Abstract
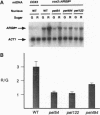
Genetic code differences prevent expression of nuclear genes within Saccharomyces cerevisiae mitochondria. To bridge this gap a synthetic gene, ARG8m, designed to specify an arginine biosynthetic enzyme when expressed inside mitochondria, has been inserted into yeast mtDNA in place of the COX3 structural gene. This mitochondrial cox3::ARG8m gene fully complements a nuclear arg8 deletion at the level of cell growth, and it is dependent for expression upon nuclear genes that encode subunits of the COX3 mRNA-specific translational activator. Thus, cox3::ARG8m serves as a mitochondrial reporter gene. Measurement of cox3::ARG8m expression at the levels of steady-state protein and enzymatic activity reveals that glucose repression operates within mitochondria. The levels of this reporter vary among strains whose nuclear genotypes lead to under- and overexpression of translational activator subunits, in particular Pet494p, indicating that mRNA-specific translational activation is a rate-limiting step in this organellar system. Whereas the steady-state level of cox3::ARG8m mRNA was also glucose repressed in an otherwise wild-type strain, absence of translational activation led to essentially repressed mRNA levels even under derepressing growth conditions. Thus, the mRNA is stabilized by translational activation, and variation in its level may be largely due to modulation of translation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Berry J. O., Breiding D. E., Klessig D. F. Light-mediated control of translational initiation of ribulose-1, 5-bisphosphate carboxylase in amaranth cotyledons. Plant Cell. 1990 Aug;2(8):795–803. doi: 10.1105/tpc.2.8.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. G., Costanzo M. C., Fox T. D. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Feb;14(2):1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. A point mutation in the 5'-untranslated leader that affects translational activation of the mitochondrial COX3 mRNA. Curr Genet. 1995 Jun;28(1):60–66. doi: 10.1007/BF00311882. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986 Nov;6(11):3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Suppression of a defect in the 5' untranslated leader of mitochondrial COX3 mRNA by a mutation affecting an mRNA-specific translational activator protein. Mol Cell Biol. 1993 Aug;13(8):4806–4813. doi: 10.1128/mcb.13.8.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Seaver E. C., Fox T. D. At least two nuclear gene products are specifically required for translation of a single yeast mitochondrial mRNA. EMBO J. 1986 Dec 20;5(13):3637–3641. doi: 10.1002/j.1460-2075.1986.tb04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Staples R. R. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int Rev Cytol. 1994;152:145–181. doi: 10.1016/s0074-7696(08)62556-5. [DOI] [PubMed] [Google Scholar]
- Dowhan W., Bibus C. R., Schatz G. The cytoplasmically-made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO J. 1985 Jan;4(1):179–184. doi: 10.1002/j.1460-2075.1985.tb02334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Folley L. S., Mulero J. J., McMullin T. W., Thorsness P. E., Hedin L. O., Costanzo M. C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- Gallwitz D., Seidel R. Molecular cloning of the actin gene from yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Mar 11;8(5):1043–1059. doi: 10.1093/nar/8.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham N. W., Boynton J. E., Hauser C. R. Translational regulation of gene expression in chloroplasts and mitochondria. Annu Rev Genet. 1994;28:71–93. doi: 10.1146/annurev.ge.28.120194.000443. [DOI] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit Rev Biochem Mol Biol. 1995;30(2):121–164. doi: 10.3109/10409239509085141. [DOI] [PubMed] [Google Scholar]
- Haffter P., Fox T. D. Suppression of carboxy-terminal truncations of the yeast mitochondrial mRNA-specific translational activator PET122 by mutations in two new genes, MRP17 and PET127. Mol Gen Genet. 1992 Oct;235(1):64–73. doi: 10.1007/BF00286182. [DOI] [PubMed] [Google Scholar]
- Haffter P., McMullin T. W., Fox T. D. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991 Feb;127(2):319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg H., Boyen A., Crabeel M., Glansdorff N. Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferase: evolutionary relationship with ornithine aminotransferase. Gene. 1990 May 31;90(1):69–78. doi: 10.1016/0378-1119(90)90440-3. [DOI] [PubMed] [Google Scholar]
- Jauniaux J. C., Urrestarazu L. A., Wiame J. M. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J Bacteriol. 1978 Mar;133(3):1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman K., Puccini C. J. A PCR-mediated gene synthesis strategy involving the assembly of oligonucleotides representing only one of the strands. Biotechniques. 1992 Mar;12(3):392–398. [PubMed] [Google Scholar]
- Marykwas D. L., Fox T. D. Control of the Saccharomyces cerevisiae regulatory gene PET494: transcriptional repression by glucose and translational induction by oxygen. Mol Cell Biol. 1989 Feb;9(2):484–491. doi: 10.1128/mcb.9.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin T. W., Fox T. D. COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J Biol Chem. 1993 Jun 5;268(16):11737–11741. [PubMed] [Google Scholar]
- McMullin T. W., Haffter P., Fox T. D. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990 Sep;10(9):4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D. M., Getz G. S. Steady state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisiae under catabolite repression and derepression. J Biol Chem. 1986 Sep 5;261(25):11816–11822. [PubMed] [Google Scholar]
- Pinkham J. L., Dudley A. M., Mason T. L. T7 RNA polymerase-dependent expression of COXII in yeast mitochondria. Mol Cell Biol. 1994 Jul;14(7):4643–4652. doi: 10.1128/mcb.14.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutre C. G., Fox T. D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987 Apr;115(4):637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Chen X., Kindle K. L., Stern D. B. Function of the Chlamydomonas reinhardtii petd 5' untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 1994 Oct;6(4):503–512. doi: 10.1046/j.1365-313x.1994.6040503.x. [DOI] [PubMed] [Google Scholar]
- Staub J. M., Maliga P. Translation of psbA mRNA is regulated by light via the 5'-untranslated region in tobacco plastids. Plant J. 1994 Oct;6(4):547–553. doi: 10.1046/j.1365-313x.1994.6040547.x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D. Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature. 1990 Jul 26;346(6282):376–379. doi: 10.1038/346376a0. [DOI] [PubMed] [Google Scholar]
- Ulery T. L., Jang S. H., Jaehning J. A. Glucose repression of yeast mitochondrial transcription: kinetics of derepression and role of nuclear genes. Mol Cell Biol. 1994 Feb;14(2):1160–1170. doi: 10.1128/mcb.14.2.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger G., Costanzo M. C., Fox T. D. Analysis of the Saccharomyces cerevisiae mitochondrial COX3 mRNA 5' untranslated leader: translational activation and mRNA processing. Mol Cell Biol. 1995 Jun;15(6):3291–3300. doi: 10.1128/mcb.15.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. P. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- Zerges W., Rochaix J. D. The 5' leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. Mol Cell Biol. 1994 Aug;14(8):5268–5277. doi: 10.1128/mcb.14.8.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]