Abstract
Objective
Examine the unique and congruent findings between multiple raters in a genome-wide association studies (GWAS) in the context of understanding individual differences in treatment response during antipsychotic therapy for schizophrenia.
Methods
We performed GWAS to search for genetic variation affecting treatment response. The analysis sample consisted of 738 patients with schizophrenia, successfully genotyped for ~492K SNPs from the Clinical Antipsychotic Trial of Intervention Effectiveness (CATIE). Outcomes included both clinician and patient report of illness severity on global impression scales, the CGI-S and PGI, respectively. Our criterion for genome-wide significance was a pre-specified threshold ensuring that, on average, only 10% of the significant findings are false discoveries.
Results
Thirteen SNPs reached genome-wide significance. The top findings indicated three SNPs in PDE4D, 5q12.1 (p =4.2×10−8, p =1.6×10−7, p =1.8×10−7), mediated the effects of quetiapine on patient reported severity and an additional three SNPs in TJP1, 15q13.1 (p = 2.25×10−7, p = 4.86×10−7, p = 4.91×10−7), mediated the effects of risperidone on patient reported severity. For clinician reported severity, two SNPs in PPA2, 4q24 (p = 3.68×10−7, p = 5.05×10−7), were found to reach genome-wide significance.
Conclusion
We found evidence of both novel and consistent association when examining the results from the patient and clinician ratings suggesting that different raters may capture unique facets of schizophrenia. Although our findings require replication and functional validation, this study demonstrates the potential of GWAS to discover genes that potentially mediate treatment response of antipsychotic medication.
Keywords: CATIE, CGI, pharmacogenomics, personalized medicine, schizophrenia, GWAS, single nucleotide polymorphism, PANSS
Introduction
Antipsychotic drugs represent the most commonly used treatment approach for managing psychotic symptoms in schizophrenia (1). However, variation in individual response to antipsychotics remains a critical problem, with a substantial proportion of patients with schizophrenia experiencing significant residual symptoms or adverse effects with antipsychotic treatment. Unfortunately, there are currently no robust methods to predict treatment response. Thus, treatment often proceeds by trial and error in order to determine the medication and dose that maximize response and minimize adverse reactions. Pharmacogenomic studies offer the possibility of identifying genetic variants associated with antipsychotic response that may facilitate the development of personalized treatment allowing efficient selection of the optimal medication and dose for each patient. Toward this end, we conducted a genome-wide association study (GWAS) to detect genetic variation related to antipsychotic treatment response in the Clinical Antipsychotic Trial of Intervention Effectiveness (CATIE).
The Clinical Global Impression (CGI) (2) severity scale is one of the most widely used measures in schizophrenia research and clinical practice. In CATIE, both the patients and clinicians rated illness severity using the Patient Global Impression (PGI) or CGI-S, respectively, at each assessment. While traditionally clinician ratings have been considered the “gold standard” in evaluating symptom severity and therapeutic progress in schizophrenia (3), recently, compelling arguments have been advanced for considering multiple raters, particularly the combination of patient and clinician ratings (4–5). Arguments for clinician ratings are well-established—50–80% of patients with schizophrenia suffer some degree of impaired “insight” into the presence and implications of their disorder (6). Given that insight is prerequisite for accurate evaluation of disease status and improvement, this research implies that a substantial subgroup of patients requires external evaluation, ideally by a psychiatric professional (3).
Balancing this line of reasoning, however, are several perspectives noting the unique benefits of patient ratings and the potential biases characterizing clinician evaluation. For instance, longitudinal research has shown that patient evaluations may outperform clinician’s in predicting long-term psychiatric outcomes, such as negative symptom exacerbation (7). Findings of prognostic value in patient-ratings are particularly common in studies using simple, global instruments such as the PGI, which do not require the cognitive skills necessary to disaggregate disease features into symptom domains (5). While it is not entirely clear why self-ratings sometimes show superior prognostic performance, it has been theorized that patients may be unable to adequately communicate their phenomenological experience of the disorder to clinicians due to flattened affect or other expression impairments (7-8). Another potential explanation stems from research into schizophrenia symptom q–estionnaires showing that patients and clinicians generally differ in their conceptualization of disease severity (5, 9). This research has shown systematic psychometric differences between patients and clinicians, with patients generally giving more weight to affective features in evaluating disease severity and clinicians tending to focus more on positive/psychotic symptoms (5). This has led to a growing sense that patient and clinician ratings capture partially distinct constructs, each containing unique, relevant clinical information (4–5, 9).
Beyond issues specific to clinician and patient rating, psychometric theory is clear that the use of multiple raters generally enhances measurement accuracy and, thus, improves statistical power to detect associations (10). This is due to the capacity of multiple raters to reduce or eliminate sources of noise/error including random errors, such as misinterpreting a question, and systematic rater biases caused by different questionnaire response styles, normative standards or incentive structures (11). In sum, a confluence of schizophrenia assessment research and psychometric theory support the value of considering both patient and clinician ratings in evaluating schizophrenia severity. This conclusion is especially relevant to the current context of pharmacogenomic GWAS, where statistical power is generally modest; thus, the use of multiple raters may increase confidence in results when there is a convergence of evidence across raters.
In addition to using assessments from multiple raters of the same instrument, the consideration of multiple instruments may facilitate more comprehensive approaches to the study of schizophrenia. Clinically, the consideration of multiple instruments is primarily motivated by the vulnerability of any single instrument to symptom bias, in which specific clusters of symptoms/disease features are better captured than others. For example, the Positive and Negative Syndrome Scale (PANSS) (12), a commonly used instrument in schizophrenia research, is often considered superior for assessing specific features of schizophrenia, such as positive and negative symptoms, while the CGI may be viewed as offering a better overall clinical assessment of the disorder (13). Further, comparing the results of GWAS studies using two different measures of treatment response may yield additional support for putative susceptibility loci or suggest unique relationships between specific clinical features and variants. Thus, when the same variant is implicated by two different measures there is increased confidence in the finding validity. Conversely, although they require replication, unique findings may indicate genetic variants more relevant to various facets of the disease and its symptoms, which may not be well-captured in a different scale.
Here, we engage the issues of multiple raters and assessments in the context of antipsychotic pharmacogenomics—conducting a GWAS of antipsychotic response in CATIE as measured by the patient rated PGI and clinician rated CGI in order to investigate if there is any convergence across raters and eliminate potential rater bias. We then compare these findings with previous CATIE GWAS assessing efficacy using the Positive and Negative Syndrome Scale (PANSS) (14) and neurocognitive measures (15), an endophenotype of schizophrenia.
Methods
Subjects
We used data from CATIE, which has been described in detail elsewhere (16). In short, CATIE was a multiphase randomized control trial of antipsychotic medications, including five primary treatments (i.e., olanzapine, perphenazine, quetiapine, risperidone and ziprasidone), which followed patients for up to 18 months. To maximize representativeness, the participants were recruited from 57 clinical settings around the United States. Patients were diagnosed with schizophrenia using the Structured Clinical Interview for DSM-IV (17). All three CATIE phases were used in this study (n = 738). Table 1 presents descriptive statistics for the sample. The mean age was 40.9 years (S.D. = 11.0) and the average durations of treatment for schizophrenia was 16.7 years (S.D. = 11.2).
Table 1.
Descriptive Sample Statistics
| Visit | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| Baseline | 1 | 3 | 6 | 9 | 12 | 15 | 18 | |||||||||
| n | % | Mean (SD) | n | Mean (SD) | n | Mean (SD) | n | Mean (SD) | n | Mean (SD) | n | Mean (SD) | n | Mean (SD) | n | |
| Age (Mean, SD) | 40.9 | 11.03 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Male | 559 | 73.7 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| European American (EA) | 437 | 57.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| African American (AA) | 222 | 29.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Other Race | 106 | 14.0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Years in Treatment (Mean, SD) | 16.7 | 11.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Olanzapine | 173 | 24.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Perphenazine | 119 | 15.6 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Quetiapine | 160 | 20.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Risperidone | 159 | 20.8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Ziprasidone | 90 | 11.8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PGI (Mean, SD) | 3.55 | 1.51 | 3.13 (1.51) | 668 | 3.16 (1.45) | 614 | 3.19 (1.42) | 608 | 3.02 (1.42) | 571 | 3.07 (1.41) | 558 | 3.09 (1.43) | 535 | 3.36 (1.31) | 90 |
| CGI-S (Mean, SD) | 3.92 | 0.95 | 3.63 (0.95) | 667 | 3.52 (0.91) | 613 | 3.51 (0.95) | 603 | 3.39 (0.98) | 572 | 3.35 (0.98) | 557 | 3.33 (0.97) | 534 | 3.45 (0.90) | 90 |
Measures
Two global impression severity scores were used in this study: patient self-report, PGI, and clinician, CGI-S. The PGI and CGI-S rate the severity of schizophrenia symptoms on a scale from 1 to 7, with higher scores indicating a greater severity of illness (2). The bottom of Table 1 displays the average PGI and CGI-S score by visit. Across all visits, the CGI-S has a higher average score than the PGI suggesting that clinicians consider the patients to have a more severe disorder than the patients themselves do. Both scores also tend to decrease as the number of visits increases.
Estimating Treatment Effects
In previous research we have developed (18), and applied to CATIE (14–15, 19–20) a novel method for estimating treatment effects in clinical trial data. Using mixed modeling, our method first determines the optimal functional form of over-time drug response, then screens many possible covariates to select those that improve the precision of the treatment effect estimates, and finally generates the individual treatment effect estimates based on the best-fitting model using best linear unbiased predictors (BLUPs) (21). As our approach condenses all information collected during the trials in an optimal, empirical fashion, it results in more precise estimates than traditional approaches that estimate treatment effects using only two assessments (e.g., subtracting pre- from post-treatment observations) (22).
Specifically, to determine the optimal drug response trajectory for each outcome we fit a series of models specifying linear change for a given number of days on drug and flat thereafter. This series began with a model assuming that maximal drug response was achieved at day one. Each subsequent model specified an incrementally longer duration until maximal drug response, with the final model assuming that the drug effect did not plateau (i.e. linear change throughout trial). The best-fitting model of the series was selected to determine the average number of days until maximal drug response (through optimizing the likelihood across the series). After determining the optimal functional form of the drug response trajectories, 31 covariates were screened to identify those that improved the precision of the treatment effect estimates. Covariates included design characteristics, socio-demographic measures, clinical information, confounding medications and baseline antipsychotic treatment. For both CGI-S and PGI only a single covariate, related to trial design, was found to improve treatment effect precision (the specific covariate was “phase 1b”, which indicates that the subject was initially randomized to the perphenazine arm, but was then switched into one of the second generation arms). Finally, treatment effects were generated as random effects. To elaborate, mixed models estimates two types of parameters, coefficients that describe the predictors’ sample average effects, and deviations from the average effects for each subject (i.e., random effects). Thus, for each of the five trial drugs investigated, treatment effects were generated by specifying random effects for the drug response (23). Intuitively, these treatment effects quantify how much, for instance, each subject’s global impression severity score phenotype change in response to a given drug, relative to the average effect for all subjects taking the drug. The treatment effect estimates were generated separately for the CGI-S and PGI, for each drug, and were treated as different outcomes in the GWAS analysis. Treatment effects estimated with this method have been previously analyzed in several published genome-wide association studies of CATIE (14–15, 19–20).
Genotyping, Quality Control and Ancestral Background
DNA sampling, genotyping and genotype quality control have been described elsewhere (24). Briefly, single nucleotide polymorphisms (SNPs) were genotyped using the Affymetrix 500K ’A’ chipset (665K SNPs) (Affymetrix, Santa Clara, CA, USA) and a custom fill-in chip (164K SNPs) (Perlegen, Mountain View, CA, USA). After stringent quality control, 492,900 SNP genotypes remained for analysis.
Approximately 57% of the CATIE subjects self-identified as white/European-American (EA), 29% as black/African-American (AA), and 14% consider themselves to have ‘other’ ancestral origins or to belong to multiple ancestral categories. To avoid false positives due to population stratification, Sullivan et al. (24) performed an extensive evaluation of multiple methods to control for ancestral heterogeneity in CATIE and concluded that the principal component and multi-dimensional scaling (MDS) approach worked best. We, therefore, proceeded with the MDS approach as implemented in PLINK (25). For our analyses, we used five MDS dimensions that appeared to capture the majority of the genetic substructure in CATIE.
Association Testing and False Discovery Rate Control
All association testing was conducted in PLINK using a linear regression model of additive SNP effects with five population stratification MDS dimensions as covariates to control for ancestry. We used a false discovery rate (FDR) (26) based approach to declare significance. In comparison to controlling a family-wise error rate (e.g., Bonferroni correction) FDR a) provides a better balance between finding true effects versus controlling false discoveries, b) results in comparable standards for declaring significance across studies because it does not directly depend on the number of tests, and c) is relatively robust against having correlated tests (27). FDR is commonly used in many high-dimensional applications and has been successfully applied in the context of GWAS (28–30). As motivated previously (31), we set a FDR threshold of 0.10 for declaring genome-wide significance. This means that on average 10% of the SNPs declared significant are expected to be false discoveries. Operationally (32), the FDR was controlled using q-values. Q-values are FDRs calculated using the p-value of the markers as thresholds for declaring significance. It is important to note that performing multiple GWAS analyses does not present a problem for the FDR because it controls the expected ratio of false discoveries to all discoveries. Thus, when many GWAS are performed, the number of false positives will increase and so will the number of true positives. The expected ratio of false to all discoveries will, therefore, remain 0.10 with our threshold for declaring genome-wide significance regardless of how many GWAS are performed.
For the most promising SNPs, we performed a variety of additional analyses to examine the robustness of the signal. First, we tested the SNPs separately in the subjects who self-identified as European Americans (EA) only and African American (AA) only. Those individuals that reported being neither EA nor AA, ~14% of the sample, were excluded from these stratified analyses because the sample size was too small to detect any effects. The sample sizes in the GWAS for the EA and AA stratified samples are rather small for a GWAS and the results of these analyses should be considered suggestive. For each SNP, we also performed haplotype (proxy) analyses that incorporate information from other SNPs in that region. Such analyses may provide a technical validation of the single SNP result or point to a particularly informative haplotype.
After identifying genome-wide significant markers, we leverage the two global impression measures and multiple drug study design to examine whether genome-wide significant markers show association to other related outcomes at less stringent significant levels (p < 0.05). Although it is possible that SNP effects are outcome specific, observing associations with multiple outcomes excludes the possibility of significant effects due to outcome-specific outliers and may be informative from a clinical perspective. Specifically, we examined whether significant SNPs from this study and other CATIE GWAS studies that use alternate definitions of treatment response (i.e., PANSS and neurocognitive measures) were associated with either of the CGI severity treatment effects. We only considered subjects under the same drug treatment for these tests.
Results
Genome-Wide Significant Signals
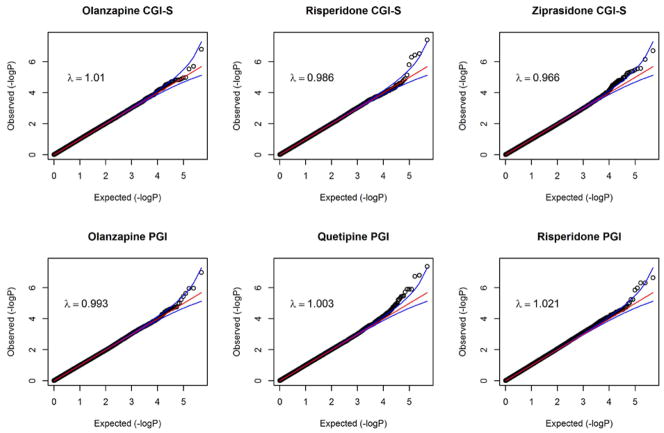
Quantile-Quantile (QQ) plots are shown in the Figure 1 for all of the drug-outcome combinations that had significant SNPs (q < 0.10). The plots show that the distribution of p-values from the GWAS are generally on a straight line, indicating the expected p-value distribution under the null hypothesis assuming no effects of the markers. However, in each of the plots, there is also evidence that markers in the right upper corner have p-values smaller than would be expected under the null hypothesis, suggesting true association between these markers and the outcome variable. The plots also display λ values (that is, the ratio of the median observed p-value of the distribution to the expected p-value under the null hypothesis), approximately equal to 1, indicating no systematic test statistic inflation and that population stratification was adequately controlled.
Figure 1.
QQ Plots of Genome-Wide Significant Associations with Clinician (CGI-S) and Patient (PGI) Rated Global Impression Severity
Table 2 provides details on those SNPs that were genome-wide significant (q < 0.10). One of the top findings included three SNPs, rs17382202, rs17742120, and rs2164660, which exhibited a negative association between PGI self-report and minor allele count for quetiapine treatment. These SNPs are located in an intron of PDE4D, 5q12.1, as described in Table 1 (p =4.2×10−8, q = 0.02; p =1.6×10−7, q = 0.03; p =1.8×10−7, q = 0.03). Examination of the linkage disequilibrium (LD) in the surrounding region showed that all three SNPs were in high LD (R2 > 0.80) with each and were in moderate LD (0.30 < R2 <0.60) with several other SNPs in the region. This suggests that these SNPs may tag the same locus. To further investigate this signal, we divided our sample by ancestry. The direction and magnitude of effects were similar in the two groups, with the AA sample showing slightly higher absolute coefficient values, but larger p values due to smaller AA sample sizes (rs17382202: βAA = −1.71, pAA = 0.008, βEA = −1.07, pEA = 1.85×10−6; rs17742120: βAA = −1.24, pAA = 0.03, βEA = −1.07, pEA = 1.85×10−6; rs2164660: β AA = −1.36, p AA = 0.02, βEA = −1.11, pEA = 4.24×10−7). Stratified haplotype and overall haplotype testing did not improve the association signal.
Table 2.
Genome-Wide Association Results with q-values < 0.10
| Phenotype | Antipsychotic | Gene symbol | Cytogenetic location | Psn(bp) | SNP number | MAF | MA | Eff | n | p-value | q-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CGI-S | Olanzapine | ATP1A2 | 1q23.2 | 156890927 | rs6688363 | 0.166 | T | + | 263 | 1.59E-07 | 0.078 |
| Risperidone | 16q22.1 | 64716237 | rs8050896 | 0.161 | T | − | 267 | 3.85E-08 | 0.019 | ||
| TNFRSF11A | 18q21.3 | 58231092 | rs2980976 | 0.168 | A | + | 268 | 2.97E-07 | 0.060 | ||
| PPA2 | 4q24 | 106655117 | rs2636697 | 0.038 | G | + | 268 | 3.68E-07 | 0.060 | ||
| 106710973 | rs2636719 | 0.039 | A | + | 265 | 5.05E-07 | 0.062 | ||||
| Ziprasidone | 11q14.1 | 79736127 | rs7395555 | 0.145 | C | − | 188 | 1.98E-07 | 0.098 | ||
| PGI | Olanzapine | SPOPL | 2q22.1 | 139112654 | rs10170310 | 0.213 | G | + | 264 | 1.03E-07 | 0.051 |
| Quetiapine | PDE4D | 5q12.1 | 59015703 | rs17382202 | 0.153 | T | − | 263 | 4.21E-08 | 0.021 | |
| 59034799 | rs17742120 | 0.161 | G | − | 265 | 1.58E-07 | 0.031 | ||||
| 59047264 | rs2164660 | 0.161 | A | − | 257 | 1.87E-07 | 0.031 | ||||
| Risperidone | TJP1 | 15q13.1 | 27980760 | rs711355 | 0.330 | T | − | 267 | 2.25E-07 | 0.081 | |
| 27980608 | rs785423 | 0.402 | A | − | 265 | 4.86E-07 | 0.081 | ||||
| 27961668 | rs813676 | 0.401 | T | − | 264 | 4.91E-07 | 0.081 |
Abbreviations: Psn, position; MAF, minor allele frequency; MA, minor allele; Eff, direction of the effect of minor allele; n, sample size. For each test, we report n, Eff (where a ‘+’ indicates global impression severity score positively associated with minor allele count), and the p- and q-values of the test. Shaded rows indicate SNPs in high linkage disequilibrium (r2 > 0.8) with each other.
Among the other significant signals was a trio of SNPs in TJP1, 15q13.1. These SNPs showed strong negative associations between PGI and minor allele count (rs711355: p = 2.25×10−7, q = 0.08; rs785423: p = 4.86×10−7, q = 0.08; rs813676: p = 4.91×10−7, q = 0.08). These three SNPs were in high LD with each other (R2 > 0.80), but not with other SNPs in the region. Minor allele frequency (MAF) was relatively high for all three SNPs (MAF > 0.33), and exhibited negligible differences by ancestry. But, the significance of the associations was inconsistent in racial\ethnic stratified reanalysis. The AA sample had less significant associations (rs711355: p = 0.02; rs785423: p = 0.19; rs813676: p = 0.19) than the EA sample (rs711355: p = 8.70×10−5; rs785423: p = 1.09×10−5; rs813676: p = 5.73×10−6). Haplotype testing improved the signal for a specific high risk haplotype and in overall haplotype tests for EAs but did not in the AA subsample. The stratified analysis results suggest that the association signal was mainly driven by the EA subsample – thus the association should be regarded as tentative, pending further evidence.
One of the strongest signals related to CGI-S comprised two highly proximate SNPs (~1 kb apart) at PPA2, 4q24. Both of these SNPs showed strong positive associations between CGI-S during risperidone treatment and minor allele count (rs2636697: p = 3.68×10−7, q = 0.06; rs2636719: p = 5.05×10−7, q = 0.09). These SNPs were in high LD (in AA R2 = 0.83 and in EA R2 = 0.84) and had fairly low MAFs (0.01/0.04 in AA/EA). Haplotype testing did not improve the association signal in either subsample.
As shown in Table 2, three additional genome-wide significant SNPs were located within annotated genes and two genome-wide significant SNPs were located in intergenic regions. Secondary analyses of these findings showed that the proportion of variance explained by each SNP was similar in the AA and EA subsamples, that haplotype tests did not improve p-values and that the detected signals were unlikely the results of genotyping errors.
Cross-outcome Analysis of Genome-Wide Significant Markers
In the top portion of Table 3, we evaluated whether variants that were associated with one global impression measure were also nominally associated with the other global impression measure. Four of the 13 SNPs showed cross-outcome association: two SNPs during risperidone treatment and other two during olanzapine treatment. In the bottom of Table 3, we extended our cross-outcome analysis using markers that reached q < 0.25 in two previously published treatment response GWAS in the CATIE sample (14–15). SNP rs17727261 had been previously associated with negative symptom change assessed using the PANSS during risperidone treatment (p = 5.41x10−7; q = 0.134). Here, we found that this SNP was also nominally associated with the both the PGI and CGI-S during risperidone treatment. Three other SNPs (rs11214606, rs6856328 and rs16865258) were nominally associated with global impression severity and were also associated with neurocognitive domain changes, specifically working memory, during olanzapine and quetiapine treatments. The column on the right-hand side of Table 3 shows that the correlations between the outcomes is low to moderate, ranging from −0.10 to 0.38. This suggests that these nominal associations are not merely reflecting the correlation among the outcomes.
Table 3.
Genetic variants that reached q-value < 0.25 with treatment response in CATIE GWAS and also reached nominal significant association with a global impression score (p-value < 0.05). We only considered patients under the same antipsychotic treatment.
| GWAS Result | Secondary Association | Correlationb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Antipsychotic | Phenotype | SNP | Gene | p-value | q-value | Phenotype | p-value | |
| Current | Risperidone | CGI-S | rs8050896 | 3.85E-08 | 0.019 | PGI | 0.009 | 0.18 | |
| PGI | rs785423 | TJP1 | 4.86E-07 | 0.081 | CGI-S | 0.024 | |||
| Olanzapine | CGI-S | rs6688363 | ATP1A2 | 1.59E-07 | 0.078 | PGI | 0.019 | 0.38 | |
| PGI | rs10170310 | SPOPL | 1.03E-07 | 0.051 | CGI-S | 0.009 | |||
| McClay et al. 2011c | Risperidone | PANSSa negative | rs17727261 | CNTNAP5 | 5.41E-07 | 0.134 | PGI | 0.034 | 0.19 |
| CGI-S | 0.002 | −0.08 | |||||||
| McClay et al. 2011d | Olanzapine | Working memory | rs11214606 | DRD2 | 4.84E-07 | 0.081 | PGI | 0.018 | 0.13 |
| Quetiapine | Working memory | rs6856328 | LPHN3 | 8.26E-07 | 0.133 | PGI | 0.010 | −0.10 | |
| rs16865258 | CLDN1 | 1.74E-06 | 0.169 | CGI-S | 0.029 | −0.09 | |||
PANSS – Positive and Negative Syndrome Scale.
Correlation of treatment effect estimates.
McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, et al. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Mol Psychiatry. 2011;16(1):76–85.
McClay JL, Adkins DE, Aberg K, Bukszar J, Khachane AN, Keefe RS, et al. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology. 2011;36(3):616–26.
Discussion
Understanding the genetic variation affecting response to antipsychotic drugs is important to develop novel diagnostic tests to match individual patients with schizophrenia to the most effective and safe medication. In this study, we performed a GWAS of antipsychotic response using the patient rated PGI and clinician rated of the CGI-S severity scales. In total, 13 SNPs achieved genome-wide significance according to our pre-identified criteria (FDR controlled at 0.10 level). Several of the significant SNPs were associated with both the PGI and CGI-S. In comparison to a previous GWAS we carried out in CATIE that used schizophrenia symptoms as measured by the positive and negative syndrome scale (PANSS) as the outcome variable, this study with global impression phenotypes would appear to be more successful. The study using the PANSS found only a single SNP to be significant at a genome-wide level and only three SNP’s in genes with q-values < 0.5 (14).
Our top findings involved PDE4D, PPA2 and TJP1. PDE4D is a cAMP-specific phosphodiesterase widely expressed in human brain. PDE4 inhibitors enhance dopamine D1 receptor signaling (33) and have been suggested to have antipsychotic potential activity (34–35). PDE4D knockout mice show decreased prepulse inhibition, decreased baseline motor activity, and an exaggerated locomotor response to amphetamine supporting a role for PDE4D in psychiatric diseases and striatal function (36). Less information is available for the other top findings: PPA2 and TJP1. The protein encoded by PPA2 (pyrophosphatase (inorganic) 2) is localized to the mitochondrion and catalyzes the hydrolysis of pyrophosphate to inorganic phosphate, which is important for the phosphate metabolism of cells (37). The PPA2 protein has recently been shown to be downregulated in schizophrenia cases (38). TJP1 encodes for a protein located on the cytoplasmic membrane surface of intercellular tight junctions (39) and it may be involved in signal transduction at cell-cell junctions (40).
When examining GWAS results from multiple raters in our study, patient and clinician rated global impression, confidence in the results increases when there is overlap in findings between the raters. In total, 4 SNPs were associated with both patient and clinician rated global impression in their respective GWAS. A SNP in TJP1 and rs8050896 were associated with risperidone treatment efficacy. TJP1 was discussed in the previous paragraph while rs8050896 is over 200kb from the nearest gene. SNPs in SPOPL and ATP1A2 were associated with olanzapine treatment efficacy. SPOPL is considered a protein-coding gene, though little has been published about its function. ATP1A2 encodes for a protein that is responsible for establishing and maintaining the electrochemical gradients of Na+ and K+ ions across the cell plasma membrane. The gradients are essential for electrical excitability of nerve and muscle tissue. ATP1A2 has been previously associated with migraines (41).
The cross-outcome analyses showed that there are both overlapping and unique findings associated with different definitions of treatment response. The overlap in findings using different definitions of treatment response gives us confidence that the signals are truly associated with antipsychotic response. A SNP located in CNTNAP5 was shown to be associated with PANSS negative symptoms and moderately associated with both the PGI and CGI-S severity scores during risperidone treatment. CNTNAP5 is involved in cell adhesion and intercellular communication in the central nervous system (42). CNTNAP5 has previously been associated with bipolar disorder (43) and autism (44). Three genes, DRD2, LPHN3 and CLDN1 had a cross association between working memory, a neurocognitive endophenotype of schizophrenia, and either the CGI-S or the PGI. DRD2 is a well-known candidate for antipsychotic response and is a drug target for olanzapine (45), the antipsychotic in which we see the effects. LPHN3 is known to be involved in cell adhesion and signal transduction, and has been found to be associated with ADHD (46). CLDN1, claudin 1, is recognized to have a major function in cellular permeability (47), specifically in controlling permeability of the blood-brain barrier tight junctions (48). Genetic variants at CLDN1 could conceivably affect drug availability in the brain.
Although there were overlapping GWAS results, most of the associated candidate genes were different for clinician versus patient rating. This can be explained by the fact that our CGI-S and PGI based estimated treatment effects showed low correlations. A previous study has reported high concordance between CGI-S and PGI scores (49) but these authors considered symptom levels rather than changes in symptom levels over the course of treatment. Indeed, we also found high concordance between the two scales across measurement occasions (See Table 1). Our finding that treatment effects were not highly correlated across measurement occasions is consistent with another study comparing the congruence of CGI-S and PGI treatment effects in depression (50). One potential explanation for the low rater agreement is that patients with schizophrenia, especially those with higher symptom severity, may show reduced insight in evaluating improvement in symptom levels after treatment and therefore have different self-rated scores when compared with clinician ratings. Another explanation is that the construct of global improvement is multi-dimensional and captures many different aspects such as social functioning, symptom improvement, etc. It is not uncommon that clinicians and patients focus on different aspects when evaluating treatment effectiveness (e.g. whereas clinicians may be focused on disease symptoms, a patient’s evaluation may be more driven by aspects of well-being). Thus, because the treatment effects show very modest correlations, we would expect to find unique associations for each of the raters.
One potential way to ascertain the overlap in genetic findings for the CGI-S and PGI would be to combine them into a composite score. In order to form a combined measure, one would have to assume that clinicians and patients are assessing similar aspects of the disease. Research has shown, however, that patients and clinicians differ in how they conceptualize the disorder (5, 9). The secondary association analyses essentially replaces a composite score approach. These analyses do not suffer from the same problems as composite score approaches as they do not assume that the clinician and patient ratings assess the same clinical construct. Thus, rather than assuming that SNPs effect CGI-S and PGI measures through a shared underlying clinical construct, the secondary analyses would detect any “pleiotropic” effects of SNPs impacting the different aspects of clinical functions assessed by CGI-S and PGI measures.
Another potential limitation is the use of a threshold of p-value less than 0.05 as the criterion for determining whether there is a secondary association of a SNP with another outcome (See Table 3). Although such a threshold is too liberal for declaring significance in a GWAS study because of the many tests that are performed, we would argue that this threshold is acceptable for a secondary analysis because the two measures assess a similar phenotype, the p-values from the secondary association are expected to be more significant than expected under the null. Also, a higher p-value threshold can be used because of the much smaller number of tests and/or the high prior probability that the SNP under consideration has an effect. Furthermore, due to the winner’s curse phenomenon, the effect sizes of significant findings from the GWAS will be overestimated (51–52). In a secondary association, however, these estimates will “shrink” toward the true effect size, and the same SNP will yield much more modest p-values. Because of these reasons, it is reasonable to use a smaller p-value threshold for declaring significance in the secondary association.
As with any genetic associations, our findings require replication in an independent sample and functional validation. Thus, it is premature to suggest direct clinical applications of our findings for prescribing antipsychotics. Rather, actualizing the promise of pharmacogenomics and translating academic findings into clinical applications will require a cumulative process of aggregating and jointly considering large bodies of evidence using meta-analytic and data integration techniques. To facilitate this process we provide all p-values (www.pharmacy.vcu.edu/biomarker) as a resource for investigators with the requisite samples to carry out replication or meta-analysis.
The present study demonstrates the potential of GWAS to discover genes associated with antipsychotic treatment response. A better understanding of these mechanisms and the role of specific polymorphisms may eventually help to personalize antipsychotic medication in order to more rationally and efficiently determine the optimal treatment for each patient.
Acknowledgments
Grant support: S.L.C. was supported by NIDA grant DA026119 and NIAAA grant K01AA021266. D.E.A. was supported by NIMH grant K01MH093731.
Footnotes
Disclosures: Authors have nothing to disclose.
References
- 1.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374(9690):635–45. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 2.Guy W. DHEW Publ No ADM 76-338. National Institute of Mental Health; Rockville, MD: 1976. Clinical Global Impressions ECDEU Assesment Manual fo Psychopharmacology, Revised; pp. 218–22. [Google Scholar]
- 3.Bowie CR, Twamley EW, Anderson H, Halpern B, Patterson TL, Harvey PD. Self-assessment of functional status in schizophrenia. Journal of Psychiatric Research. 2007;41(12):1012–8. doi: 10.1016/j.jpsychires.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray EJ. Measurement issues in the evaluation of psychopharmacological therapy. In: Fisher S, Greenberg RP, editors. The limits of biological treatments for psychological distress: Comparisons with psychotherapy and placebo. Hillsdale, NJ, England: Lawrence Erlbaum Associates, Inc; 1989. pp. 39–67. [Google Scholar]
- 5.Lindstrom E, Jedenius E, Levander S. A symptom Self-rating Scale for Schizophrenia (4S): Psychometric properties, reliability and validity. Nordic Journal of Psychiatry. 2009;63(5):368–U4. doi: 10.1080/08039480902807298. [DOI] [PubMed] [Google Scholar]
- 6.Lincoln TM, Lullmann E, Rief W. Correlates and long-term consequences of poor insight in patients with schizophrenia. A systematic review. Schizophrenia Bulletin. 2007;33(6):1324–42. doi: 10.1093/schbul/sbm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard JJ, Mueser KT, Bellack AS. SELF-RATED AND INTERVIEW-RATED NEGATIVE MOOD STATES IN SCHIZOPHRENIA - THEIR CONVERGENCE AND PREDICTION OF THOUGHT DISTURBANCE. Journal of Psychopathology and Behavioral Assessment. 1992;14(3):277–91. [Google Scholar]
- 8.Berenbaum H, Oltmanns TF. Emotional experience and expression in schizophrenia and depression. J Abnorm Psychol. 1992;101(1):37–44. doi: 10.1037//0021-843x.101.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung HY, Hwang SSH, Yi JS, Kim Y, Kim YS. Clinician-rated functioning and patient-rated quality of life in schizophrenia: Implications of their correspondence for psychopathology and side effects. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2010;34(1):225–30. doi: 10.1016/j.pnpbp.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Perkins DO, Wyatt RJ, Bartko JJ. Penny-wise and pound-foolish: The impact of measurement error on sample size requirements in clinical trials. Biological Psychiatry. 2000;47(8):762–6. doi: 10.1016/s0006-3223(00)00837-4. [DOI] [PubMed] [Google Scholar]
- 11.Judd C, Ryan CS, Park B. Accuracy in the judgement of in-group and out-group variability. Journal of Personality and Social Psychology. 1991;61:366–79. doi: 10.1037//0022-3514.61.3.366. [DOI] [PubMed] [Google Scholar]
- 12.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13(2):261–76. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 13.Levine SZ, Rabinowitz J, Engel R, Etschel E, Leucht S. Extrapolation between measures of symptom severity and change: an examination of the PANSS and CGI. Schizophr Res. 2008;98(1–3):318–22. doi: 10.1016/j.schres.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 14.McClay JL, Adkins DE, Aberg K, Stroup S, Perkins DO, Vladimirov VI, et al. Genome-wide pharmacogenomic analysis of response to treatment with antipsychotics. Molecular Psychiatry. 2011;16(1):76–85. doi: 10.1038/mp.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClay JL, Adkins DE, Aberg K, Bukszar J, Khachane AN, Keefe RS, et al. Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology. 2011;36(3):616–26. doi: 10.1038/npp.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, et al. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353(12):1209–23. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 17.First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV Axis I Disorders--Administration Booklet. Washington D.C: American Psychiatric Press, Inc; 1994. [Google Scholar]
- 18.van den Oord EJ, Adkins DE, McClay J, Lieberman J, Sullivan PF. A systematic method for estimating individual responses to treatment with antipsychotics in CATIE. SchizophrRes. 2009;107(1):13–21. doi: 10.1016/j.schres.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aberg K, Adkins DE, Bukszar J, Webb BT, Caroff SN, Miller del D, et al. Genomewide association study of movement-related adverse antipsychotic effects. Biol Psychiatry. 2010;67(3):279–82. doi: 10.1016/j.biopsych.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adkins DE, Aberg K, McClay JL, Bukszar J, Zhao Z, Jia P, et al. Genomewide pharmacogenomic study of metabolic side effects to antipsychotic drugs. Mol Psychiatry. 2011;16(3):321–32. doi: 10.1038/mp.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinheiro JC, Bates DM. Mixed-effects models in S and S-plus. New York, NY: Springer; 2000. [Google Scholar]
- 22.Willett JB, Singer JD, Martin NC. The design and analysis of longitudinal studies of development and psychopathology in context: Statistical models and methodological recommendations. Development and Psychopathology. 1998;10(2):395–426. doi: 10.1017/s0954579498001667. [DOI] [PubMed] [Google Scholar]
- 23.Robinson GK. That BLUP is good thing: the estimation of random effects. Statistical Science. 1991;6(1):15–32. [Google Scholar]
- 24.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13(6):570–84. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 27.Brown BW, Russell K. Methods correcting for multiple testing: operating characteristics. Statistics in Medicine. 1997;16(22):2511–28. doi: 10.1002/(sici)1097-0258(19971130)16:22<2511::aid-sim693>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Lei SF, Yang TL, Tan LJ, Chen XD, Guo Y, Guo YF, et al. Genome-wide association scan for stature in Chinese: evidence for ethnic specific loci. Hum Genet. 2009;125(1):1–9. doi: 10.1007/s00439-008-0590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu YZ, Guo YF, Wang L, Tan LJ, Liu XG, Pei YF, et al. Genome-wide association analyses identify SPOCK as a key novel gene underlying age at menarche. PLoS Genet. 2009;5(3):e1000420. doi: 10.1371/journal.pgen.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, et al. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009;84(1):35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19(10):537–42. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Black MA. A note on the adaptive control of false discovery rates. J R Stat Soc B. 2004;66:297–304. [Google Scholar]
- 33.Kuroiwa M, Snyder GL, Shuto T, Fukuda A, Yanagawa Y, Benavides DR, et al. Phosphodiesterase 4 inhibition enhances the dopamine D1 receptor/PKA/DARPP-32 signaling cascade in frontal cortex. Psychopharmacology (Berl) 2011 doi: 10.1007/s00213-011-2436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanes SJ, Tokarczyk J, Siegel SJ, Bilker W, Abel T, Kelly MP. Rolipram: a specific phosphodiesterase 4 inhibitor with potential antipsychotic activity. Neuroscience. 2007;144(1):239–46. doi: 10.1016/j.neuroscience.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halene TB, Siegel SJ. Antipsychotic-like properties of phosphodiesterase 4 inhibitors: evaluation of 4-(3-butoxy-4-methoxybenzyl)-2-imidazolidinone (RO-20-1724) with auditory event-related potentials and prepulse inhibition of startle. J Pharmacol Exp Ther. 2008;326(1):230–9. doi: 10.1124/jpet.108.138586. [DOI] [PubMed] [Google Scholar]
- 36.Siuciak JA, McCarthy SA, Chapin DS, Martin AN. Behavioral and neurochemical characterization of mice deficient in the phosphodiesterase-4B (PDE4B) enzyme. Psychopharmacology (Berl) 2008;197(1):115–26. doi: 10.1007/s00213-007-1014-6. [DOI] [PubMed] [Google Scholar]
- 37.Curbo S, Lagier-Tourenne C, Carrozzo R, Palenzuela L, Lucioli S, Hirano M, et al. Human mitochondrial pyrophosphatase: cDNA cloning and analysis of the gene in patients with mtDNA depletion syndromes. Genomics. 2006;87(3):410–6. doi: 10.1016/j.ygeno.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Mistry M, Gillis J, Pavlidis P. Genome-wide expression profiling of schizophrenia using a large combined cohort. Mol Psychiatry. 2012 doi: 10.1038/mp.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohandas TK, Chen XN, Rowe LB, Birkenmeier EH, Fanning AS, Anderson JM, et al. Localization of the tight junction protein gene TJP1 to human chromosome 15q13, distal to the Prader-Willi/Angelman region, and to mouse chromosome 7. Genomics. 1995;30(3):594–7. doi: 10.1006/geno.1995.1281. [DOI] [PubMed] [Google Scholar]
- 40.Ebnet K, Schulz CU, Meyer Zu Brickwedde MK, Pendl GG, Vestweber D. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J Biol Chem. 2000;275(36):27979–88. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- 41.Santoro L, Manganelli F, Fortunato MR, Soldovieri MV, Ambrosino P, Iodice R, et al. A new Italian FHM2 family: clinical aspects and functional analysis of the disease-associated mutation. Cephalalgia. 2011;31(7):808–19. doi: 10.1177/0333102411399351. [DOI] [PubMed] [Google Scholar]
- 42.Traut W, Weichenhan D, Himmelbauer H, Winking H. New members of the neurexin superfamily: multiple rodent homologues of the human CASPR5 gene. Mamm Genome. 2006;17(7):723–31. doi: 10.1007/s00335-005-0157-1. [DOI] [PubMed] [Google Scholar]
- 43.Djurovic S, Gustafsson O, Mattingsdal M, Athanasiu L, Bjella T, Tesli M, et al. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J Affect Disord. 2010;126(1–2):312–6. doi: 10.1016/j.jad.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Pagnamenta AT, Bacchelli E, de Jonge MV, Mirza G, Scerri TS, Minopoli F, et al. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol Psychiatry. 2010;68(4):320–8. doi: 10.1016/j.biopsych.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapur S, Zipursky RB, Remington G. Clinical and theoretical implications of 5-HT2 and D2 receptor occupancy of clozapine, risperidone, and olanzapine in schizophrenia. Am J Psychiatry. 1999;156(2):286–93. doi: 10.1176/ajp.156.2.286. [DOI] [PubMed] [Google Scholar]
- 46.Domene S, Stanescu H, Wallis D, Tinloy B, Pineda DE, Kleta R, et al. Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(1):11–8. doi: 10.1002/ajmg.b.31141. [DOI] [PubMed] [Google Scholar]
- 47.Tsukita S, Furuse M. Pores in the wall: claudins constitute tight junction strands containing aqueous pores. J Cell Biol. 2000;149(1):13–6. doi: 10.1083/jcb.149.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeiffer F, Schafer J, Lyck R, Makrides V, Brunner S, Schaeren-Wiemers N, et al. Claudin-1 induced sealing of blood-brain barrier tight junctions ameliorates chronic experimental autoimmune encephalomyelitis. Acta Neuropathol. 2011;122(5):601–14. doi: 10.1007/s00401-011-0883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinert T, Eisele F, Langle G, Albani C, Flammer E, Borbe R. PGI-I (patient’s global impression) as an outcome and quality indicator of psychiatric in-patient treatment: results and concordance with doctor’s assessments. Psychiatr Prax. 2010;37(7):343–9. doi: 10.1055/s-0030-1248444. [DOI] [PubMed] [Google Scholar]
- 50.Forkmann T, Scherer A, Boecker M, Pawelzik M, Jostes R, Gauggel S. The Clinical Global Impression Scale and the influence of patient or staff perspective on outcome. BMC Psychiatry. 2011;11:83. doi: 10.1186/1471-244X-11-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 52.Goring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69(6):1357–69. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]