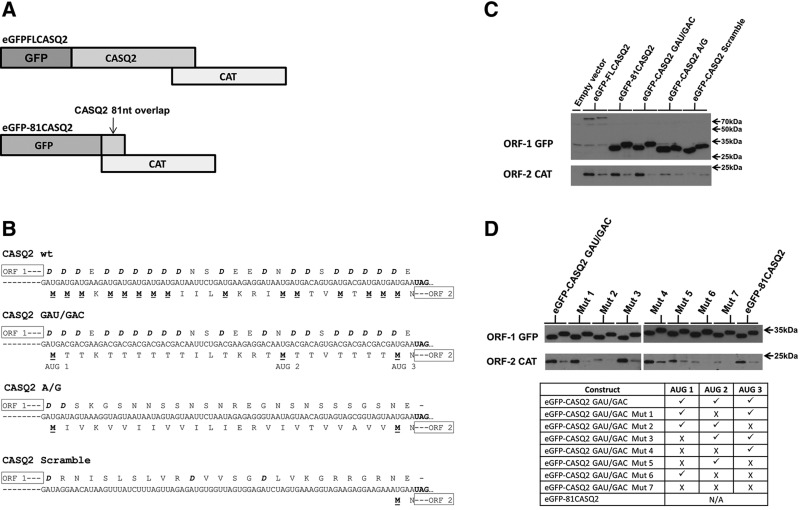
FIGURE 3.
(A) Diagrammatic representation of the constructs used to investigate the effect of mutations on the expression of ORF-2. In all constructs the coding region of ORF-2 was replaced with that of the CAT gene. See text for details. (B) The sequences of the calsequestrin 2 (CASQ2) 81-nt sequence from the region of overlap between ORF-1 and ORF-2 subjected to mutation. The multiple aspartate residues of ORF-1 are shown in bold and italics and the potential initiation methionine residues of ORF-2 are shown in bold and underlined. The wild-type unaltered sequence is shown (CASQ2 wt) together with the CASQ2 GAU/GAC mutant that retains the multiple aspartate residues but lacks the multiple ORF-2 initiation codons, the CASQ2 A/G mutant that lacks the multiple ORF-1 aspartate and ORF-2 methionine residues, and the CASQ2 Scramble mutant that retains the nucleotides from the overlap region that have been randomly distributed to remove the multiple aspartate residues from ORF-1 and the multiple methionine residues from ORF-2. These mutants replaced the 81-nt CASQ2 overlap shown in A. (C) Detection of GFP and CAT protein in lysates of cells transfected with expression plasmids by Western blot. GFP-specific and CAT-specific monoclonal antibodies were used to detect protein expression. The protein encoded by the eGFP-FLCASQ2 construct and detected by the anti-GFP antibody is 73.5 kDa because it is a fusion product of eGFP and CASQ2. Cells were transfected with the plasmid construct indicated. Lanes on the left of each pair are the constructs identified and lanes on the right of each pair are the appropriate STOP mutant. (D) Detection of GFP and CAT protein in cells transfected with expression plasmids as in C. Cells were transfected with the eGFP-CASQ2 GAU/GAC plasmid construct shown in B and with mutants derived from it in which one or more of the three AUG codons of ORF-2 labeled 1, 2, and 3 in the eGFP-CASQ2 GAU/GAC construct in B were mutated to ACG, AGC, and AGA, respectively. Lanes on the left of each pair are the parental construct and lanes on the right of each pair are the appropriate STOP mutant. The table below summarizes the presence (✓) and/or absence (X) of each of the three AUG codons in the mutants.