Ribosomal proteins often consist of a globular domain as well as a nonglobular extension region that snakes its way into the subunit to stabilize the tightly packed RNA. The functionally important S12 protein of the small subunit has a globular C-terminal domain that binds near the decoding center and an N-terminal extension that traverses from the solvent to the interface surface of the subunit. Here the authors examine the role of this extension domain in SSU assembly and function using both in vivo and in vitro approaches. Partial N-terminal deletions of S12 result in slow growth and cold sensitivity; at a molecular level, these strains have increased levels of free SSUs and immature 16S rRNA, both hallmarks of a defect in SSU biogenesis.
Keywords: ribosome, small subunit, biogenesis, assembly, S12, intrinsically disordered proteins
Abstract
The small subunit (SSU) of the ribosome of E. coli consists of a core of ribosomal RNA (rRNA) surrounded peripherally by ribosomal proteins (r-proteins). Ten of the 15 universally conserved SSU r-proteins possess nonglobular regions called extensions. The N-terminal noncanonically structured extension of S12 traverses from the solvent to intersubunit surface of the SSU and is followed by a more C-terminal globular region that is adjacent to the decoding center of the SSU. The role of the globular region in maintaining translational fidelity is well characterized, but a role for the S12 extension in SSU structure and function is unknown. We examined the effect of stepwise truncation of the extension of S12 in SSU assembly and function in vitro and in vivo. Examination of in vitro assembly in the presence of sequential N-terminal truncated variants of S12 reveals that N-terminal deletions of greater than nine amino acids exhibit decreased tRNA-binding activity and altered 16S rRNA architecture particularly in the platform of the SSU. While wild-type S12 expressed from a plasmid can rescue a genomic deletion of the essential gene for S12, rpsl; N-terminal deletions of S12 exhibit deleterious phenotypic consequences. Partial N-terminal deletions of S12 are slow growing and cold sensitive. Strains bearing these truncations as the sole copy of S12 have increased levels of free SSUs and immature 16S rRNA as compared with the wild-type S12. These differences are hallmarks of SSU biogenesis defects, indicating that the extension of S12 plays an important role in SSU assembly.
INTRODUCTION
Universally conserved small subunit (SSU, 30S) ribosomal protein (r-protein) S12 is composed of two distinct structural elements in all domains of life: an N-terminal nonglobular extension of varying length and sequence, and a conserved C-terminal globular region (see Fig. 1; Smith et al. 2008; Ben-Shem et al. 2010). S12 is essential for growth in organisms ranging from E. coli (Sharma et al. 2007; Shoji et al. 2011) to Danio rerio (zebrafish) (Zhang and Lin 2009) and is a key mediator of fidelity of translation in both prokaryotes and eukaryotes (Alksne et al. 1993). Examination of the crystal structure of E. coli SSUs (Schuwirth et al. 2005) reveals that the N-terminus of the protein binds the solvent surface of the SSU, with the extension contacting the rRNA dense regions to end in a C-terminal globular region localized at the intersubunit face of the SSU (see Fig. 1A). The length and localization of the extension as well as hints of its possible functional significance (Agarwal et al. 2011) led us to examine the role of the extension of S12 in establishing SSU structure during assembly.
FIGURE 1.
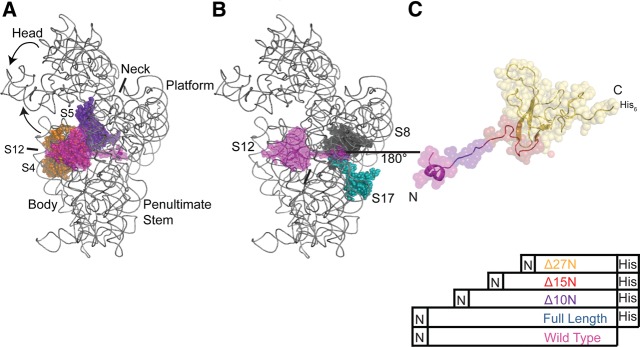
Structural relationship of S12 to other SSU components. (A) Positions of S12 (magenta), S4 (orange), and S5 (violet) in the three-dimensional structure of the E. coli 30S subunit. 16S rRNA is gray and other r-proteins omitted for clarity. All figures containing three-dimensional structures were prepared using PyMOL and Protein Data Bank file 2AW7 (Schuwirth et al. 2005). (B) Location of S12 (magenta), S8 (black), and S17 (teal) in the three-dimensional structure of the 30S subunit. 16S rRNA is gray and other r-proteins omitted for clarity. (C) Three-dimensional structure of S12 as found in the 30S subunit, rotated 180° with respect to B, with the variants color coded. S12WT is magenta, S12Full Length is blue, S12▵10N is purple, S12▵15N is red, and S12▵27N is orange.
The region of the SSU that functions in tRNA binding and decoding lies at the intersection of three structural domains: the body, platform, head, and penultimate and ultimate stems (Woese et al. 1980; Moazed and Noller 1990; Powers and Noller 1991; Alksne et al. 1993; Lee et al. 1997; Lodmell and Dahlberg 1997; Clemons et al. 1999; Yusupov et al. 2001). The localization of the globular region of S12 proximal to the decoding center is unique among SSU r-proteins, and has a well-characterized role in maintaining fidelity of translation (Gorini and Kataja 1964b; Lodmell and Dahlberg 1997; Clemons et al. 1999; Ogle et al. 2002; Schuwirth et al. 2005). Most of the contacts between the N-terminal segment of S12 and 16S rRNA lie within the platform and it also interacts with the body of the 30S subunit (see Fig. 1A; Allers and Shamoo 2001; Schuwirth et al. 2005). In addition to binding rRNA, the extension also interacts with S8 and S17, and these three r-proteins participate in the assembly of the body, shoulder, and platform of the SSU (see Fig. 1B; Stern et al. 1988a,b; Svensson et al. 1988; Schuwirth et al. 2005; Calidas and Culver 2011). Little is known of the role of the extension in the structure and function of the SSU. The widespread contacts made by the extension with 16S rRNA and other r-proteins led us to hypothesize that it plays a role in aligning distant structural elements of the SSU.
More than one-third of all r-proteins possess extensions (Brodersen et al. 2002; Schuwirth et al. 2005) and it has been hypothesized that extensions scaffold tertiary structure (Ban et al. 2000; Wimberly et al. 2000; Wilson and Nierhaus 2005). However, the exact role of the extensions in bacterial ribosome function and assembly is unclear. Of the r-proteins with extensions that have been studied, complete truncation of the extension of E. coli L20 has the most profound effect, leading to a dominant lethal phenotype, while partial deletions resulted in slow growth due to defects in large subunit (LSU or 50S) assembly (Guillier et al. 2005). In contrast, deletion of the long loops of E. coli L4 and L22 had no effect on the assembly of the 50S subunit (Zengel et al. 2003). In yeast, mutations in the C-terminal extension of r-protein S14, a homolog of bacterial r-protein S11 blocked maturation of precursor SSU particles (Jakovljevic et al. 2004). Thus, a common role and significance of r-protein extensions in ribosomal subunit biogenesis and function remains unclear.
In this work we examined the role of the extension of S12 in SSU assembly and function in vitro and in vivo. We created multiple truncations of the extension of S12 starting from the N-terminus (Figs. 1C, 4A, below): truncation within the two-turn helix (S12▵6N), truncation past the two turn helix (S12▵10N, S12▵11N), truncation of half the extension (S12▵15N), truncation of three-fourths of the extension (S12▵24N), and truncation of the entire extension (S12▵27N). SSUs reconstituted in vitro with the full complement of recombinant r-proteins and different S12 variants exhibited diminishing functionality with a decrease in length of the extension (Fig. 2). In addition, minimal ribonucleoprotein particles (RNPs) reconstituted in vitro in the presence of S12▵15N and S12▵27N show altered structure of 16S rRNA when compared with RNPs containing full-length S12, notably in the vicinity of the central pseudoknot (Fig. 3). Thus, truncation of the extension alters SSU structure and function in vitro.
FIGURE 4.
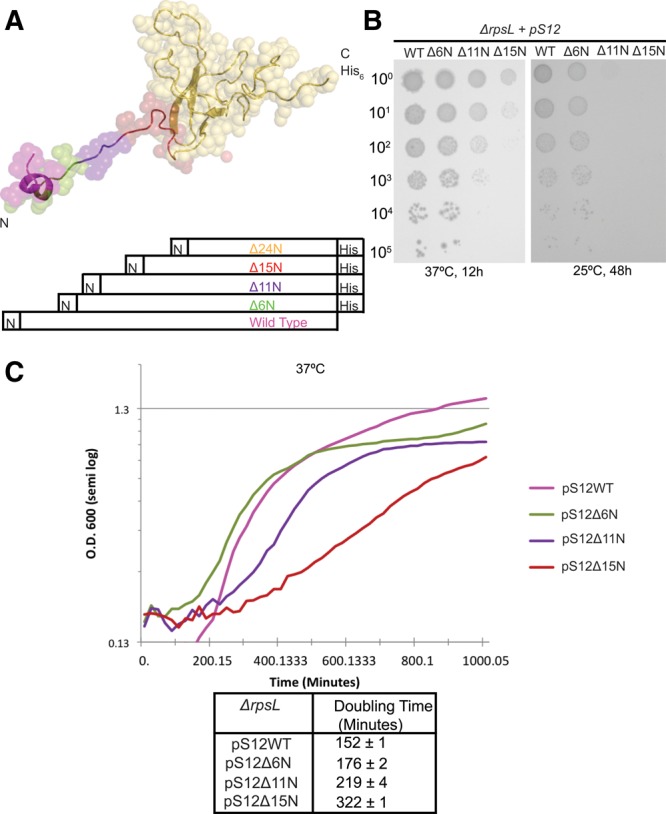
In vivo analysis of strains bearing truncations of S12. (A) Three-dimensional structure of S12 as found in the 30S subunit, with the positions of truncations color coded as in Figure 2, except that S12▵6N is green. (B) Serial dilution analysis of the growth of ▵rpsl strains dependent upon WT and truncated proteins at 37°C and 25°C. Overnight cultures were diluted to equal OD600 and then serially diluted by 10-fold increments. These were spotted on LB supplemented with kanamycin (50 µg/mL), ampicillin (100 µg/mL), and arabinose (0.2%). (C) Growth of ▵rpsl strains in liquid media was measured in a Bioscreen automatic growth analyzer (Growth Curves) and plotted on a semi-log plot. Doubling times are presented in tabular form.
FIGURE 2.
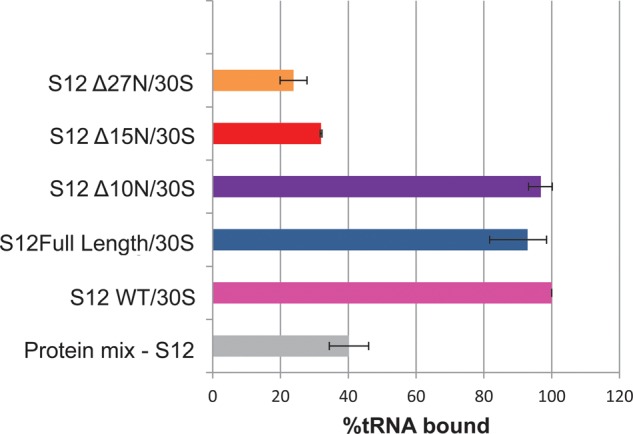
Analysis of the effect of truncation of S12's extension in vitro. Truncation of the extension by more than 10 amino acids affects the function of 30S particles in vitro. Activity of particles is measured by the ability to bind labeled tRNA. Results were normalized to tRNA binding by 30S subunits reconstituted with S12WT.
FIGURE 3.
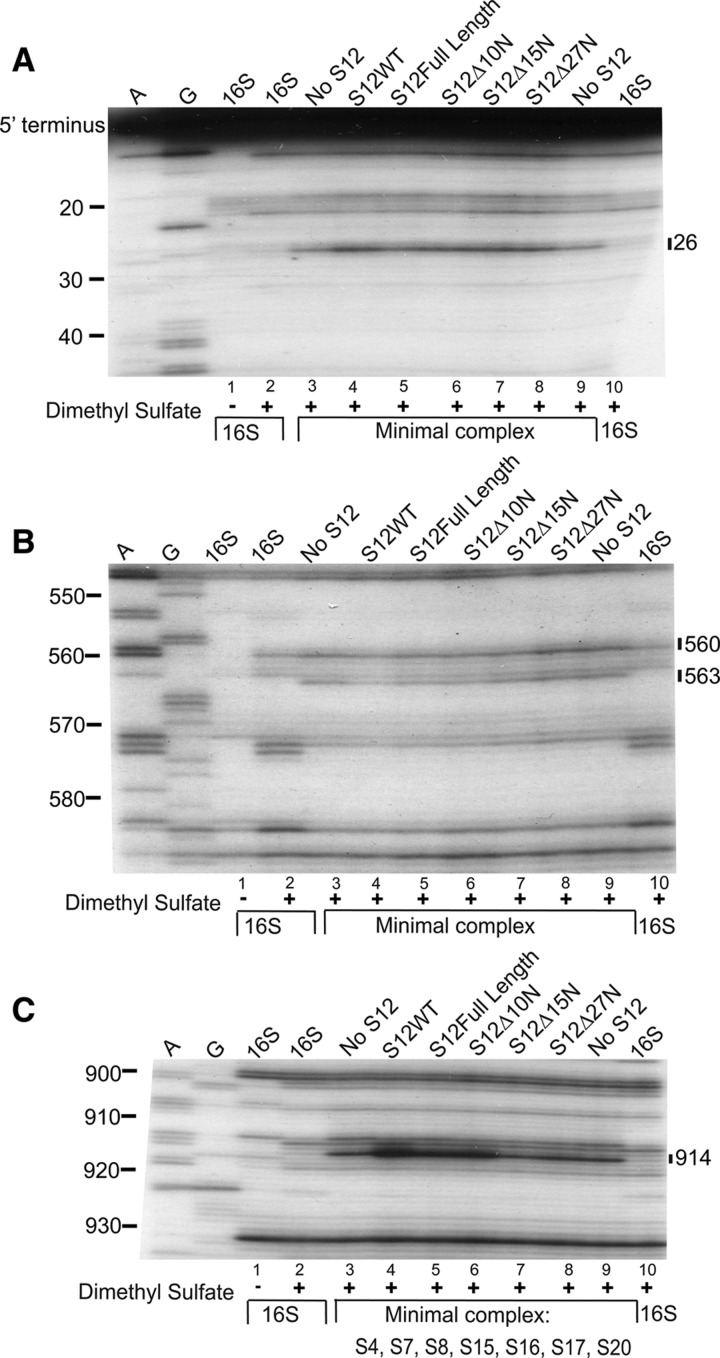
Primer extension analysis of RNPs containing S12 variants. Sequencing gel of 16S rRNA in the following reactions: untreated 16S rRNA (lane 1); dimethyl sulfate treated 16S rRNA (lanes 2,10); dimethyl sulfate-treated minimal complex (S4, S7, S8, S15, S16, S17, S20, 16S rRNA) (lanes 3,9); dimethyl sulfate-treated minimal complex with S12WT (lane 4), S12 full-length (lane 5), S12▵10N (lane 6), S12▵15N (lane 7), and S12▵27N (lane 8). A and G are sequencing lanes. Bars indicate changes in methylation patterns compared with complexes lacking S12 (lanes 3,9). The primers utilized in A, B, and C are 232, 683, and 1046, respectively.
We further examined whether the extension played a role in SSU assembly in vivo. As the gene encoding S12 (rpsL) is essential (Sharma et al. 2007; Shoji et al. 2011), we used a genomic deletion strain (▵rpsl pSTR) whose viability is dependent upon plasmid expression of wild-type S12 (pSTR). The wild-type plasmid was then replaced by plasmids expressing S12 variants (pS12▵6N, pS12▵11N, pS12▵15N, pS12▵24N) by plasmid shuffle to create ▵rpsl pS12▵6N, ▵rpsl pS12▵11N, and ▵rpsl pS12▵15N (Fig. 4A). Diminution of the N-terminal segment correlated with increasing growth defects and subunit profile abnormalities (Figs. 4B,C, 5A,B). Precursor 16S rRNA (17S rRNA) is observed in SSU fractions from the Δrpsl pS12▵15N strain, also to a lesser degree in ▵rpsl pS12▵6N and ▵rpsl pS12▵11N (Fig. 5C). Data from both in vitro and in vivo studies suggest that the N-terminal extension of S12 plays a critical role in assembly of E. coli SSUs and indicates that r-protein extensions can greatly alter rRNA maturation and architecture.
FIGURE 5.
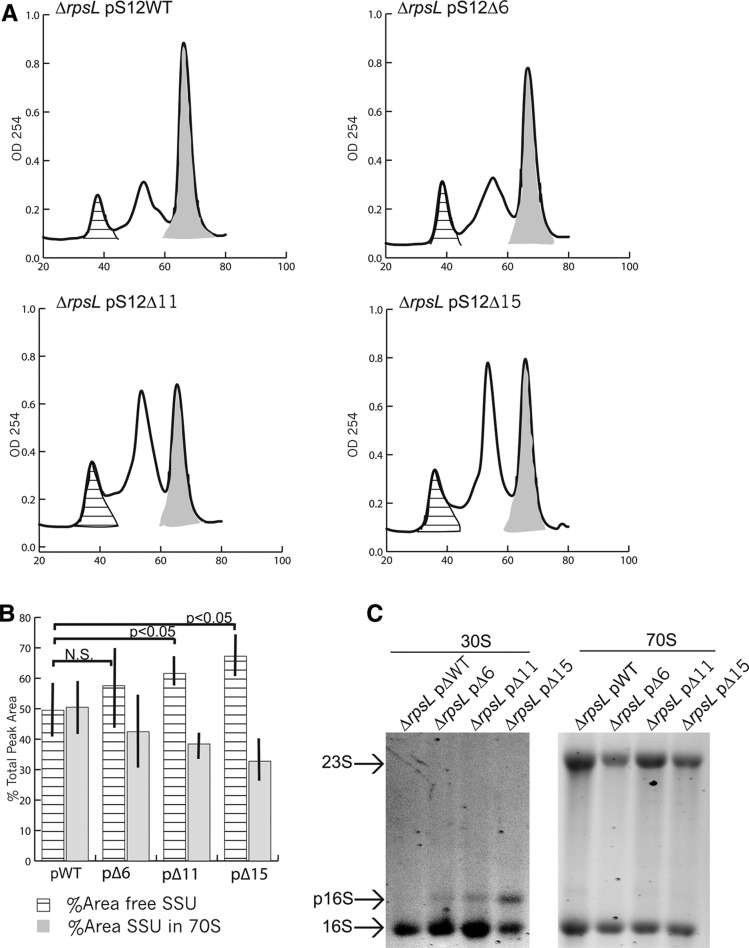
In vivo analysis of ribosomes formed in strains bearing truncations of S12. (A) Representative sucrose gradient sedimentation profiles of ribosomal subunits from ▵rpsl strains bearing pS12WT or pS12ΔXN. The 10%–40% gradients were pumped from the top to the bottom using a Biocomp Piston Gradient Fractionator and elution of the small and large subunits followed by 70S ribosomes was detected by monitoring at OD254. Area under the curve was calculated from the cross-hatched (free SSU) and gray-shaded regions (70S) in each profile. (B) Comparison between strains of percent area occupied by free subunits (cross-hatched) and subunits within ribosomes (gray) is represented by a bar graph. (C) Agarose gel analysis of RNA content of free SSUs and ribosomes.
RESULTS
The N-terminal extension of S12 plays a role in SSU tRNA binding in vitro
We wished to determine whether the extension of S12 modulates SSU formation, its function, and 16S rRNA folding. To achieve this end, we generated a series of N-terminal truncations of S12. Four different C-terminal His-tagged constructs were made for overexpression and purification (S12Full Length, S12▵10N, S12▵15N, and S12▵27N) (Fig. 1C). The C-terminal His-tag was included to aid in purification of the mutant proteins from possible contamination with endogenous wild-type S12. Wild-type untagged (S12WT) S12 was used as a control to monitor whether the C-terminal His-tag altered S12 function. The overexpressed, purified S12 variants were then assembled with 16S rRNA in the presence of different r-proteins to examine the role of S12 in assembly and function of the 30S subunit in vitro.
Functional, active SSUs bind tRNAs in an message-dependent manner in vitro making this is an ideal assay to assess the activity of SSUs reconstituted with S12 variants (Fig. 2). In this assay, labeled tRNA is supplied in excess, so that each tRNA-binding site of the SSU should be saturated. S12WT, S12Full Length, and truncated proteins (S12▵10N, S12▵15N, and S12▵27N) were reconstituted with the remaining SSU r-proteins and 16S rRNA (S12WT/30S, S12Full Length/30S, S12▵10N/30S, S12▵15N/30S, S12▵27N/30S). S12WT/30S and S12Full Length/30S have comparable binding activity, demonstrating that the C-terminal tag does not affect S12 activity or the incorporation of the resulting SSU. Complexes assembled with S12▵10N also bind tRNA comparable to S12WT/30S and S12Full Length/30S. On the other hand, S12▵15N/30S and S12▵27N/30S are as deficient at binding tRNA as RNPs reconstituted in the absence of S12 (Protein mix-S12). This change in functional capacity could result from reduced incorporation of shorter S12 into RNPs and/or the improper assembly or folding of particles when the extension is reduced in length by half or more.
N-terminal truncation of S12 results in changes in 16S rRNA conformation
Given differences in function of 30S subunits with different S12 constructs, we next examined the effect of the truncation of S12 on the architecture of 16S rRNA using chemical probing and primer extension. Differences in 16S rRNA reactivity were observed when S12 was added in vitro to complexes containing 16S rRNA and early binding r-proteins S4, S7, S8, S15, S16 S17, and S20 (Stern et al. 1988b); however, upon addition of later binding r-proteins to these complexes, the changes attributed to S12 are no longer observed. Thus, probing of these minimal complexes (16S rRNA and S4, S7, S8, S15, S16 S17, and S20) ±S12 allows structural changes associated with S12 binding to be examined. RNPs were assembled with minimal complex and different variants of S12 (WT, Full-Length, ▵10N, ▵15N, and ▵27N), subsequently probed with dimethyl sulfate, and 16S rRNA was analyzed by primer extension.
Addition of S12WT to the minimal complex (Fig. 3A, cf. lanes 3,9 with lane 4) results in an enhancement of the reactivity of A26, proximal to the 5′ terminus of 16S rRNA as previously observed (Stern et al. 1988b). Similar levels of enhancement are observed upon addition of S12Full-Length, S12▵10N, S12▵15N, and S12▵27N (Fig. 3A, lanes 5,6,7,8, respectively). This suggests that truncation of the N-terminal region does not cause global misfolding of the entire population of protein molecules or complete inability of these variants to associate with 16S rRNA. In contrast, within the central domain, at residues A560 and A563 (Fig. 3B), the protection observed upon addition of S12WT (Fig. 3B, lane 4) or S12Full-Length (Fig. 3B, lane 5), is reduced upon truncation of the protein by 10 amino acids (Fig. 3B, lane 6) and is absent upon further truncation (Fig. 3B, lanes 7,8). Indeed, in the presence of S12▵15N (Fig. 3B, lane 7) or S12▵27N (Fig. 3B, lane 8) the pattern appears to revert back to that seen in the absence of S12 (Fig. 3B, lanes 3,9; Stern et al. 1988b). Analysis of contacts between S12 and 16S rRNA in the crystal structure of the 30S subunit indicates that the loss of protection at nucleotide 563 is most likely due to deletion of residue 11 of the protein (Fig. 6D–F).
FIGURE 6.
Modification data from Figure 3 mapped as spheres on the three-dimensional structure of the 30S ribosomal subunit. 16S rRNA is gray, the various S12 variants are color coded as in Figure 2, and other r-proteins are omitted for clarity. (A) S12WT and 16S rRNA in the 30S subunit. (B–F) Data from complexes containing S12WT, S12Full-Length, S12▵10N, S12▵15N, and S12▵27N plotted as spheres on 16S rRNA. Black spheres indicate nucleotides protected upon addition of S12WT, while green spheres indicate positions of enhancement of reactivity. Strength of modification is represented by the size of the sphere. The view is rotated 180° and zoomed in for greater clarity.
Addition of S12WT (Fig. 3C, lane 4) to the minimal complex (Fig. 3C, lanes 3,9) enhances the reactivity of A914, and this change is observed to a lesser extent upon incorporation of both S12Full-Length (Fig. 3C, lane 5) and S12▵10N (Fig. 3C, lane 6). Truncation of the extension by 15 amino acids (Fig. 3C, lane 7) or more (Fig. 3C, lane 8) abolishes this change in reactivity, indicating that the conformation of 16S rRNA is more similar to that seen in the absence of S12.
Our results indicate that truncation of the N-terminus of S12 by 10 amino acids or more does not abolish contact between the globular region of S12 and 16S rRNA. However, a change is observed that correlates with loss of a direct contact between the N-terminal extension and nucleotide A563. The changes in reactivity at nucleotides A560 and A914 are more likely due to long-range structural changes associated with the absence of the N-terminus of S12. These findings are consistent with the N-terminal extension of S12 playing a role in shaping the architecture of the assembling SSUs by orchestrating changes in both proximal and distant structural elements.
The N-terminal extension of S12 plays a role in vivo
The gene encoding S12 (rpsL) is essential in E. coli (Sharma et al. 2007; Shoji et al. 2011). Therefore, to analyze the effect of changes in the N-terminus of S12 we used a genomic deletion of rpsL (ΔrpsL pSTR) where growth is supported by expression of S12 from a covering plasmid (pSTR) (Link et al. 1997; Sharma et al. 2007). Plasmid shuffle with negative selection was used to introduce plasmids expressing S12WT, S12▵6N, S12▵11N, S12▵15N, and S12▵24N (Fig. 4A) under the control of an inducible promoter (Shoji et al. 2011). We generated four strains using this technique: ▵rpsL pS12WT, ▵rpsL pS12▵6N, ▵rpsL pS12▵11N, and ▵rpsL pS12▵15N. To aid in plasmid shuffle, a specific plasmid for expression of the truncated S12 proteins was used that simultaneously allowed negative selection of the full-length rpsL gene and S12 mutant expression (Link et al. 1997). As a consequence, the set of truncation mutants is as similar as possible, but not identical, to those S12 truncations studied in vitro. Despite multiple efforts and reoptimized conditions, we were unable to generate ▵rpsL pS12▵24N, leading us to speculate that the presence of some amino acids between residues 15 and 24 of S12 are necessary for cellular viability.
The growth of strains dependent on wild-type and truncated forms of S12 was examined on plates (Fig. 4B) and in liquid media (Fig. 4C). In both conditions, it is clear that truncation of S12 by more than six amino acids at its N-terminus is deleterious to cell growth even at the optimum growth temperature (37°C); truncation of three-fourths of the extension of S12 (S12▵24N) does not appear to support growth. Also, strains dependent on truncated forms of S12 (▵rpsL pS12▵6N, ▵rpsL pS12▵11N, and ▵rpsL pS12▵15N) exhibit cold sensitivity (Fig. 4B), a hallmark of ribosome biogenesis defects.
This is consistent with data from the initial plasmid shuffle to generate the strains. The success rate of each shuffle was defined as the percent colonies observed after shuffling in plasmids expressing S12 variants (pS12WT, pS12▵6N, pS12▵11N, pS12▵15N, and pS12▵24N) that were phenotypically consistent with the loss of the first plasmid (pSTR). We had an average success rate (out of four experiments) of 100% for ▵rpsL pS12WT, 86% for ▵rpsL pS12▵6N, 83% for ▵rpsL pS12▵11N, 36% for ▵rpsL pS12▵15N, and 0% for ▵rpsL pS12▵24N (data not shown).
At 37°C, ▵rpsL pS12WT and ▵rpsL pS12▵6 exhibit similar growth properties (Fig. 4B,C) consistent with in vitro analysis indicating that deletion of up to 10 amino acids at the N-terminus does not affect assembly significantly. ▵rpsL pS12▵11N and ▵rpsL pS12▵15N demonstrate increased doubling and lag times in liquid culture (Fig. 4C). Expression of S12▵6N, S12▵11N, S12▵15N, and S12▵24N in a wild-type background had no significant effect on growth (data not shown). Therefore, the mutant proteins are likely not dominant, although the stability and expression of each variant is unknown. In conclusion, truncation of half or more of the extension has deleterious effects on the growth of the cell, especially at low temperatures, suggesting a role for the N-terminal extension of S12 in SSU biogenesis.
Truncation of the N-terminus of S12 alters ribosome profiles
One explanation for the growth defects observed in ▵rpsL pS12▵11N and ▵rpsL pS12▵15N as well as the lethal phenotype conferred by ▵rpsL pS12▵24N is improper SSU assembly. Consequently, we examined the sucrose gradient sedimentation profiles of free subunits and ribosomes in all strains (▵rpsL pS12WT, ▵rpsL pS12▵6N, ▵rpsL pS12▵11N, and ▵rpsL pS12▵15N) at 37°C (Fig. 5A). There is no significant change in the percent area of free SSUs upon comparing ▵rpsL pS12WT and ▵rpsL pS12▵6N (P = 0.34) (Fig. 5A,B). There is a significant increase in free SSUs in ▵rpsL pS12▵11N (P = 0.03) and ▵rpsL pS12▵15N (P = 0.013) strains compared with the wild-type S12 control strain (Fig. 5A,B). Thus, these SSUs appear to be somewhat defective in subunit association, strongly suggesting a defect in initiation and/or biogenesis. Consistent with the latter hypothesis, we observe presence of precursor 16S rRNA (p16S rRNA) in free SSUs from various rpsL truncation strains (Fig. 5C). To summarize, there is a strong correlation between changes in growth, changes in ratio of free SSUs to ribosomes, presence of p16S rRNA, and increasing truncation of S12.
DISCUSSION
We created partially overlapping subsets of N-terminal truncation mutants of S12 to analyze the role of the noncanonically structured segment, both in vitro and in vivo (Figs. 1C, 4A). Changes in tRNA binding and architecture of 16S rRNA were observed in vitro when the length of the N-terminal extension of S12 was reduced by half or more. In vivo truncation of the N-terminus of S12 can change growth at normal and cold temperatures, the ratio of free SSUs to 70S ribosomes, and 16S rRNA maturation. These data suggest a critical role of the N-terminal extension of S12 in SSU biogenesis and function.
In vitro analyses offer initial information for a role of the N-terminal segment of S12 in 30S subunit function and structuring the SSU platform. Truncation of the extension by half or more (S12▵15N and S12▵27N) caused a marked reduction in the ability of RNPs to bind tRNA and the reduction resulted in binding data comparable to particles lacking S12 (Fig. 2). However, it is unlikely that this is due to a failure of truncated S12 to incorporate into SSUs as probing data reveal a reactivity change in 16S rRNA residues that are correlated with S12WT association (Figs. 3A, 6). Moreover, this change can be attributed to its globular region, indicating that absence of the extension does not cause global unfolding of S12 and that these deletions do assemble into RNPs. However, we did observe changes in reactivity in other regions of 16S rRNA associated with systematic truncation of S12. For example, the loss of protection at A563 could be directly attributed to truncating S12 past amino acid 11 (Figs. 3B, 6E), as Arginine 11 interacts with A563 in SSUs (see Fig. 6D). Our previous work suggested that alternative conformations of 16S rRNA existed at nucleotides 566–568 in a minimal complex lacking all 5′ domain binding proteins and that the N-terminal segment of S12 played a role in assembly of the interdomain region between the SSU body and platform (Calidas and Culver 2011). In addition to this direct effect, upon deletion of the N-terminus of S12, differences in reactivity have been observed that are distal to S12 in SSUs (see Fig. 6). Nucleotides 560 and 914 exhibit differential reactivity in the presence of S12▵15N and S12▵27N (Figs. 3B,C, 6E,F) as compared with S12Full Length (Figs. 3C, lane 5, 6C) or S12▵10N (Figs. 3C, lane 6, 6D). Changes in 16S rRNA architecture associated with truncation of S12 appear to be attributable to both direct and long-range effects. Since the main aim of this publication is to determine the role of the extension of S12 in stabilizing 16S rRNA architecture, this work has been carried out on minimal particles. It is possible that the changes in 16S rRNA architecture might affect r-proteins binding other regions of the particle; however, it is striking that no reactivity differences were observed in regions directly associated with translational fidelity. This is consistent with the vast majority of fidelity mutations mapping to the globular region of S12 (Gorini and Kataja 1964a; Sharma et al. 2007; Agarwal et al. 2011). Our probing experiments indicate that the S12 protein variants bind to 16S rRNA (at least minimally), and changes are consistent with previous structural (Schuwirth et al. 2005), probing (Stern et al. 1988b), and assembly (Powers and Noller 1995; Talkington et al. 2005; Adilakshmi et al. 2008) data.
After plasmid shuffle, such that the cloned truncations were the sole source of S12 in vivo, three major changes could be observed. First, growth defects were observed with all tested strains expressing truncated S12 (Fig. 4B,C). The changes were exacerbated at low temperature and with decreasing length of the N-terminal segment, such that deletion of N-terminal 15 amino acids affected growth more than deletion of 11 amino acids, which grew more slowly than the variant lacking six residues. Moreover, we were unable to obtain colonies expressing deletions of 24 amino acids. Thus, the N-terminal extension of S12 likely contributes to the essential role of S12. In contrast, we see no significant changes in growth in the presence of antibiotics that alter decoding (data not shown), implying that the N-terminus of S12 plays a predominant role in SSU assembly. Second, we observe that upon truncation of half the extension in vivo, there is a significant increase in the population of free SSUs (Fig. 5A,B). This phenotype is similar to that observed upon mutation of the C-terminal extension of yeast S14, which results in an accumulation of 20S pre rRNA in 43S pre-ribosomes (Jakovljevic et al. 2004). Finally, we observed an increase in p16S rRNA in strains bearing truncated variants of S12. In E. coli, an increase in free SSUs associated with p16S rRNA has been found in strains with perturbed ribosome biogenesis cascades such as era-depleted cells, ΔyjeQ and ΔksgA (Inoue et al. 2003; Campbell and Brown 2008; Connolly et al. 2008). Changes in growth and aberrant subunit/ribosome ratios as well as increased p16S rRNA content strongly implies a role for the extension of S12 in SSU biogenesis.
While it would have been ideal to directly compare the same truncated S12 proteins in vitro and in vivo, this was complicated by the requirement that the plasmid allowing expression of the truncated rpsL gene also encode a gene that allows for selection against the plasmid expressing wild-type full-length rpsL. This compromise allowed the most similar comparison while ensuring that possible wild-type S12 expression was minimized. While our in vivo and in vitro data are in general agreement, there is a difference in the phenotype observed in vivo for the ▵rpsL pS12▵11N strain and the in vitro data from S12▵10N. While in vitro-formed particles incorporating S12▵10N have tRNA-binding activity and structures similar to those containing S12WT, the ▵rpsL pS12▵11N strain exhibits growth defects and an aberrant subunit profile. The differences between in vivo SSU biogenesis and in vitro reconstitution of SSUs or minimal 16S rRNA-containing particles could account for the observed differences. Notably, there is an increase in pre-16S rRNA in the ▵rpsL pS12▵11N strain, thus suggesting that elements of the in vivo biogenesis cascade are influenced by this truncation and this involvement would not be evident in our in vitro experiments. Additionally, the 11th residue of S12 is an arginine, and mutation of this residue has previously been implicated in changes in SSU function in vivo (Agarwal et al. 2011). In fact, this is also consistent with data from in vitro structural analysis of particles incorporating S12▵15N; in which protection at nucleotide 563, in direct contact with R11 in the X-ray crystal structure is lost. Thus, the observed importance of this residue in SSU function and biogenesis would be consistent with our data.
These observations suggest a model for the role of the noncanonically structured region of S12 in SSU biogenesis. Deletion of the extension results in structural perturbations of the platform of 16S rRNA in vitro, suggesting that the extension may be important in the architecture of this region of the assembled SSU (Figs. 3, 6). Previous in vitro studies suggest that stabilization of 16S rRNA architecture in the platform occurs late in SSU assembly (Talkington et al. 2005; Adilakshmi et al. 2008), and this work supports such a role for the S12 N-terminal extension in scaffolding 16S rRNA structure.
Roles for the extended loops and unstructured regions of r-proteins have been the subject of speculation, but little is known of their function in assembly and translation (see Ban et al. 2000; Wimberly et al. 2000; Wilson and Nierhaus 2005; Timsit et al. 2009). One hypothesis is that r-proteins with long extensions represent a subset of intrinsically disordered proteins whose function depends upon a structural transition from a disordered form to an ordered structure upon the binding of its partner (Uversky et al. 2000; Iakoucheva et al. 2002; Timsit et al. 2009). For example, the N-terminal extension of r-protein L20 undergoes a helix–coil transition upon binding 23S rRNA, and is essential for large subunit assembly (Guillier et al. 2005; Timsit et al. 2006). Similar data for S4 in the in vitro assembly of the 30S subunit also lend credence to the idea that r-proteins with long noncanonically structured regions play an important role in assembly by cofolding with 16S rRNA (Mayerle and Woodson 2013). Our study suggests that the N-terminal extension of S12 also plays a role in stabilizing SSU structure during biogenesis.
MATERIALS AND METHODS
Plasmids
Primers introducing a translation initiation codon at desired sites within rpsL were utilized for PCR amplification from MG1655 genomic DNA to generate full-length and truncation variants of S12. The truncations are designated by S12▵XN, where X signifies the number of truncated residues, while N indicates that the protein has been truncated at the N-terminus. C-terminal His-tagged versions of S12 were expressed and purified for use in vitro. Full-length and truncated variants (▵10N, ▵15N, and ▵27N) of S12 were cloned into the NdeI and HindIII sites of pET24 (Novagen, kanamycin resistant) without a stop codon at the C-terminus of the gene. S12WT, which is the full-length untagged protein cloned in pET21b is a gift from the laboratory of Dr. Noller.
The str operon including the genes rpsL, rpsG, fusA, and tufA was cloned into the NotI site of pKOV to generate pSTR. pKOV expresses sacB, a gene that causes the loss of the host plasmid in the presence of sucrose (Link et al. 1997). Primers introducing a translation initiation codon at desired sites within rpsL were utilized for PCR amplification from MG1655 genomic DNA to generate full-length and truncation variants of S12. The wild-type (S12WT) and truncated (S12▵6N, S12▵11N, S12▵15N, and S12▵24N) rpsl genes were cloned into the NcoI and HindIII sites of pBAD HisB (Invitrogen, ampicillin resistant) with a C-terminal stop codon, to express untagged protein in vivo. The protein was truncated at a different set of residues due to the constraints of restriction enzyme choice.
Purification of proteins
Strains expressing S12WT from pET21b and S12Full Length, S12▵10N, S12▵15N, and S12▵27N from pET24 were grown to OD600 = 0.7, then induced with 1 mM IPTG for 3 h. The cells were pelleted at 7000 rpm for 10 min, then lysed in Buffer E (20 mM K+-HEPES at pH 7.6, 20 mM KCl, and 6 mM βME) in an Avestin Emulsiflex-C3 cell lyser. The lysate was cleared by spinning at 10,000 rpm in a JA-17 rotor in a Beckman high-speed centrifuge for 5 min at 4°C.
S12WT supernatant was dialyzed overnight into Buffer B (20 mM sodium acetate at pH 5.6, 20 mM KCl, 6 M urea, and 6 mM βME). Proteins were purified by fast protein liquid chromatography using a Resource-S cation exchange column (GE) and eluted with a 35-mL salt gradient from 100 to 350 mM KCl in Buffer C (20 mM sodium acetate at pH 5.6, 1 M KCl, 6 M urea, and 6 mM βME). The proteins were eluted at ∼230 mM KCl. Purified proteins were dialyzed into Buffer D (80 mM K+ -HEPES at pH 7.6, 1 M KCl, and 6 mM βME), aliquoted, and frozen in liquid nitrogen.
S12▵XN and S12Full Length pellets were resuspended in Buffer BH (100 mM sodium dihydrogen phosphate, 330 mM KCl, 6 M urea, 20 mM Imidazole, 6 mM βME at pH 7.4) by rocking the pellet overnight at 4°C. The solution was cleared at 7000 rpm for 20 min, twice, before loading sample for affinity purification on a HiTrap Chelating HP column (GE). The column was washed with Buffer CH (100 mM sodium dihydrogen phosphate, 330 mM KCl, 6 M urea, 50 mM imidazole, 6 mM βME at pH 7.4). The proteins were eluted by a gradient of from 125 mM to 250 mM Imidazole in 20 mL Buffer EH (100 mM sodium dihydrogen phosphate, 330 mM KCl, 6 M Urea, 250 mM imidazole, 6 mM βME at pH 7.4). The proteins eluted at ∼163 mM imidazole concentration. The proteins were step dialyzed with gradations of 0.5 M urea and 50 mM imidazole into Buffer DH (80 mM K+-HEPES at pH 7.6, 1 M KCl, 3 M urea, and 6 mM βME), aliquoted, and stored in liquid nitrogen.
Assembly of ribonucleoprotein particles in vitro
16S rRNA was isolated as described (Moazed et al. 1986), heat activated by incubating at 42°C for 15 min in Buffer A (80 mM K+-Hepes at pH 7.6, 330 mM KCl, 20 mM MgCl2, and 0.01% Nikkol), and then incubated on ice for 10 min. Ribonucleoprotein particles were reconstituted essentially as described (Culver and Noller 2000) except that the reconstitution was carried out in the presence of 30 mM urea. 30S particles were formed by mixing 280 pmol of any variant of S12 (wild-type, full-length, ▵10, ▵15, or ▵27) with 240 pmol of total r-protein mix (all r-proteins except S12) and 40 pmol of 16S rRNA in a final volume of 100 µL with a final KCl concentration of 330 mM. In minimal particles, 240 pmol of minimal r-protein mix (S4, S7, S8, S15, S16, S17, and S20) was substituted for total r-protein mix. In order to form the RNPs, the reactions were incubated at 42°C for 20 min. These were then cooled on ice for 10 min before proceeding to the next experiment.
In vitro transfer RNA (tRNA) binding
A poly-U-dependent tRNA-binding experiment was performed essentially as described (Culver and Noller 1999) with a saturating excess of tRNA such that the assay monitored factor-independent, multi-site tRNA-binding activity of the SSU. The assay was carried out in the presence of 30 mM urea. The results presented are from three independent experiments.
Chemical probing of RNP conformation
Chemical probing of the minimal RNPs by dimethyl sulfate was carried out essentially as described (Moazed et al. 1986). The complexes were assembled, chilled on ice for 10 min, and then treated with dimethyl sulfate at a final concentration of 33.8 mM and incubated for 1 h on ice. The reactions were visualized by primer extension as described (Moazed et al. 1986) using primers initiating cDNA synthesis at nucleotides 232, 683, and 1046. Results are representative of three independent experiments.
Generation of E. coli strains
A strain lacking a genomic copy of S12 (S12koBW) was created by Sharma et al. (2007). It was used as a donor strain in P1 transduction experiments to replace the rpsL gene in the strain MG1655str with a kanamycin resistance marker, generating ▵rpsl pSTR. ▵rpsl pSTR is temperature and sucrose sensitive. It was transformed with pBAD expressing WT and ▵X rpsl and the transformants grown overnight in 5 mL of LB. The overnight culture was then plated on LB agar plates containing kanamycin (50 µg/mL), ampicillin (100 µg/mL), arabinose (2%), and sucrose (5%) to replace pSTR with pS12, generating ▵rpsL pS12WT, ▵rpsL pS12▵6N, ▵rpsL pS12▵11N, and ▵rpsL pS12▵15N. In order to avoid picking suppressors or revertants, slow growing (≈48 h) colonies expressing truncated forms of S12 were chosen for further analysis. The presence of compensatory mutations in the strains dependent on truncated proteins cannot be ruled out, but their slow growth phenotype leads us to believe that this is unlikely.
Analysis of growth properties
All overnight cultures were grown at 37°C in the presence of kanamycin (50 µg/mL), ampicillin (100 µg/mL), and arabinose (0.2%) with shaking (200 rpm). For growth on solid media, overnight cultures were diluted to OD600 = 0.6, then serially diluted by a factor of 10. The dilutions were spotted on LB-agar plates containing kanamycin (50 µg/mL), ampicillin (100 µg/mL), and arabinose (0.2%) and the plates incubated at 37°C and 25°C to examine growth at optimal and nonpermissive temperatures.
Growth curves at 37°C were obtained by two methods. In one method overnight cultures were diluted to equal OD600 in 50 mL LB media and the OD600 was determined from aliquots taken at 30-min intervals. In the second technique, two overnight cultures of each strain were diluted to equal OD600 in 200 µL of media in a microtiter plate. This was replicated in five wells for each overnight culture. The plates were grown in a Bioscreen automatic growth analyzer with shaking to periods of 20 sec of maximum intensity per minute. Growth was monitored at the wideband setting every 20 min (Weichart and Kell 2001). The data were collected during a single run of a plate at 37°C. Growth in microtiter plates yielded longer doubling times than growth in flasks for each strain. However, there was no significant difference in the ratio of the doubling time of mutants to wild-type strains, therefore only data from five independent growths within the Bioscreen analyzer are presented.
Analysis of ribosome sedimentation profiles
All of the strains were inoculated to equal OD600 in 500 mL LB. All strains were grown to an OD600 of 0.6 or for a maximum of 8 h. The cells were treated as described (Connolly et al. 2008) to obtain subunit profiles. The data are derived from four independent growths. The area under the peak for each profile was calculated using the program QTiPlot. Ribosome containing peak intensity was corrected to represent only SSUs as in Connolly et al. (2008). Data are representative of three independent experiments.
Analysis of rRNA from SSUs and ribosomes
SSUs and ribosomes were collected after sucrose gradient centrifugation of cell lysate. RNA was extracted as described in Culver and Noller (2000), and analyzed by agarose gel electrophoresis. Data are representative of two independent experiments.
ACKNOWLEDGMENTS
The donor strain for P1 transduction, S12koBW, was a gift from Dr. Rachel Green. We thank Drs. George Church for pKOV plasmids, Scott Butler and David Goldfarb and their laboratories for help with equipment and materials, and Drs. Elaine Sia and Martin Pavelka as well as their laboratories for technical advice.
REFERENCES
- Adilakshmi T, Bellur DL, Woodson SA 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal D, Gregory ST, O'Connor M 2011. Error-prone and error-restrictive mutations affecting ribosomal protein S12. J Mol Biol 410: 1–9 [DOI] [PubMed] [Google Scholar]
- Alksne LE, Anthony RA, Liebman SW, Warner JR 1993. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci 90: 9538–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers J, Shamoo Y 2001. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol 311: 75–86 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M 2010. Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V 2002. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16S RNA. J Mol Biol 316: 725–768 [DOI] [PubMed] [Google Scholar]
- Calidas D, Culver GM 2011. Interdependencies govern multidomain architecture in ribosomal small subunit assembly. RNA 17: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TL, Brown ED 2008. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J Bacteriol 190: 2537–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons WM Jr, May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V 1999. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature 400: 833–840 [DOI] [PubMed] [Google Scholar]
- Connolly K, Rife JP, Culver G 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, Noller HF 1999. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA 5: 832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, Noller HF 2000. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol 318: 446–460 [DOI] [PubMed] [Google Scholar]
- Gorini L, Kataja E 1964a. Phenotypic repair by streptomycin of defective genotypes in E. coli. Proc Natl Acad Sci 51: 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L, Kataja E 1964b. Streptomycin-induced oversuppression in E. coli. Proc Natl Acad Sci 51: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Allemand F, Graffe M, Raibaud S, Dardel F, Springer M, Chiaruttini C 2005. The N-terminal extension of Escherichia coli ribosomal protein L20 is important for ribosome assembly, but dispensable for translational feedback control. RNA 11: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK 2002. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 323: 573–584 [DOI] [PubMed] [Google Scholar]
- Inoue K, Alsina J, Chen J, Inouye M 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol Microbiol 48: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Leger-Silvestre I, Gas N, Woolford JL Jr 2004. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell 14: 331–342 [DOI] [PubMed] [Google Scholar]
- Lee K, Varma S, SantaLucia J Jr, Cunningham PR 1997. In vivo determination of RNA structure-function relationships: Analysis of the 790 loop in ribosomal RNA. J Mol Biol 269: 732–743 [DOI] [PubMed] [Google Scholar]
- Link AJ, Phillips D, Church GM 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J Bacteriol 179: 6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodmell JS, Dahlberg AE 1997. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277: 1262–1267 [DOI] [PubMed] [Google Scholar]
- Mayerle M, Woodson SA 2013. Specific contacts between protein S4 and ribosomal RNA are required at multiple stages of ribosome assembly. RNA 19: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1990. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J Mol Biol 211: 135–145 [DOI] [PubMed] [Google Scholar]
- Moazed D, Stern S, Noller HF 1986. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J Mol Biol 187: 399–416 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Powers T, Noller HF 1991. A functional pseudoknot in 16S ribosomal RNA. EMBO J 10: 2203–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF 1995. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA 1: 194–209 [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R 2007. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol 374: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG 2011. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol 413: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Lee JC, Gutell RR, Hartman H 2008. The origin and evolution of the ribosome. Biol Direct 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Changchien LM, Craven GR, Noller HF 1988a. Interaction of proteins S16, S17 and S20 with 16S ribosomal RNA. J Mol Biol 200: 291–299 [DOI] [PubMed] [Google Scholar]
- Stern S, Powers T, Changchien LM, Noller HF 1988b. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16S rRNA. J Mol Biol 201: 683–695 [DOI] [PubMed] [Google Scholar]
- Svensson P, Changchien LM, Craven GR, Noller HF 1988. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16S ribosomal RNA. J Mol Biol 200: 301–308 [DOI] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit Y, Allemand F, Chiaruttini C, Springer M 2006. Coexistence of two protein folding states in the crystal structure of ribosomal protein L20. EMBO Rep 7: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit Y, Acosta Z, Allemand F, Chiaruttini C, Springer M 2009. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50S ribosomal subunit assembly. Int J Mol Sci 10: 817–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41: 415–427 [DOI] [PubMed] [Google Scholar]
- Weichart DH, Kell DB 2001. Characterization of an autostimulatory substance produced by Escherichia coli. Microbiology 147: 1875–1885 [DOI] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH 2005. Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol 40: 243–267 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, et al. 1980. Secondary structure model for bacterial 16S ribosomal RNA: Phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res 8: 2275–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zengel JM, Jerauld A, Walker A, Wahl MC, Lindahl L 2003. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA 9: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lin Y 2009. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res 37: D455–D458 [DOI] [PMC free article] [PubMed] [Google Scholar]