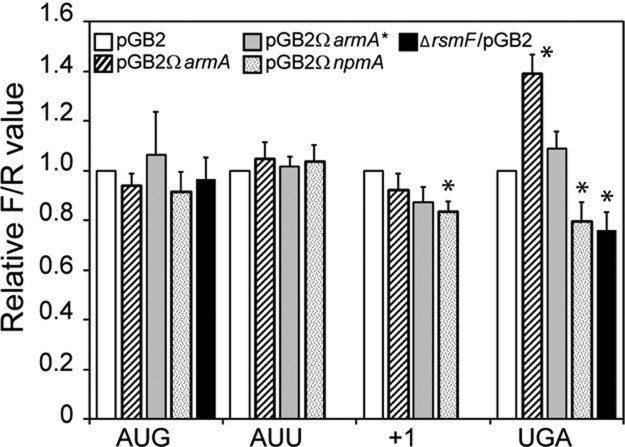
FIGURE 5.
Translational accuracy in E. coli MM294 in the presence of ArmA, ArmA*, or NpmA or in the absence of RsmF. Strain MM294 harboring pGB2 (white), pGB2ΩarmA (strips), pGB2ΩarmA* (gray), or pGB2ΩnpmA (dotted) was transformed with plasmids encoding Renilla luciferase (RLuc) starting with the AUG codon and Firefly luciferase (FLuc) starting with codon AUG or AUU on a single transcript. Translation initiation efficiency was measured by dividing the chemiluminescence of FLuc by that of RLuc and normalizing the ratio to that of MM294/pGB2 (relative F/R value). To measure the efficiency of +1 reading frame maintenance and UGA readthrough in the presence of ArmA or NpmA, fusion constructs containing the RLuc and FLuc open reading frames separated by short windows containing a stop codon (UGA) or a frameshift site (+1) were used. The efficiency of translation initiation at AUG and the accuracy of UGA readthrough in the absence of RsmF (black) were also determined. In this case, the ratio FLuc/RLuc was normalized to the ratio FLuc/RLuc obtained for E. coli MM294. The values are the means of three independent experiments carried out in quintuplates with error deviations. (*) Significant difference (P< 0.01) of the mean value.