Summary
It has been suggested that the transcription factor Nanog is essential for the establishment of pluripotency during the derivation of embryonic stem cells and induced pluripotent stem cells (iPSCs). However, successful reprogramming to pluripotency with a growing list of divergent transcription factors, at ever-increasing efficiencies, suggests that there may be many distinct routes to a pluripotent state. Here, we have investigated whether Nanog is necessary for reprogramming murine fibroblasts under highly efficient conditions using the canonical-reprogramming factors Oct4, Sox2, Klf4, and cMyc. In agreement with prior results, the efficiency of reprogramming Nanog−/− fibroblasts was significantly lower than that of control fibroblasts. However, in contrast to previous findings, we were able to reproducibly generate iPSCs from Nanog−/− fibroblasts that effectively contributed to the germline of chimeric mice. Thus, whereas Nanog may be an important mediator of reprogramming, it is not required for establishing pluripotency in the mouse, even under standard conditions.
Highlights
-
•
Nanog is not required for canonical reprogramming of murine fibroblasts to iPSCs
-
•
Nanog-deficient iPSCs contribute to the germline of chimeric mice
Here, Eggan et al. show that using highly efficient reprogramming conditions employing canonical KSOM factors enables derivation of chimera- and germline-competent iPSCs from Nanog-deficient MEFs. This counters the previous notion that Nanog is an essential gatekeeper to the pluripotent state and highlights that there are multiple routes to pluripotency.
Introduction
The transcription factor Nanog was identified based on its ability to support embryonic stem cell (ESC) self-renewal in the absence of leukemia inhibitory factor (LIF) (Chambers et al., 2003). Further studies demonstrated that Nanog helps to maintain pluripotency in ESCs by promoting Oct4 and Sox2 expression, while inhibiting a gene expression program leading to primitive endoderm differentiation (Niakan et al., 2010). Nanog levels have been shown to fluctuate greatly within ESC cultures (Chambers et al., 2007; Mitsui et al., 2003). It has been proposed that allelic regulation of the Nanog gene could contribute to its heterogeneous expression pattern (Mitsui et al., 2003; Chambers et al., 2007; Singh et al., 2007; Kalmar et al., 2009; MacArthur et al., 2012; Miyanari and Torres-Padilla, 2012). However, more recent single-cell studies using single-molecule mRNA fluorescence in situ hybridization or allelic reporters suggest that allelic regulation of Nanog may not strongly contribute to variable Nanog expression (Faddah et al., 2013; Filipczyk et al., 2013).
Although the mechanisms that regulate the expression of Nanog continue to be intensively studied, it has been shown that both alleles of Nanog can be eliminated in ESCs without interfering with their differentiation capacity or somatic engraftment in chimeric animals after blastocyst injection (Chambers et al., 2007). Thus, although Nanog may help to maintain pluripotency, it is not strictly required.
Subsequent reports argued that although Nanog is not essential for ESC maintenance, it is absolutely necessary for the establishment of new ESC lines from blastocyst-stage embryos (Silva et al., 2009). Similarly, it was found that induced pluripotent stem cell (iPSC) lines could not be derived from Nanog−/− somatic cells (Silva et al., 2009). These observations led to the conclusion that Nanog is an essential “gate keeper,” which must be expressed before a cell can transit to a pluripotent “ground state.”
Given the important role of Nanog in the maintenance of pluripotency, it is perhaps surprising that addition of Nanog to iPSC-reprogramming cocktails does not necessarily increase the efficiency of reprogramming (Zhao et al., 2008). Moreover, it has been shown that several distinct combinations of transcription factors can reprogram fibroblasts into iPSCs (Buganim et al., 2012) and that the Nanog target gene Esrrb can compensate for Nanog deficiency in some contexts (Festuccia et al., 2012; Martello et al., 2012). Finally, single-cell analysis of the reprogramming process suggests that it occurs through a stochastic series of events in which transcription factor binding and downstream transcriptional cascades can occur at random (Buganim et al., 2012; Polo et al., 2012). Consistent with the mounting evidence that there are many independent pathways to pluripotency, we report here that although eliminating Nanog decreases the efficiency of reprogramming, Nanog is not required for the generation of iPSCs, even under canonical conditions utilizing the expression of Klf4, Sox2, Oct4, and cMyc (KSOM).
Results
We previously found that greatly increasing the titer and promoter strength of retroviral elements encoding reprogramming factors can dramatically improve the efficiency of iPSC generation (Dimos et al., 2008; Ichida et al., 2009). We reasoned that the relatively high efficiency enabled by these modifications might provide a larger window of opportunity than that available in earlier experiments (Silva et al., 2009) for determining whether Nanog was truly necessary for the establishment of pluripotency.
In order to test this hypothesis, we first derived Nanog−/− somatic cells to use as a target population for reprogramming experiments. We utilized Nanog−/− ESCs engineered to express GFP under the control of the ubiquitously expressed CAGGS promoter through random integration of the CAGGS::GFP transgene. These cells were injected into embryonic day 3.5 (E3.5) blastocysts, transfered into recipient females, and resulting embryos were allowed to develop to E1.5 (Chambers et al., 2007). We then prepared mouse embryonic fibroblasts (MEFs) from the resulting chimeric embryos and purified Nanog−/− MEFs via FACS based on their expression of GFP (Figure S1 available online).
As a component of the gene-targeting strategy used to delete Nanog, a neomycin-resistance gene was placed under control of its endogenous promoter (Chambers et al., 2007). Thus, selection with the neomycin analog G418 could be used to rule out the unlikely possibility that undifferentiated pluripotent cells, capable of activating the Nanog promoter, were present in our Nanog−/− MEF cultures. We found that no cells in our MEF preparations survived G418 selection. Thus, there were no undifferentiated cells remaining in these MEF cultures, and we concluded that they were an appropriate substrate for determining whether Nanog was indeed required for the establishment of pluripotency (Figure 1D, top panel).
Figure 1.

Nanog Null MEFs Are Reprogrammed Using KSOM and Activate the Pluripotency Transcriptional Network
(A) The number of iPSC colonies generated from Nanog−/− MEFs with four factors (KSOM) or three factors (KSO). Colonies were scored at day 21 post transduction with reprogramming factors. Error bars represent the SD between two biological replicates.
(B) Primary Nanog−/− iPSC colony 17 days post transduction with KSOM. Scale bars represent 500 μm.
(C) Nanog−/− iPSCs growing on gelatin (top panels) or on irradiated feeder cells (bottom panels). Scale bars represent 500 μm.
(D) Nanog−/− iPSCs activate the endogenous Nanog locus. Cells were treated with 400 ng/ml G418 for 4 days, and representative images were taken at days 0 and 4. Scale bars represent 500 μm.
See also Figure S1.
To ask whether Nanog−/− MEFs could be reprogrammed, we transduced them with high-titer retroviruses encoding either Klf4, Sox2, and Oct4 (KSO) or KSOM (Figure S1). After 21 days, we reproducibly observed an average of five colonies with an iPSC morphology per 180,000 Nanog−/− MEFs transduced with KSOM, representing a reprogramming efficiency 100-fold lower than obtained using control Nanog+/+ MEFs (Figures 1A, 1B, and S2B). The oncogene c-Myc is dispensable for reprogramming, and iPSCs generated in its absence are less tumorigenic in vivo. We therefore next sought to reprogram Nanog−/− MEFs using only KSO. We reproducibly observed two to three putative iPSC colonies emerge per 180,000 MEFS using these three factors. Although the efficiency of apparent reprogramming was lower without c-Myc, we were able to generate iPSC lines using either KSO or KSOM (Figure 1A).
To test whether these Nanog−/− cells were indeed reprogrammed, we isolated GFP+, putative iPSC colonies and expanded them in 2i media (Silva et al., 2008). We designated two putative KSOM Nanog−/− iPSC lines, G2 and G5, whereas two KSO iPSC lines were dubbed 3.1 and 3.2 (Figure 1C). These putative iPSCs maintained an ESC-like morphology over more than ten passages on both gelatin and irradiated feeders (Figure 1C). Like Nanog−/− ESCs, they grow more slowly than control Nanog+/+ ESCs (Figure 1C).
Consistent with the notion that these putative Nanog−/− iPSCs had been fully reprogrammed to ground state pluripotency, we found that they had silenced viral reprogramming transgenes and induced endogenous KSO expression (Figures S2 and 2A). Endogenous Oct4 was expressed in these putative Nanog−/− iPSCs at levels similar to both control Nanog+/+ ESCs and iPSCs as well as Nanog−/− ESCs. Sox2 and Klf4 were expressed in putative Nanog−/− iPSCs at levels similar to Nanog−/− ESCs but slightly lower than control Nanog+/+ ESCs and iPSCs (Figure 2A).
Figure 2.

Nanog Null iPSCs Express Endogenous Pluripotency Genes
(A) qPCR for expression of endogenous KSO. Levels are normalized to GAPDH and plotted relative to control V6.5 mESCs (=1). y axis shows the fold change in expression as determined by the comparative CT method. qPCR was performed in duplicate. Error bars represent the SD between two biological replicates.
(B) qPCR for expression of pluripotency-related genes. Levels are normalized to GAPDH and plotted relative to control V6.5 mESCs (=1). y axis shows the fold change in expression as determined by the comparative CT method. qPCR was performed in duplicate. Error bars represent the SD between two biological replicates.
See also Figure S2.
To ask if the endogenous pluripotency network was activated in these putative Nanog−/− iPSCs, we performed drug selection with G418. As mentioned above, because Nanog−/− cells express the neomycin-resistance gene under control of the Nanog promoter, G418 can be used as a proxy for Nanog promoter activity (Chambers et al., 2007). After 4 days of G418 treatment, putative Nanog−/− iPSC lines G2 and G5 grew without disturbance, whereas control V6.5 ESCs were drug sensitive (Figure 1D).
We next proceeded to further characterize gene expression in putative Nanog−/− iPSCs. As expected, putative Nanog−/− iPSCs did not express exon 2–4 of the Nanog transcript, consistent with the gene-targeting strategy used to generate the knockout line (Chambers et al., 2007). Conversely, high expression of Nanog was detected in control Nanog+/+ ESCs and iPSCs, but not in partially reprogrammed iPSCs (piPS B1) (Figures 2B and S3). Convergent expression of Utf1, Dppa2, Lin28, and Esrrb has been demonstrated to be a stringent indicator of the pluripotent state (Buganim et al., 2012). Thus, we measured expression of Utf1, Lin28, and Esrrb in putative Nanog−/− iPSC lines G2 and G5 and found that they were expressed at levels similar to those found in Nanog−/− ESCs and control Nanog+/+ V6.5 ESCs. On the other hand, a partially reprogrammed Nanog+/+ cell line (piPS B1), which is composed of cells that are not pluripotent, did not express these genes (Figure 2B).
Having confirmed that the Nanog−/− iPSCs expressed key markers of pluripotency, we sought to determine the extent to which the global expression profile of Nanog−/− iPSCs recapitulated that of ESCs. To this end, we performed RNA sequencing (RNA-seq) of two replicates each of control Nanog+/+ ESC, iPSC, MEFs, and partially reprogrammed iPSC as well as Nanog−/− ESC and two iPSC clones, G2 and G5. We observed RNA-seq reads aligning to Nanog exon1, but not exons 2–4, in both the Nanog−/− ESC and iPSC clones (Figure S3). This confirms the absence of Nanog expression and indicates that the endogenous Nanog promoter is activated in these cells. As expected, we observed many RNA-seq reads mapping to all exons in control Nanog+/+ ESCs and iPSCs, but not in control MEFs or partially reprogrammed iPSCs (Figure S3).
Unsupervised hierarchical clustering of the samples based on the expression of all genes revealed that all pluripotent cells clustered together and apart from both MEFs and partially reprogrammed iPSCs. As expected, both Nanog−/− iPSC lines showed a high degree of similarity to Nanog−/− ESCs (Figure 3A). Pairwise comparisons further revealed that relative to MEFs, Nanog−/− iPSCs were as similar to Nanog−/− ESCs as control, Nanog+/+ iPSCs were to control Nanog+/+ ESCs (Figure 3C).
Figure 3.

Nanog Null iPSCs Recapitulate the Global Transcriptome Profile of ESCs
(A) Unsupervised hierarchical clustering of global gene expression obtained by RNA-seq. Biological replicates were analyzed for each sample, and the composite result is shown. piPSC, partially reprogrammed iPSC line B1. JS, Jenson Shannon.
(B) The expression of selected pluripotency-associated factors, as well as early lineage and fibroblast markers, is shown. FPKM, fragments per kilobase of transcript per million fragments mapped. In the case of Oct4 (Pou5f1) and Sox2, RNA-seq does not distinguish between endogenous and exogenous viral transcripts.
(C) The overlap of genes significantly altered (FDR <0.05) more than 2-fold between indicated pluripotent stem cells and MEFs is shown. Genes altered in either independent Nanog−/− iPSC clone (G2 or G5) are included in the Nanog−/− iPSC category.
See also Figure S3.
Analysis of a wide range of reported pluripotency markers revealed that Nanog−/− iPSCs expressed all markers with a high degree of similarity to both Nanog−/− ESCs and control ESCs and iPSCs (Figure 3B). Moreover, Nanog−/− iPSCs expressed low levels of ectoderm, mesoderm, and fibroblast markers similar to Nanog−/− ESCs. Interestingly, as previously reported in Nanog−/− ESCs (Chambers et al., 2007, Niakan et al., 2010), each of the Nanog−/− iPSC lines expressed increased levels of early endoderm markers including Sox17, Gata4, and Gata6 when compared to Nanog+/+ ESCs or Nanog+/+ iPSCs.
Finally, to definitively test whether these putative Nanog−/− iPSCs were indeed pluripotent, we asked whether they could colonize chimeric embryos and contribute differentiated progeny to the three embryonic germ layers. We injected cells from putative Nanog−/− iPSC lines G5, 3.1, and 3.2 into blastocysts and found that they contributed to E12.5 embryos by green fluorescence and to resulting chimeric adults by green fluorescence and coat color (Figures 4A–4C). In the case of the Nanog−/− iPSC lines reprogrammed with KSO, 12 out of 16 and 3 out of 8 embryos recovered were chimeric, and for the Nanog−/− iPSC line made with KSOM (G5), 3 out of 14 embryos were chimeric. Coat-color analysis of adult mice revealed that for the KSO Nanog−/− iPSC lines, 8 out of 15 and 8 out of 14 animals were chimeric, and for the KSOM Nanog−/−, 3 out of 14 animals were chimeric. Importantly, Nanog−/− cells contributed substantially to tissues from the three germ layers in adult chimeras, including the brain, heart, lung, and liver (Figures 4A and S4A).
Figure 4.
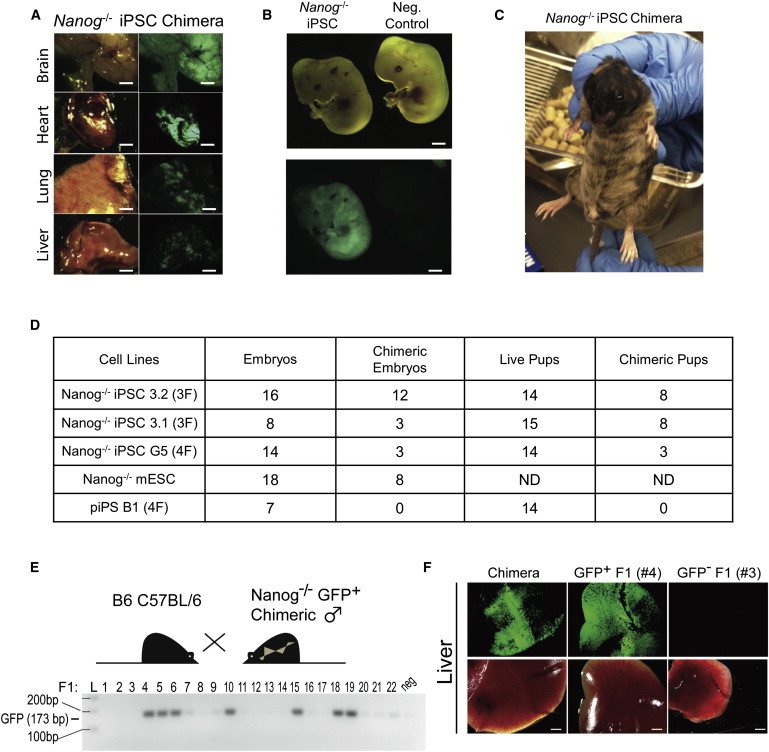
Nanog Null iPSCs Are Pluripotent and Extensively Contribute to Chimeras
(A) Representative images of brain, heart, lung, and liver from postnatal day 33 (P33) Nanog−/− chimeras generated from injection of Nanog−/− iPSC line 3.1 into WT blastocysts. Scale bars represent 5 mm.
(B) Representative E12.5 chimeras generated from injection of Nanog−/− iPSC line 3.1 into WT blastocysts. Scale bars represent 2 mm.
(C) Four-week-old chimera generated from injection of Nanog−/− iPSC line 3.1 into WT blastocysts.
(D) Summary of chimera generation results from three and four factor lines (3.1, 3.2, G5). Numbers in boxes represent number of chimeras and number of embryos or animals recovered and total number of injected embryos. ND, not determined in this experiment. F, female.
(E) Genotyping results of adult progeny (P90) from chimeric and C57BL/6 cross to determine germline transmission of Nanog−/− iPSCs. neg, negative control GFP expression (C57BL/6 uncrossed animal).
(F) Representative images of tissues from adult chimera progeny (P90) genotyped as positive (#4) or negative (#3) GFP transgene, as well as a chimera parent as a positive control. Scale bars represent 2 mm.
See also Figure S4.
To evaluate if the Nanog−/− iPSCs could contribute to the germline and generate mature germ cells, we crossed chimeric Nanog−/− GFP+ iPSC males with C57BL/6 females. Genotyping for the GFP transgene in the resulting adult progeny revealed 7 out of 22 positive animals (Figure 4E). The genotyping strategy was further confirmed by detection of GFP expression in the tissues of transgene-positive animals, for example, F1 #4, but not their transgene-negative littermates (F1 #3, Figure 4F). Partially reprogrammed cells (piPS B1), on the other hand, did not contribute to embryonic or adult chimeras (Figure 4D). These experiments confirmed that unlike the partially reprogrammed Nanog−/− cell lines previously derived (Silva et al., 2009), the Nanog−/− iPSC lines we report here were pluripotent and fully reprogrammed.
Discussion
Although our results seem to contradict previous reports (Silva et al., 2009), we believe that these incongruences are likely explained by a higher efficiency of reprogramming in our hands, which allowed us to observe relatively rare Nanog-independent reprogramming events that were previously undetected. Regardless, our findings underscore the redundant and pliable nature of reprogramming in vitro, further confirming that there are distinct routes to a pluripotent state. One the one hand, this is not surprising in light of recent studies showing that redundant factors within the pluripotency transcriptional network can compensate for loss of Nanog, and lineage-specific transcription factors can replace all canonical reprogramming factors when expressed in the right combinations (Festuccia et al., 2012; Martello et al., 2012; Shu et al., 2013). On the other hand, recent reports that Nanog expression within pluripotent stem cell cultures is not as heterogeneous as previously believed make the finding that it is not required for transition to or maintenance in the pluripotent state surprising (Faddah et al., 2013; Filipczyk et al., 2013).
Similar to our observation that Nanog−/− iPSCs could give rise to chimeric animals, Nanog null ESCs have been shown to contribute to the three germ layers (Chambers et al., 2007). However, in the case of ESC chimeras, Nanog−/− GFP+ cells were not detected in the germline after E12.5. Because the CAGs::GFP construct that marks Nanog−/− cells was introduced by random integration into ESCs, we reasoned that perhaps the GFP transgene might, by chance, not be expressed in cells of the germline that could hamper the ability to detect germline contribution of these cells. We therefore performed crosses using chimeras produced from Nanog null iPSCs and found that they could produce offspring carrying the GFP+ transgene originating from the injected iPSCs. Thus, these results indicate, in contrast to previous results, that Nanog−/− iPSCs can give rise to functional, mature germ cells.
Here, we provide global transcriptional analysis of both our Nanog−/− iPSCs as well as Nanog−/− mESCs (Figure 3). Although these cells have been shown to robustly colonize chimeric embryos, we show that there are still many differences in global expression profiles between Nanog-deficient and wild-type (WT) pluripotent stem cells (Figure 3) (Chambers et al., 2007). A number of genes are differentially expressed between pluripotent cells of these two genotypes, and thus, it would be interesting to further investigate both the mechanism of activation of the core pluripotency network as well as the transcriptional circuit involved in pluripotency maintenance in this context.
Although we have done this work exclusively in murine cells, interactions between members of the core pluripotency network are highly conserved between mouse and human. While the relative inefficiency of iPSC reprogramming in human cells may make rare reprogramming events difficult to detect, it is of great interest to perform similar experiments in human cells. These studies may provide insights between the so-called naive and primed ESC states in the NANOG-deficient context (Gafni et al., 2013).
Based on our results, we conclude that although reprogramming is indeed less efficient in the absence of Nanog, Nanog is not required for the establishment of a pluripotent state, as has been previously suggested. Instead, we conclude that even under standard conditions, there are Nanog-independent routes to pluripotency.
Experimental Procedures
iPSC Generation
Parental Nanog−/− mESCs were obtained from I. Chambers and were cultured on feeders in 2i plus LIF conditions as previously described by Chambers et al. (2007) and Silva et al. (2008). To obtain Nanog−/− MEFs, Nanog−/− mESCs were injected into blastocyst-stage embryos. At E12.5, MEFs were dissected out and sorted for constitutive GFP expression, indicating Nanog−/− genotype (Chambers et al., 2007). For reprogramming, MEFs were transduced with retroviruses carrying murine KSO, with or without c-Myc exactly as described by Ichida et al. (2009). On day 20 posttransduction with reprogramming transgenes, iPSC colonies were picked and passaged onto feeders and cultured in 2i plus LIF conditions with passaging every 5 days (Silva et al., 2008).
Chimera Generation
All procedures involving animal subjects were approved in advance by the Harvard University Institutional Animal Care and Use Committee. Chimeras were generated by injection of Nanog−/− iPSCs into E3.5 strain 129 blastocysts. At E12.5, embryos were dissected, and whole embryos were analyzed for GFP expression in somatic tissues. Fourteen-day-old pups were dissected to analyze chimeric contribution in adult tissues by fluorescence. To analyze the contribution of Nanog−/− iPSCs to the germline, adult male chimera animals were bred with C57BL/6 females, and resulting pups were genotyped with Jackson Laboratory GFP primers 5′-AGTTCATCTGCACCACCG-3′ and 5′- TCCTTGAAGAAGATGGTGCG-3′. Three-month-old adult progeny were analyzed for chimeric contribution to adult tissues.
qPCR
qPCR was performed using iScript cDNA Synthesis Kit and SYBR Green qPCR Supermix (Bio-Rad) according to manufacturers’ instructions on a Bio-Rad iQ5. Levels were normalized to GAPDH expression using the delta-delta CT method and plotted relative to expression in control V6.5 mESCs. Primer sequences used for qPCR: Esrrb, forward 5′-CACCTGCTAAAAAGCCATTGACT-3′, reverse 5′-CAACCCCTAGTAGATTCGAGACGAT-3′; GAPDH, forward 5′-TTCACCACCATGGAGAAGGC-3′, reverse 5′-CCCTTTTGGCTCCACCCT-3′; Klf4, forward 5′-CTATGCAGGCTGTGGCAAAACC-3′, reverse 5′-TTGCGGTAGTGCCTGGTCAGTT-3′; Lin28, forward 5′-GAAGAACATGCAGAAGCGAAGA-3′, reverse 5′-CCGCAGTTGTAGCACCTGTCT-3′; Nanog, forward 5′-AAACCAGTGGTTGAAGACTAGCAA-3′, reverse 5′-GGTGCTGAGCCCTTCTGAATC-3′; Utf1, forward 5′-GTCCCTCTCCGCGTTAGC-3′, reverse 5′-GGCAGGTTCGTCATTTTCC-3′; Sox2, forward 5′-AAGGGTTCTTGCTGGGTTTT-3′, reverse 5′-AGACCACGAAAACGGTCTTG-3′; Oct4, forward 5′-CACGAGTGGAAAGCAACTCA-3′, reverse 5′-AGATGGTGGTCTGGCTGAAC-3′.
RNA-Seq
RNA was harvested from at least two biological replicates using TRIzol (Invitrogen) according to the manufacturers’ directions. RNA quality was determined using BioAnalyzer (Aligent). RNA integrity numbers above 7.5 were deemed sufficiently high quality to proceed with library preparation. In brief, RNA-seq libraries were generated from ∼250 ng total RNA using the Illumina TruSeq RNA kit v.2, according to the manufacturers’ directions. Libraries were sequenced at the Broad Institute’s Genomics Platform on a HiSeq 2500. A total of 20–60 million 100 bp, paired end reads were obtained for each sample. Reference files of the murine genome build mm10, as well as Ensembl transcript annotations, were obtained from iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn). Reads were aligned to the genome using the split read aligner TopHat (v.2.0.7) and Bowtie2 (v.2.0.5) using default parameters as previously described by Trapnell et al. (2012). Transcript assembly, isoform-specific quantitation and differential expression analysis was performed using Cufflinks (v.2.1.1) (Trapnell et al., 2012). A genome-wide corrected false discovery rate (FDR) of less than 0.05 was considered significant. Computations were performed on the Odyssey cluster supported by the FAS Science Division Research Computing Group at Harvard University.
Acknowledgments
We thank I. Chambers for generously sharing Nanog−/− ESC lines, L. Rubin for providing piPSC lines, and members of the Eggan Laboratory for helpful discussion. This work was supported by R01 and P01 grants from NIGMS to K.E. B.N.D.-D. is a Milton Safenowitz Post Doctoral Fellow of the ALS association.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
RNA-seq data have been deposited to the Gene Expression Omnibus (accession number GSE53212).
Supplemental Information
References
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Faddah D.A., Wang H., Cheng A.W., Katz Y., Buganim Y., Jaenisch R. Single-cell analysis reveals that expression of nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell Stem Cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S.R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipczyk A., Gkatzis K., Fu J., Hoppe P.S., Lickert H., Anastassiadis K., Schroeder T. Biallelic expression of nanog protein in mouse embryonic stem cells. Cell Stem Cell. 2013;13:12–13. doi: 10.1016/j.stem.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur B.D., Sevilla A., Lenz M., Müller F.J., Schuldt B.M., Schuppert A.A., Ridden S.J., Stumpf P.S., Fidalgo M., Ma’ayan A. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat. Cell Biol. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Göttgens B., Niwa H., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Miyanari Y., Torres-Padilla M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Niakan K.K., Ji H., Maehr R., Vokes S.A., Rodolfa K.T., Sherwood R.I., Yamaki M., Dimos J.T., Chen A.E., Melton D.A. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W., Tang X., Yang H., Shen L., Zuo X. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Hamazaki T., Hankowski K.E., Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yin X., Qin H., Zhu F., Liu H., Yang W., Zhang Q., Xiang C., Hou P., Song Z. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


