]
- 26. Sherman L, Sleeman J, Herrlich P, Ponta H (1994) Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr Opin Cell Biol 6: 726–733. [DOI] [PubMed] [Google Scholar]
- 27. Santamaria JA, Becerra J (1991) Tail fin regeneration in teleosts: cell-extracellular matrix interaction in blastemal differentiation. J Anat 176: 9–21. [PMC free article] [PubMed] [Google Scholar]
- 28. Ghatak S, Misra S, Toole BP (2005) Hyaluronan constitutively regulates ErbB2 phosphorylation and signaling complex formation in carcinoma cells. J Biol Chem 280: 8875–8883. [DOI] [PubMed] [Google Scholar]
- 29. Jiang D, Liang J, Noble PW (2007) Hyaluronan in tissue injury and repair. Annu Rev Cell Dev Biol 23: 435–461. [DOI] [PubMed] [Google Scholar]
- 30. Misra S, Toole BP, Ghatak S (2006) Hyaluronan constitutively regulates activation of multiple receptor tyrosine kinases in epithelial and carcinoma cells. J Biol Chem 281: 34936–34941. [DOI] [PubMed] [Google Scholar]
- 31. Iovine MK, Johnson SL (2000) Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics 155: 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westerfield M (1993) The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio). University of Oregon Press, Eugene, OR.
- 33. Li Y, Toole BP, Dealy CN, Kosher RA (2007) Hyaluronan in limb morphogenesis. Dev Biol 305: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, et al. (2009) Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development 136: 2825–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun GW, Kobayashi H, Suzuki M, Kanayama N, Terao T (2002) Production of cartilage link protein by human granulosa-lutein cells. J Endocrinol 175: 505–515. [DOI] [PubMed] [Google Scholar]
- 36. Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J (2003) Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn 226: 190–201. [DOI] [PubMed] [Google Scholar]
- 37. Poss KD, Keating MT, Nechiporuk A (2003) Tales of regeneration in zebrafish. Dev Dyn 226: 202–210. [DOI] [PubMed] [Google Scholar]
- 38. Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD (2005) Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132: 5173–5183. [DOI] [PubMed] [Google Scholar]
- 39. Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, et al. (1996) Solution structure of the link module: a hyaluronan-binding domain involved in extracellular matrix stability and cell migration. Cell 86: 767–775. [DOI] [PubMed] [Google Scholar]
- 40. Lohmander LS, De Luca S, Nilsson B, Hascall VC, Caputo CB, et al. (1980) Oligosaccharides on proteoglycans from the swarm rat chondrosarcoma. J Biol Chem 255: 6084–6091. [PubMed] [Google Scholar]
- 41. Gardell S, Baker J, Caterson B, Heinegard D, Roden L (1980) Link protein and a hyaluronic acid-binding region as components of aorta proteoglycan. Biochem Biophys Res Commun 95: 1823–1831. [DOI] [PubMed] [Google Scholar]
- 42. Ripellino JA, Margolis RU, Margolis RK (1989) Immunoelectron microscopic localization of hyaluronic acid-binding region and link protein epitopes in brain. J Cell Biol 108: 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Binette F, Cravens J, Kahoussi B, Haudenschild DR, Goetinck PF (1994) Link protein is ubiquitously expressed in non-cartilaginous tissues where it enhances and stabilizes the interaction of proteoglycans with hyaluronic acid. J Biol Chem 269: 19116–19122. [PubMed] [Google Scholar]
- 44. Stirpe NS, Dickerson KT, Goetinck PF (1990) The chicken embryonic mesonephros synthesizes link protein, an extracellular matrix molecule usually found in cartilage. Dev Biol 137: 419–424. [DOI] [PubMed] [Google Scholar]
- 45. Kang JS, Kawakami Y, Bekku Y, Ninomiya Y, Izpisua Belmonte JC, et al. (2008) Molecular cloning and developmental expression of a hyaluronan and proteoglycan link protein gene, crtl1/hapln1, in zebrafish. Zoolog Sci 25: 912–918. [DOI] [PubMed] [Google Scholar]
- 46. Urano T, Narusawa K, Shiraki M, Sasaki N, Hosoi T, et al. (2011) Single-nucleotide polymorphism in the hyaluronan and proteoglycan link protein 1 (HAPLN1) gene is associated with spinal osteophyte formation and disc degeneration in Japanese women. Eur Spine J 20: 572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wirrig EE, Snarr BS, Chintalapudi MR, O′Neal J L, Phelps AL, et al. (2007) Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development. Dev Biol 310: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Evanko SP, Johnson PY, Braun KR, Underhill CB, Dudhia J, et al. (2001) Platelet-derived growth factor stimulates the formation of versican-hyaluronan aggregates and pericellular matrix expansion in arterial smooth muscle cells. Arch Biochem Biophys 394: 29–38. [DOI] [PubMed] [Google Scholar]
- 49. Sheng W, Wang G, Wang Y, Liang J, Wen J, et al. (2005) The roles of versican V1 and V2 isoforms in cell proliferation and apoptosis. Mol Biol Cell 16: 1330–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wight TN (2002) Versican: a versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol 14: 617–623. [DOI] [PubMed] [Google Scholar]
- 51. Czipri M, Otto JM, Cs-Szabo G, Kamath RV, Vermes C, et al. (2003) Genetic rescue of chondrodysplasia and the perinatal lethal effect of cartilage link protein deficiency. J Biol Chem 278: 39214–39223. [DOI] [PubMed] [Google Scholar]
- 52. Prince CW (2004) Roles of hyaluronan in bone resorption. BMC Musculoskelet Disord 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang L, Cheng YY, Koo PL, Lee KM, Qin L, et al. (2003) The effect of hyaluronan on osteoblast proliferation and differentiation in rat calvarial-derived cell cultures. J Biomed Mater Res A 66: 880–884. [DOI] [PubMed] [Google Scholar]
- 54. Nagahata M, Tsuchiya T, Ishiguro T, Matsuda N, Nakatsuchi Y, et al. (2004) A novel function of N-cadherin and Connexin43: marked enhancement of alkaline phosphatase activity in rat calvarial osteoblast exposed to sulfated hyaluronan. Biochem Biophys Res Commun 315: 603–611. [DOI] [PubMed] [Google Scholar]
- 55. Slevin M, Krupinski J, Kumar S, Gaffney J (1998) Angiogenic oligosaccharides of hyaluronan induce protein tyrosine kinase activity in endothelial cells and activate a cytoplasmic signal transduction pathway resulting in proliferation. Lab Invest 78: 987–1003. [PubMed] [Google Scholar]
- 56. Slevin M, West D, Kumar P, Rooney P, Kumar S (2004) Hyaluronan, angiogenesis and malignant disease. Int J Cancer 109: 793–794 author reply 795–796. [DOI] [PubMed] [Google Scholar]
- 57. Taylor KR, Trowbridge JM, Rudisill JA, Termeer CC, Simon JC, et al. (2004) Hyaluronan fragments stimulate endothelial recognition of injury through TLR4. J Biol Chem 279: 17079–17084. [DOI] [PubMed] [Google Scholar]
- 58.West DC, Kumar S (1989) Hyaluronan and angiogenesis. Ciba Found Symp 143: : 187–201; discussion 201–187, 281–185. [DOI] [PubMed] [Google Scholar]
- 59. West DC, Kumar S (1989) The effect of hyaluronate and its oligosaccharides on endothelial cell proliferation and monolayer integrity. Exp Cell Res 183: 179–196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cx43 knock down results in reduced expression levels of hapln1a . Whole mount in situ hybridization shows reduced expression of hapln1a in WT fins treated for cx43 knock-down (Cx43KD) fins compared to WT fins treated for cx43 mismatch control (Cx43MM) control fins.
(TIF)
The hapln1a -KD was effective in alf dty86 . Immunostaining for Hapln1a or HA (Red) and HOECHST staining for DNA (blue). The green reveals the location of the targeting and control MOs, which are fluroescein tagged (A) Longitudinal section of an alf dty86 fin ray treated with Hapln1a control morpholino (MM). (B) Longitudinal section of an alf dty86 fin ray knocked down for Hapln1a with a targeting morpholino (KD). Compared to the control MM fins, Hapln1a knock-down (KD) fins exhibit reduced staining for Hapln1a. HA was detected by biotinylated-HABP followed by streptavidin-Alexa 546 conjugate. (C) Longitudinal section of an alf dty86 fin ray treated with Hapln1a control morpholino (MM). (D) Longitudinal section of an alf dty86 fin ray knocked down for Hapln1a with a targeting morpholino (KD). Compared to the control MM fins, Hapln1a knock-down (KD) fins exhibit reduced staining for HA. Arrow identifies the basal layer of the epidermis; m, mesenchyme; e, epithelium. Scale bar is 50 µm.
(TIF)