Abstract
The human BAP1 deubiquitinating enzyme is a chromatin-bound transcriptional regulator and tumor suppressor. BAP1 functions in suppressing cell proliferation, yet its role in the DNA damage response pathway is less understood. In this study we characterized DNA damage-induced phosphorylation of BAP1 at Serine 592 (pS592) and the cellular outcomes of this modification. In contrast to the majority of BAP1, pS592-BAP1 is predominantly dissociated from chromatin. These findings support a model whereby stress induced phosphorylation functions to displace BAP1 from specific promoters. We hypothesize that this regulates the transcription of a subset of genes involved in the response to DNA damage.
Keywords: BAP1, Deubiquitinating enzyme, DNA Damage, Phosphorylation, Ubiquitin
1. Introduction
The human BAP1 (BRCA1-Associated Protein 1) gene encodes a 729 amino acid deubiquitinating enzyme (DUB) that is nuclear localized and chromatin-associated [1, 2]. In the BAP1-null NCI-H226 cell line BAP1 is a tumor suppressor [2], and in uveal melanoma (UM) BAP1 unequivocally adheres to the classical ‘two-hit’ tumor suppressor model [3]. Genetic studies have uncovered somatic BAP1 mutations in numerous cancers, and germline BAP1 mutations cause a cancer susceptibility disorder that predisposes BAP1+/- individuals to multiple cancers (reviewed in [4]). The tumor suppressor properties of BAP1 derive in part from its ability to regulate cell cycle progression. Restoration of BAP1 inhibits cell growth in cancer cell lines that lack BAP1 (NCI-H226 cells) or express defective BAP1(769-P cells) [2, 5].
BAP1 is thought to function in transcriptional complexes where it deubiquitinates proteins and co-regulates gene expression. The best studied binding partner, HCF-1, is a transcriptional regulator that functions with numerous transcription factors, assembling in chromatin modifying complexes associated with both gene activation and repression (reviewed in [6, 7]). BAP1, HCF-1 and the YY1 transcription factor form a ternary complex regulating gene expression [8], and it is reasonable to suspect similar complexes are formed with other known transcription cofactors that are known to bind BAP1 (such as FoxK1/2, ASXL1/2, CBX1/3, etc.) [8-10]. A large fraction of BAP1 is bound to HCF-1 [5, 9], and they co-occupy >3700 gene promoters in mice [11]. An intact BAP1/HCF-1 interaction is required for BAP1-mediated growth suppression in the 769-P clear cell renal cell carcinoma (ccRCC) line [5]. BAP1 deubiquitinates poly-ubiquitinated HCF-1, yet depletion of BAP1 has shown mixed effects on the stability of HCF-1 protein levels [5, 9, 11, 12].
A second transcriptional complex is formed with a polycomb group (PcG) protein ASXL1. The purified BAP1/ASXL1 complex was shown to deubiquitinate Histone H2A but not Histone H2B in reconstituted nucleosomes [13]. The drosophila orthologs of BAP1 and ASXL1 (Calypso and ASX) co-localize to 879 genomic sites, including the PcG target gene Ubx where the DUB activity of Calypso was required for transcriptional repression [13]. Loss of ASX in fly embryos reduces Calypso levels and leads to a modest increase in ubiquitinated Histone H2A levels [13]. Changes to ubiquitin-Histone H2A levels have also been observed when BAP1 levels are altered in human and mouse cancer cell lines [5, 14]. The BAP1/ASXL1 complex could be a critical component of hematopoiesis as ASXL1 mutations and dysfunction are linked to human myeloproliferative and myelodysplastic disorders [15], and BAP1 knockout mice develop hematological features characteristic of these diseases [11].
BAP1 has also been implicated in the cellular response to DNA damage. Depletion of BAP1 using shRNA in HeLa cells led to reduced cell viability following exposure to ionizing radiation (IR) [16]. A similar result was observed in two ccRCC cell lines that express mutant BAP1; restoration of WT BAP1 protected cells from IR-induced cell death [5]. The loss of BAP1 does not influence the formation of IR-induced double strand break repair foci in ccRCC and mesothelioma cell lines [5, 17], however the transcription of genes involved in the DNA replication and repair pathways were amongst those deregulated following BAP1 depletion [8]. Proteomic studies have identified several sites of phosphorylation in BAP1, including five serines within a fifteen residue stretch (583, 592, 595, 596, and 597) that become modified after UV and/or IR induced DNA damage [18-20]. One of these sites S592 conforms to the canonical SQ/TQ motif recognized by the DNA damage activated phosphatidylinositol 3-kinase-related kinases ATM, ATR, and DNA-PKcs [21]. Thus, evidence suggests a growth suppressive role for BAP1 following DNA damage and post-translational modification by phosphorylation may mediate this effect.
In this study we sought to characterize BAP1 phosphorylation and its role in DNA damage response (DDR) pathway. We have characterized a commercially available antibody that recognizes BAP1 phosphorylated at S592 (pS592) and used it to probe the induction of phosphorylation and subsequent outcomes following irradiation with UV and exposure to other cellular stressors. Our findings show that a small fraction of BAP1 is rapidly phosphorylated at S592 in S-phase following replication stress. In contrast to bulk BAP1, the majority of pS592-BAP1 is not associated with chromatin. We propose a model whereby phosphorylation at S592 is a regulatory mechanism to dissociate BAP1 from chromatin and regulate specific genes involved in DNA replication and repair.
2. Materials and Methods
2.1. Cell culture and treatments
HeLa cells and NCI-H226 cells were purchased from American Type Culture Collection. HeLa cells were cultured in DMEM while NCI-H226 cells were cultured in RPMI-1640. Both media were supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were maintained in a humidified incubator at 37°C with 5% CO2.
For UV-treatment, cells were irradiated with the indicated dose of UV-C (254 nm) using a Stratalinker 1800 (Stratagene). Cells were washed with cold PBS, mock- or UV-treated, fed media, and allowed to recover for the indicated times. Hydroxyurea was added to media at a final concentration of 2 mM for 3 h or the indicated times. HeLa cells were treated with 1 mM methyl methanesulfonate, 0.5 mM sodium arsenite, or 0.125 mM H2O2 for 1 h before recovering in fresh media for 3 h prior to harvesting.
2.2. Antibodies
Antibodies used for immunoblots: BAP1 (Santa Cruz, sc-28383), pS592-BAP1 (Cell Signaling, #9373), ubiquitinated-Histone H2A (Cell Signaling, #8240), MEK1 (Bethyl, A302-140A), VCP (Cell Signaling #2648), and Actin (Sigma, A3853). Antibodies used in IPs: BAP1 (Bethyl, A302-243A), normal rabbit IgG (Santa Cruz, sc-2027). The HCF-1 antibody was used in IPs and IBs (Bethyl, A301-399A).
2.3. Plasmids and transfections
The BAP1 cDNA was subcloned into the pcDNA4 expression vector, and a modified pcDNA4 plasmid encoding an N-terminal Flag-HA epitope tag. The S592A mutation was made using overlapping PCR. Wild type and mutant BAP1 genes were confirmed by DNA sequencing. HeLa cells were transfected with pcDNA4-Flag-HA-BAP1 vectors using Lipofectamine-2000 (Life Technologies) for 24 h according to the manufacturer's protocol.
2.4. Cell lysis and fractionations
Soluble cell extracts (SCEs) were prepared by resuspending cell pellets in 1 mL/106 cells of NP40 lysis buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 0.5% Nonidet P40, protease inhibitor cocktail (Complete EDTA-free, Roche), phosphatase inhibitor cocktail (PhosStop, Roche)]. Lysates were allowed to rotate at 4 °C for 1 h before centrifuging at 16,000g to recover the SCE. The insoluble material (IM) was resuspended in 2X SDS-PAGE gel loading buffer and sonicated with a microtip (Misonix) using two 5 second pulses and an output of 3.
Cellular fractionations were performed at 4°C according to a previously described method [22]. For salt extractions of chromatin, hypotonic buffer supplemented with indicated amounts of NaCl was used to sequentially extract the P2 pellet by iteratively incubating for 10 min and centrifuging at 16,000g. The P1 and P2 chromatin-containing fractions were resuspended in 2X SDS-PAGE gel loading buffer and sonicated. SCEs and the S1 samples from fractionations were quantitated using the Bio-Rad protein assay. For Western blot analysis, 20 μg of SCEs or S1 fractions and an equal percentage of other fraction (IM, P1, S2, or P2) were separated by SDS-PAGE and transferred to nitrocellulose or PVDF membranes.
2.5. Immunoprecipitations
IP of endogenous proteins were performed using 0.3 mg of HeLa SCE, 10 μL of protein A dynabeads (Invitrogen) and 2.5 μg of antibody (except HCF-1, 1.25 μg). Antibody-bound dynabeads were incubated with whole cell extracts overnight at 4°C. Beads were was three times with NP40 lysis buffer, two times with PBS and bound proteins were eluted with 2X SDS-PAGE gel loading buffer. Anti-Flag IPs were performed with anti-Flag(M2) agarose (Sigma-Aldrich). SCEs (4 mg) were incubated with 20 μL for 2 h at 4°C, washed three times with NP40 lysis buffer, three times with PBS, and eluted seven times of PBS + 250 μg/mL flag peptide (Sigma-Aldrich). Eluates were pooled prior to analysis.
2.6. Dephosphorylation reactions
Incubations with calf intestinal phosphatase (New England BioLabs) were performed using SCEs according to the manufacturer's protocol for 30 min at 37°C.
2.7. Double thymidine block
HeLa cells were arrested in early S-phase using a double thymidine block. Cells were harvested at 2 h intervals following release from the second block. For UV-treatment of synchronized cells, a parallel set of plates were treated with 1 mJ/cm2 UV at the indicated times and allowed to recover 1 h prior to harvesting.
2.8. Flow Cytometry
Harvested cells were washed with PBS and fixed by resuspending in 0.5 mL PBS and adding 5 mL of 70% ethanol. Cells were stained with propidium iodide: 0.5 mL of 2 mg/mL RNAase (Sigma-Aldrich, R5125) and 0.5 mL propidium iodide solution [PBS, 0.1 mg/mL propidium iodide, 0.6% Triton X-100]. Cells were stained 1 hour before passing through mesh-capped tubes (Falcon, 352235) and analyzing on a Becton-Dickson LSR II flow cytometer (20,000–50,000 events). Cell cycle analysis was performed using FlowJo software. Gates were used to remove cell debris (forward scatter-area vs. side scatter-area) and cell doublets (propidium iodide-area vs. propidium iodide-width). The resulting histograms of propidium iodide-area were analyzed for cell cycle distributions using the Watson Pragmatic Model.
3. Results and Discussion
3.1. BAP1 is phosphorylated at Ser592 following UV irradiation
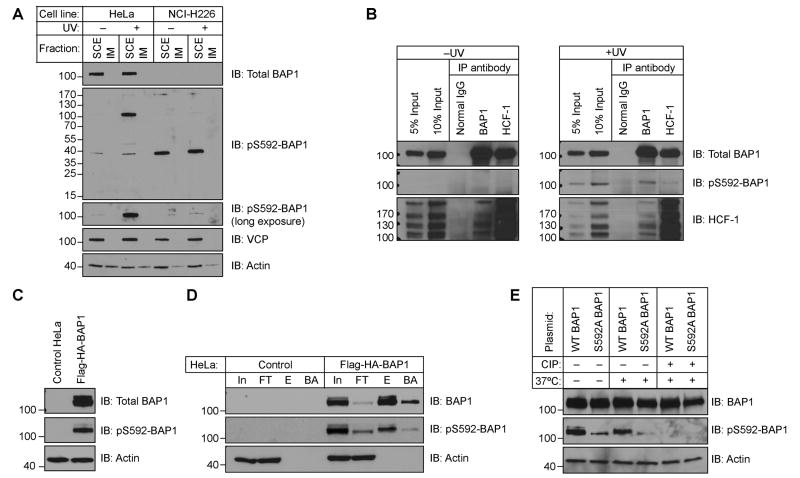
We assessed the levels of pS592-BAP1 in HeLa cells and the BAP1-null non-small cell lung carcinoma cell line NCI-H226 [1, 2] using a commercially available anti-pS592-BAP1 antibody that was raised against a phospho-peptide corresponding to the residues flanking BAP1 S592. Western blots of soluble cell extracts (SCEs) of mock- or UV-treated cells confirmed that HeLa cells contain BAP1 migrating with an apparent MW of 100 kDa, while NCI-H226 cells do not (Fig 1A). Under these lysis conditions [10], the nuclear and chromatin-associated BAP1 [8, 9] is completely extracted and separated from the insoluble material (IM). In response to UV irradiation we observe a robust induction of a band at 100 kDa detected with the pS592-BAP1 antibody. In the NCI-H226 line there is a faint but reproducible signal at 100 kDa following UV-treatment. Densitometry determined this signal to represent ∼1% of the signal observed in UV-treated HeLa. Our attempts to immunoprecipitate (IP) phospho-BAP1 with this antibody recovered BAP1, but LC-MS/MS showed it also precipitated p97/Vasolin-containing protein (VCP) (unpublished observations). VCP is an abundant protein that becomes phosphorylated at S784 following DNA damage [19, 23] and happens to co-migrate on SDS-PAGE gels at the same position as BAP1. From this we conclude the pS592-BAP1 has utility in immunoblots despite cross-reacting with what is likely pS784-VCP to a much lesser extent.
Figure 1.
BAP1 is phosphorylated at S592 following UV irradiation. (A) The pS592-BAP1 antibody recognizes a ∼100 kDa band after UV-treatment in HeLa cells but not in the BAP1-null NCI-H226 cells. Cells were mock/UV-treated with 1 mJ/cm2 and allowed to recover 3 h before analyzing soluble cell extract (SCE) and insoluble material (IM). (B) IP of endogenous BAP1 and HCF-1 retrieves pS592-BAP1. Cells were mock- or UV irradiated with a 50 mJ/cm2 dose and allowed to recover 2 h before IP from SCEs. (C) Overexpressed Flag-HA-BAP1 is phosphorylated in the absence of UV. (D) The pS592-BAP1 signal is immunoprecipitated with anti-Flag agarose (In, Input; FT, Flow thru; E, eluate; BA, boiled agarose). (E) Phospho-specificity of the pS592-BAP1 antibody.
To further assess whether the pS592-BAP1 antibody recognizes phosphorylated BAP1, we performed IPs of BAP1 and HCF-1 from endogenous HeLa SCEs (Fig 1B). Consistent with previous reports [5, 8], BAP1 only brings down a small fraction of HCF-1, while HCF-1 brings down a significant fraction of BAP1. In the absence of UV, there is no detectable pS592-signal in the IP inputs, yet after UV-treatment the pS592-BAP1 signal is induced and both BAP1 and HCF-1 co-immunoprecipitate pS592-BAP1.
We next asked whether the pS592-BAP1 antibody recognizes overexpressed BAP1. We transiently expressed Flag-HA-tagged BAP1 and found it is phosphorylated in the absence of UV irradiation (Fig 1C). We confirmed that the pS592-BAP1 signal corresponds to exogenous BAP1, and not another protein of similar mass induced by overexpression, by IP with anti-Flag agarose (Fig 1D). The specificity of the antibody for phospho-BAP1 was confirmed by treating SCEs of cells transiently expressing WT BAP1 and a S592A mutant with calf intestinal phosphatase (Fig 1E). These studies demonstrate that UV irradiation induces phosphorylation of S592, and the pS592-BAP1 antibody easily detects endogenous levels of pS592-BAP1.
3.2. Phosphorylation of BAP1 at S592 Occurs in S-phase following UV irradiation
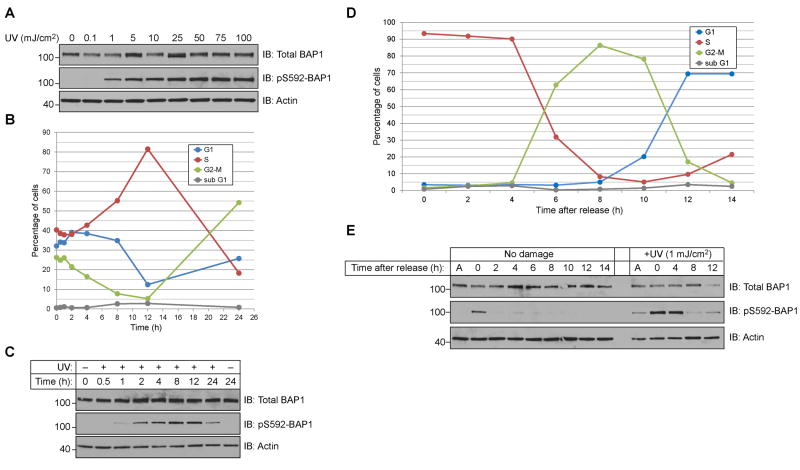
High doses of UV radiation, such as the 50 mJ/cm2 dose recommended by the antibody manufacturer, are known to induce an apoptotic response. Instead we sought to find a UV-dose that induced phosphorylation without causing cell death. We observed a dose-dependent increase in pS592-BAP1 signal that plateaued at a UV-dose of 25 mJ/cm2 (Fig 2A). To demonstrate that a 1 mJ/cm2 dose was not lethal, we assessed the cell cycle distribution of HeLa cells at various times following UV treatment. Histograms of DNA content (supplementary Fig 1) were analyzed with FlowJo software to quantify cell cycle distributions. For 12 h following irradiation cells progressed through G2-M and G1-phases but arrested in S-phase (Fig 2B). After 24 h it was apparent that the cells resumed proliferating as the majority had completed S-phase. Importantly the sub-G1 population, a measure of apoptotic cells, did not exceed 5% of the population.
Figure 2.
BAP1 phosphorylation at S592 occurs in S-phase. (A) UV-dose response of BAP1 phosphorylation in HeLa cells allowed to recover 3 h. (B) HeLa cells recover from a 1 mJ/cm2 of UV-dose and undergo minimal apoptosis. Cell cycle populations of HeLa cells at the indicated time following radiation (C) Cells from (B) were analyzed for BAP1 and pS592-BAP1 levels. (D) Cell cycle analysis of HeLa cells released from a double thymidine block. (E) Cells from (D) were either mock- or UV-treated at the indicated times and allowed to recover 1 h before harvesting.
The levels of BAP1 and pS592-BAP1 in these cells were examined by Western blotting. While BAP1 levels remained unchanged, the pS592-BAP1 signal showed a strong correlation with cells in S-phase (Fig 2C). To better assess whether BAP1 is phosphorylated in S-phase following irradiation with UV, we examined the induction of pS592-BAP1 signal in cell populations within distinct cell cycle phases. We used a double thymidine block to synchronize cells in early S-phase, and collected cells every 2 h following release. We observed a synchronous progression of cells out of S-phase, through G2 and M-phases, and into G1-phase (Fig 2D and supplementary Fig 2). A parallel set of plates were irradiated with UV in early-S (0 h), late-S (4 h), G2-M (8 h) and in G1 (12 h). Total BAP1 levels did not fluctuate during the cell cycle in UV-damaged or untreated cells. The pS592-BAP1 signal, however, showed a maximal induction when cells were in S-phase (Fig 2E). From these studies we conclude that BAP1 phosphorylation at S592 in HeLa following UV irradiation occurs in S-phase and phosphorylation is lost upon completion of replication.
3.3. BAP1 phosphorylated at S592 is predominately unassociated with chromatin
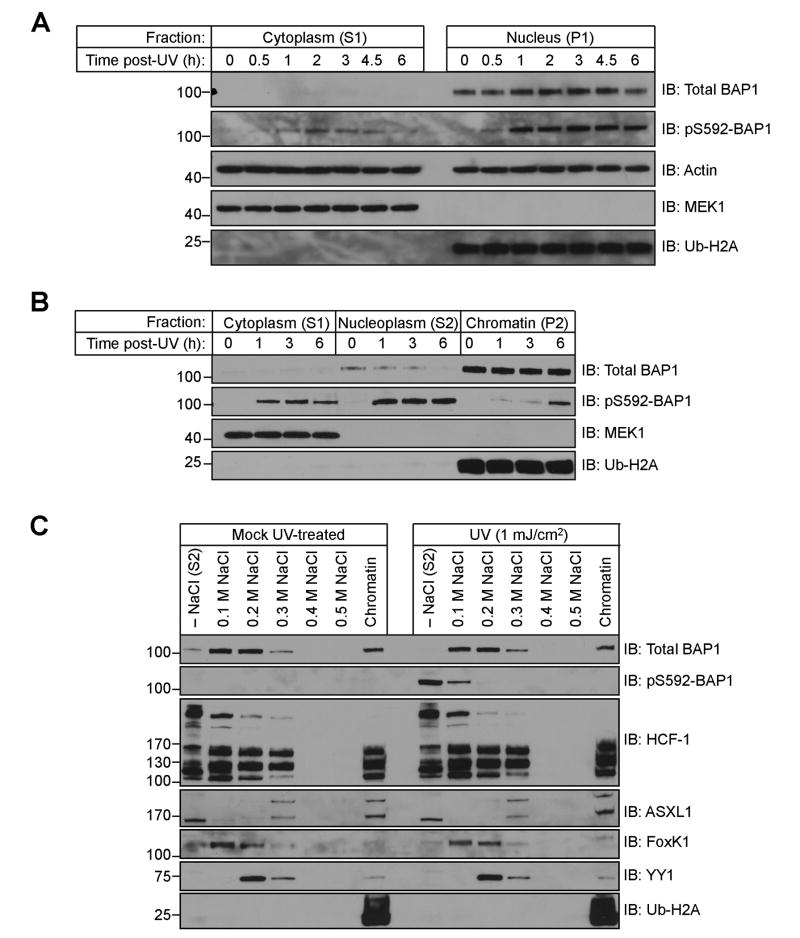
We sought to monitor the localization of pS592-BAP1 as a function of time following irradiation with UV. We used differential centrifugation and a non-ionic detergent to fractionate cells into cytosolic (S1) and nuclear (P1) fractions [22]. We mock- or UV-treated HeLa cells and allowed them to recover for various times before separating into S1 and P1 fractions. Throughout the time course, BAP1 and pS592 BAP1 are predominately localized in the nucleus (Fig 3A). A small portion of pS592-BAP1 is recovered in the cytosolic fraction and this pool increase to a maximum at 2 h, and decreases thereafter. As controls we reprobed these blots with antibodies against Actin (loading), MEK1 (cytosolic), and ubiquityl-Histone H2A (chromatin).
Figure 3.
BAP1 phosphorylated at S592 is not associated with chromatin. (A) pS592-BAP1 is predominately nuclear. Separation of HeLa cells into S1 (cytosolic) and P1 (nuclear) fractions following 1 mJ/cm2 of UV radiation and recovery for the indicated times. (B) Nuclear pS592-BAP1 is predominately nucleoplasmic. (C) Sequential extraction of a HeLa chromatin pellet with increasing concentrations of NaCl.
We further fractionated HeLa nuclei (S1) by lysing in a hypotonic buffer to separate the nucleoplasmic proteins (S2) from chromatin-associated proteins (P2). In the nuclei of untreated cells, we find the majority of BAP1 is in the chromatin-containing P2 fraction, while only a small amount appears in the S2 nucleoplasmic fraction (Fig 3B). Between 1 and 6 h, pS592-BAP1 appears largely in the nucleoplasmic (S2) fraction while the signal originating from the chromatin (P2) fraction peaks at 6 h. The pS592-BAP1 associated with chromatin could be completely eluted with 0.1 M NaCl, while the majority of BAP1 eluted between 0.1–0.3 M NaCl (Fig 3C). Following UV irradiation we did not observe an increase in the nucleoplasmic levels of known BAP1 binding partners HCF-1, FoxK1, YY1, and ASXL1 as their chromatin elution profiles were unperturbed. Thus, pS592-BAP1 binds much less tightly to chromatin than the dephosphorylated form.
The findings presented in Fig 3B were also used to estimate the fraction of total BAP1 that is phosphorylated at S592. Densitometry was used to quantitate the bands in the BAP1 and pS592-BAP1 immunoblots. After 1 h we observe 95% of the pS592-BAP1 signal in the S1 and S2 fractions, and this accounts for just 13% of the total BAP1. This puts an upper limit of 13% of the total BAP1 that is phosphorylated. Collectively these findings support a model whereby following irradiation with UV, chromatin-bound BAP1 in S-phase cells is phosphorylated at S592 and this phospho-species dissociates from chromatin.
3.4. BAP1 is Phosphorylated at S592 in Response to DNA Replication Stress
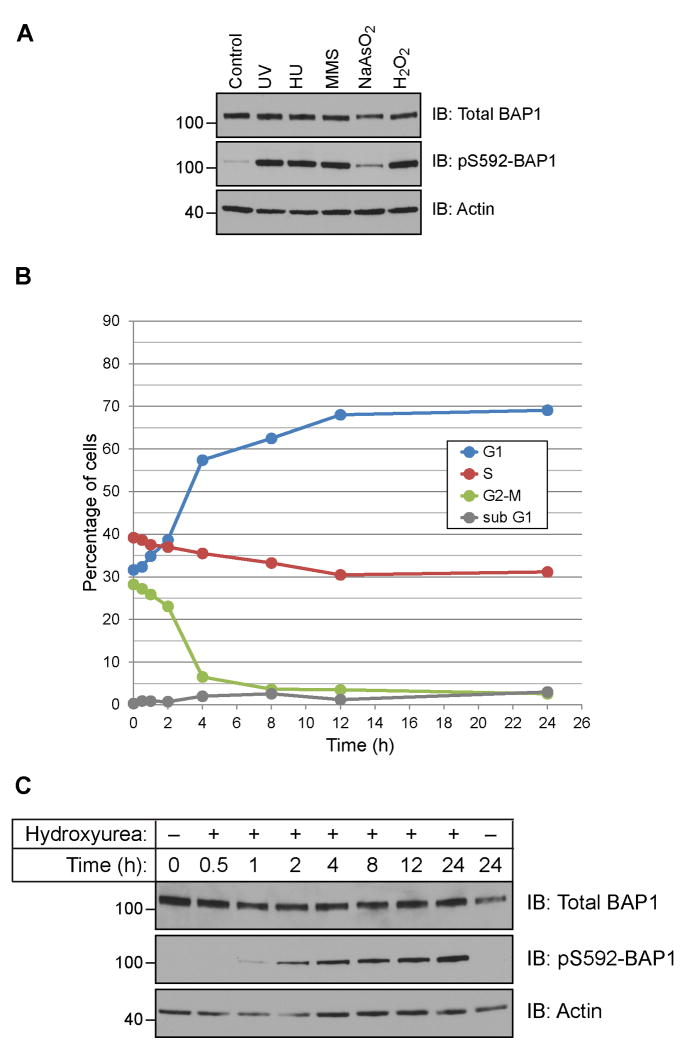
Having determined that BAP1 is phosphorylated in response to UV irradiation, we next asked whether other forms of genotoxic stress also elicit a similar response. Hydroxyurea (HU) is known to cause replication stress and induce stalled replication forks [24]. Replication stress can also be induced by methyl methanesulfonate (MMS), an alkylating agent that methylates purine bases of DNA, and oxidizing agents such as arsenite and hydrogen peroxide (H2O2) which generate reactive oxygen species that react with DNA bases. Each of these forms of stress was capable of inducing phosphorylation, though to different extents (Fig 4A). We observed a similar level of induction of phosphorylation with UV, HU, MMS and H2O2, while treatment with arsenite at levels known to induce stress granule formation had a much weaker effect.
Figure 4.
BAP1 is phosphorylated in response to DNA replication stress. (A) BAP1 is phosphorylated in HeLa cells treated with UV, hydroxyurea (HU), methyl methanesulfonate (MMS), arsenite (NaAsO2) and hydrogen peroxide (H2O2). (B) HeLa cells arrest in S-phase and at the G1/S boundary following exposure to hydroxyurea. (C) Cells from (B) were analyzed for BAP1 and pS592-BAP1 levels.
If replication stress induces phosphorylation of BAP1, we predict that prolonged exposure to HU (stalling an asynchronous cell population in S-phase and at the G1/S boundary) would concomitantly induce phosphorylation of BAP1. We treated HeLa cells with HU and monitored cell cycle progression and pS592 levels as a function of time. Cell cycle analysis found that cells in S-phase immediately arrest, while cells in G2-M progressed into G1 and stalled at the G1/S boundary along with G1 cells (Fig 4B and supplementary Fig 3). Assessing the protein levels in these cells showed little variation in total BAP1 levels, but a steady increase in pS592-BAP1 levels that reached a maximum at 24 h (Fig 4C). These finding support a model whereby replication stress is the stimulus for BAP1 phosphorylation at S592.
Our findings have demonstrated that BAP1 is phosphorylated at S592 within a consensus ATM/ATR site following DNA damage and replication stress. Phosphorylation at S592 is absent in unstressed cells, yet is rapidly triggered to modify a small fraction of BAP1 in S-phase of stressed cells. In contrast to the majority of BAP1 which is chromatin-bound, pS592-BAP1 is predominately unassociated with chromatin. These findings support a model whereby phosphorylation of BAP1 at S592 following DNA damage or stress promotes its dissociation from chromatin. An alternative model, where cytosolic and nucleoplasmic BAP1 become phosphorylated at S592 to trigger chromatin association is less attractive for a number of reasons. The majority of BAP1 is chromatin-bound (Fig 3B), and the kinase implicated under these stressors, ATR, is activated by TOPBP1at sites of damage or stalled replication [25]. Thus, both the substrate and the active kinase reside largely on chromatin. Additionally, the majority of the pS592-BAP1 signal is unassociated with chromatin, even after 6 h (Fig 3B), and chromatin-bound pS592-BAP1 is more easily eluted from chromatin by salt, suggesting it has a weaker affinity for chromatin than unphosphorylated BAP1 (Fig 3C).
Defining the functional consequence of BAP1 phosphorylation and dissociation from chromatin are beyond the scope of this report. However two scenarios can be envisioned. In the first, phosphorylation and dissociation of BAP1 could be a regulatory mechanism that allows replication or repair machinery to access DNA. In the second, BAP1 dissociation from or association with chromatin could function in regulating the transcription of genes involved in DNA replication or repair. In support of the latter model, a correlation between UV-dose and pS592-BAP1 levels was only observed up to 25 mJ/cm2; higher UV-doses and additional damage did not result in higher levels of pS592-BAP1 (Fig 2A). Depletion of BAP1 has been shown to alter the mRNA levels of several proteins, including a subset that are involved in DNA replication and DNA repair [11]. We found that only small fraction of BAP1 is phosphorylated at S592 (Fig 3B), a subset that we suspect regulates transcription from these promoters. Loss of BAP1 would compromise the cells ability to effectively repair the damage or halt cell cycle progression in response to genotoxic stress.
Supplementary Material
Highlights.
BAP1 is phosphorylated at Ser592 (pS592) following DNA damage or replication stress
Phosphorylation of BAP1 is rapid and occurs in S-phase to a small fraction of BAP1
In contrast to the majority of BAP1, pS592-BAP1 is not associated with chromatin
Acknowledgments
We thank O. Laur and the Emory University Custom Cloning Core Facility for constructing BAP1-containing plasmids. This work was supported by the American Cancer Society [grant number 121603-PF-11-144-01-DMC to Z.M.E] and the National Institutes of Health [grant number GM030308 to K.D.W.].
Footnotes
Structured summary of protein interactions: HCF-1 physically interacts with Bap1 by anti bait coimmunoprecipitation (1, 2)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 2.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Meir EG, Wilkinson KD. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer research. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peña-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44:1–12. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eletr ZM, Wilkinson KD. An emerging model for BAP1's role in regulating cell cycle progression. Cell Biochem Biophys. 2011;60:3–11. doi: 10.1007/s12013-011-9184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zargar Z, Tyagi S. Role of host cell factor-1 in cell cycle regulation. Transcription. 2012;3:187–192. doi: 10.4161/trns.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Molecular and cellular biology. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. The Journal of biological chemistry. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey A, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misaghi S, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Molecular and cellular biology. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheuermann JC, Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landreville S, Agapova OA, Matatall KA, Kneass ZT, Onken MD, Lee RS, Bowcock AM, Harbour JW. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:408–416. doi: 10.1158/1078-0432.CCR-11-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa H, Wu W, Koike A, Kojima R, Gomi H, Fukuda M, Ohta T. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer research. 2009;69:111–119. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- 17.Bott M, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nature genetics. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 19.Stokes MP, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 22.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livingstone M, et al. Valosin-containing protein phosphorylation at Ser784 in response to DNA damage. Cancer Res. 2005;65:7533–7540. doi: 10.1158/0008-5472.CAN-04-3729. [DOI] [PubMed] [Google Scholar]
- 24.Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261:16037–16042. [PubMed] [Google Scholar]
- 25.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.