Abstract
Immune control of viral infections is modulated by diverse T cell receptor (TCR) clonotypes engaging peptide-MHC class I complexes on infected cells, but the relationship between TCR structure and antiviral function is unclear. Here we apply in silico molecular modeling with in vivo mutagenesis studies to investigate TCR-pMHC interactions from multiple CTL clonotypes specific for a well-defined HIV-1 epitope. Our molecular dynamics simulations of viral peptide-HLA-TCR complexes, based on two independent co-crystal structure templates, reveal that effective and ineffective clonotypes bind to the terminal portions of the peptide-MHC through similar salt bridges, but their hydrophobic side-chain packings can be very different, which accounts for the major part of the differences among these clonotypes. Non-specific hydrogen bonding to viral peptide also accommodates greater epitope variants. Furthermore, free energy perturbation calculations for point mutations on the viral peptide KK10 show excellent agreement with in vivo mutagenesis assays, with new predictions confirmed by additional experiments. These findings indicate a direct structural basis for heterogeneous CTL antiviral function.
Chronic viral infections in mice and humans are modulated by cytotoxic T lymphocyte (CTL)-mediated immune pressure, but the level of immune control is quite variable. This is particularly apparent with HIV-1, a chronic human viral infection that undergoes rapid sequence variation under CTL-mediated immune pressure (see details in a recent review1). Immune escape is impeded by the elicitation of a diverse T cell response consisting of multiple specificities2,3, but these can vary significantly in antiviral efficacy4,5,6. Some induced responses effectively target both wild-type (WT) virus and naturally arising mutants, impeding the development of immune escape7,8,9. Others are readily detectable by tetramer staining, but exhibit little antiviral function in vitro, and appear to exert little immune selection pressure in vivo7.
These differences among CTL clonotypes in potency and cross-reactivity of recognition of HIV-1 and its variants7 are in line with the T cell receptor (TCR)-based modulation of effector cell subsets10 and suggest that the fine specificity of TCRs may modulate their antiviral function7,8,11,12,13. Although a co-crystalized X-ray structure has recently been reported for a single clonotype interacting with the human leukocyte antigen (HLA) B*2705-restricted epitope KK10 (KRWIILGLNK, Gag aa 263–272)11, the underlying molecular mechanism for clonotypic differences in antiviral efficacy against this and other epitopes has not been fully understood due to the limited availability of co-crystalized structures of the TCR-peptide-MHC complexes.
In this study, we used a combined approach employing both experimental and theoretical techniques to address the relationship between HIV-1 peptide (KK10)-HLA-TCR complex structure and function. We took advantage of unique reagents generated from persons with untreated HIV-1 infection: intrapatient CTL clones with distinct TCR clonotypes, all induced in vivo against the same HLA B*2705-restricted epitope in Gag, but differing in measures of antiviral function. Using molecular dynamics (MD) simulations coupled with functional assays, we show that specific binding patterns among KK10-HLA (B*2705)-TCR interactions are associated with enhanced antiviral efficacy and cross-reactivity of the clonotypes.
Results
Antiviral function of KK10-specific clonotypes
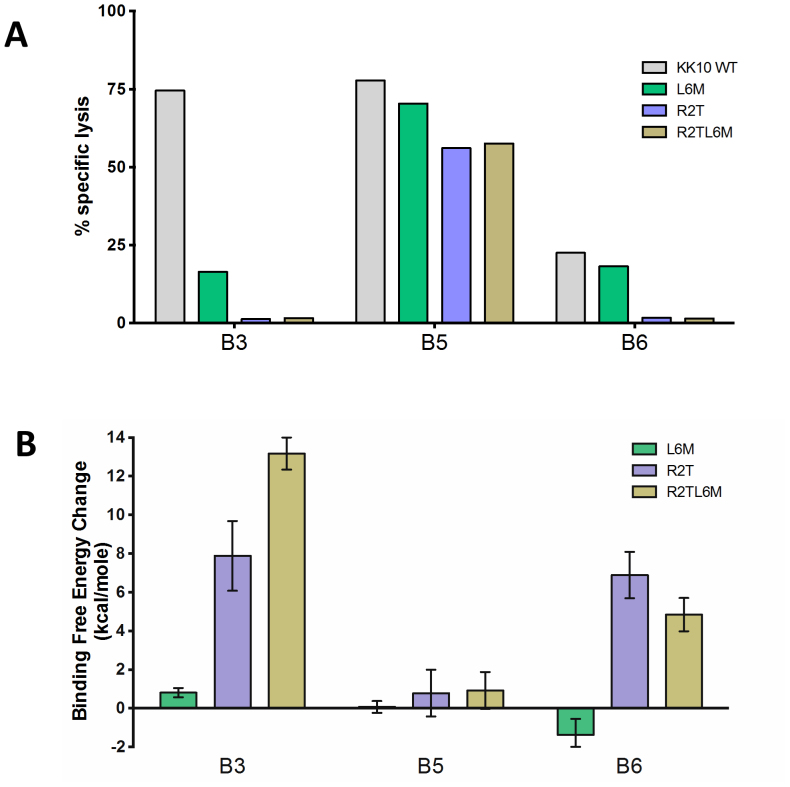
Previously established HLA-B*2705 KK10-specific CTL clonotypes B3, B5 and B6 derived from an HIV-1 controller (FW56) were used in these studies. All clonotypes had considerable sequence diversity in TCR complementarity determining regions (CDRs), and differed in Vβ and Vα gene usage7 (Table S1). For example, the sequence identities are only ~30% between clones B3, B5, and between clones B5, B6 (Table S2). These clones represented both dominant and subdominant clonotypes present in vivo, but differed markedly in their ability to inhibit virus replication in vitro7. Repeated testing using cryopreserved samples revealed that these clonotypes maintained stable differences in their ability to recognize CD4 T cells infected with HIV-1, as shown in the specific lysis assays (Fig. 1A). It is clear that clonotype B5 broadly recognized the WT and its variant viruses, whereas B3 robustly recognized WT virus but showed little cross-reactivity against variants. In contrast, B6 exhibited weak and narrow recognition. The molecular or structural basis for this diversity among different clonotypes is still largely unknown, which is the main focus of the following large scale molecular modeling.
Figure 1. Differential antiviral efficacy of B*27-KK10 specific clonotypes.
(A). The ability of KK10-specific clonotypes to recognize NL4-3 wild-type and variant viruses was tested in the standard 4-h chromium release assay with virally infected HLA-B*2705-encoding green fluorescent protein (GFP) reporter GXR cells at an effector/target cell ratio of 1:1. Viable infected (GFP-positive) GXR cells were sorted by a FACS Aria cell-sorting instrument after infection for 5 days and used as target cells. (B). The FEP simulation results for the predicted TCR-KK10 binding free energy change due to L6M and R2T single mutations, and R2TL6M double mutation in viral peptide KK10.
Structural modeling of TCR-pMHC interaction
Having defined clonotypic difference of antiviral efficacy, we next examined structural properties of effective and ineffective clonotypes in an attempt to relate function to structure. Computational methods have been widely used in structural modeling of biomolecular complexes. In particular, template-based models (homology models) have demonstrated high accuracy in protein structure predictions, as shown in recent CASP competitions14,15. Meanwhile, large-scale free energy perturbation (FEP) is regarded as the most rigorous and reliable method in predicting binding affinities, which has also achieved high accuracy in characterizing key residues and their mutational effects for many protein-protein and protein–ligand bindings, as compared with experiments16,17,18. We thus went on to investigate the structural features, dynamic properties, and mutational effects of the ternary binding complexes of KK10-HLA (B*2705)-TCR with computational techniques. Here we used two independent co-crystal structures as templates (more below) to construct homology models of the ternary binding complexes for each FW56 TCR clonotype (see Materials and Methods section for more details), which does not have a crystal structure yet. Considering the high sequence diversities among these TCR clonotypes, it is also difficult, if not impossible, to experimentally determine all KK10-HLA-TCR complex structures here. After the modeling construction, we then performed an aggregate of microsecond MD simulations of these complexes in explicit solvent using IBM Blue Gene supercomputers, which allows us to study their dynamics with atomic resolution. The sensitivity to viral peptide mutants of various clonotypes was also quantitatively measured by calculating their binding affinity changes with FEP (more below).
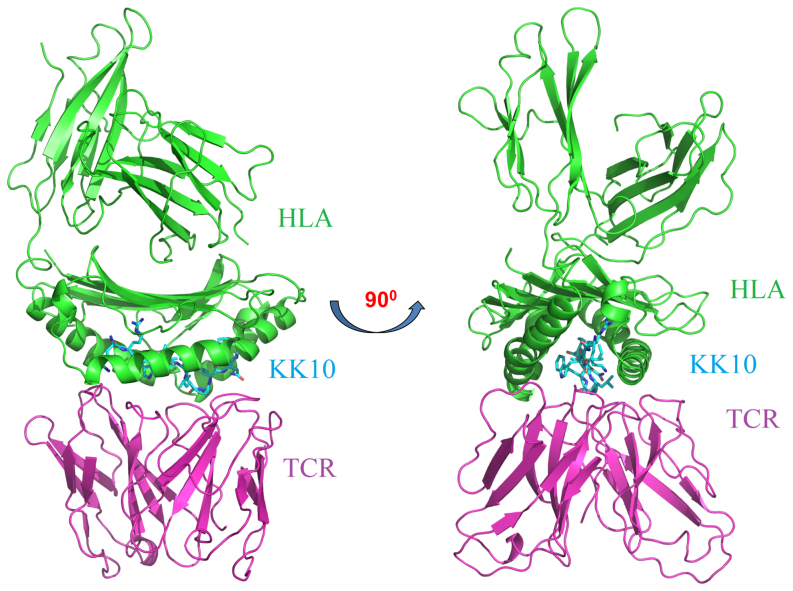
One representative model of the complex structure for the clonotype B5 is shown in Fig. 2. The viral peptide fits well into the binding groove formed at the interface of the HLA and the TCR. Extensive MD simulations also indicate that these model structures of clonotypes B3, B5 and B6 are stable, as shown by RMSDs (Fig. S1), indicating a reasonable construction of homology models.
Figure 2. Structural view of HLA B*2705-KK10-TCR 3-way binding complex.
The HLA-B*2705 is shown in green; the KK10 viral peptide is colored in cyan; and the TCR is colored in magenta. All the structural figures are generated with PyMol software.
Conserved binding features for functionally different clonotypes
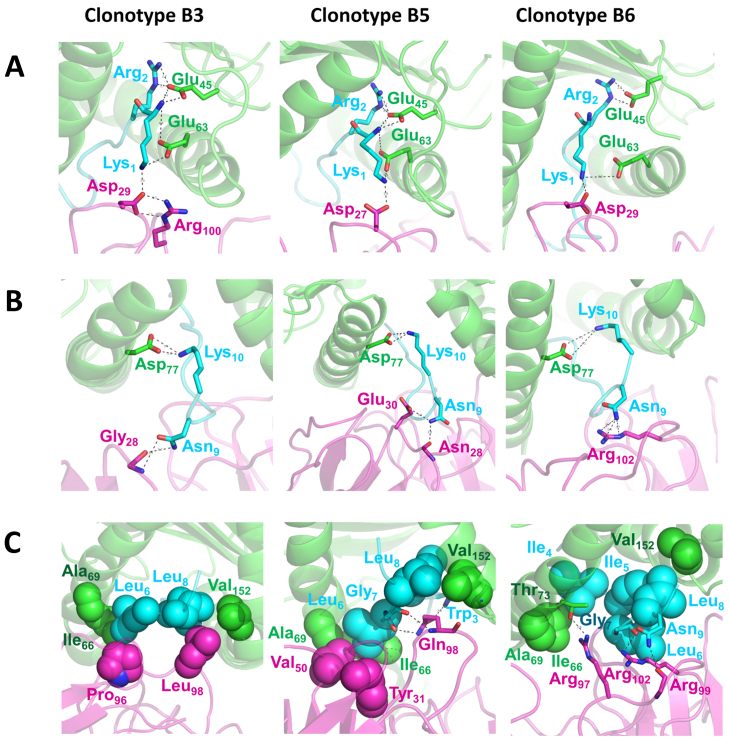
The simulated structures revealed a similar binding mode (binding pocket) on TCRs for KK10-HLA over all clonotypes examined, with more than 20 residues from both the TCR and the HLA-KK10 binary complex involved at the binding site (Fig. 3). We first examined interactions involving the N and C termini of the viral peptide KK10. Although the sequence of the TCR α chain in the clonotype B5 is different from that in clonotypes B3 and B6, we found three common salt-bridges in all clonotypes at the N-terminus of KK10: Lys1 of KK10 with Glu63 of HLA, Arg2 of KK10 with Glu45 of HLA, and Lys1 of KK10 with Asp27/29 at CDR1α loop of the TCR. In particular, for the effective clonotypes B3 and B5, KK10 is further stabilized by extra hydrogen bonds between Glu45/Glu63 of HLA and the amide group at the N-terminus of KK10 (Fig. 3A). The Glu63 at the B pocket of HLA was also recently recognized by genome-wide association analysis (GWAS) as one of the key residues in the single-nucleotide polymorphism of HIV-1 controllers and progressors19,20. Similarly, favorable electrostatic interactions contribute to the C-terminal stabilization of the viral peptide. A salt-bridge between Lys10 of KK10 and Asp77 of HLA is found across all clonotypes, while Asn9 of KK10 provides additional stability via hydrogen bonds with polar/charged residues from the CDRβ of the TCR (Fig. 3B). Thus, conserved and well-organized salt-bridges and hydrogen bonds play an important role in securing KK10 tightly into the binding pocket between the HLAs and the TCRs. These common N- and C-terminal features contribute to the overall binding affinity between KK10-HLA and TCRs in all clonotypes examined.
Figure 3. Structural comparison of different clonotypes bound to the KK10 peptide.
The binding site and interactions at the N-terminal region (A), the C-terminal region (B), and the middle region (C) of KK10 peptide are rendered with spheres (non-polar interactions) or sticks (polar interactions) (green, HLA; cyan, HIV-1 KK10 peptide; magenta, TCR). The overall complexes are represented as cartoons. The representative snapshots were taken from totally 600-ns molecular dynamic simulations.
Specific hydrophobic packing patterns determine the antiviral efficacy and sensitivity to viral peptide mutants of the clonotypes
Contrary to the highly conserved interactions with the terminal residues of KK10, the central residues are shown to have a more diverse and sophisticated interaction with the CDR loops of TCRs, thus endowing clonotypes B3, B5, and B6 with distinct interactive features. The clonotype B3, effective against WT KK10, has a well-defined hydrophobic packing between the viral peptide and the CDR3β of the TCR (Fig. 3C), where Leu6 and Leu8 of KK10 form favorable contacts with Pro96 and Leu98 of the CDR3β, respectively. The hydrophobic interaction is further enhanced by the additional contacts with hydrophobic residues Ile66, Ala69, and Val152 of the HLA. The formation of a stable hydrophobic core, accompanied by a lower desolvation penalty, makes the B3 TCR and pMHC association more specific. Furthermore, such hydrophobic core (or cavity) formed by nanoscale dewetting (drying) was suggested to provide a substantial driving force for protein complex collapse and overall stability in our previous studies21,22,23. This strong and specific hydrophobic interactions in B3 clonotype (KK10-Leu6 with B3 TCR's Pro96, KK10-Leu8 with B3 TCR's Leu98; see Fig. 3) not only indicates high antiviral efficacy of B3 to the WT virus, but also implies a potentially relatively weak cross-reactivity against variants (i.e. more sensitive to mutants; see more below), as was seen experimentally.
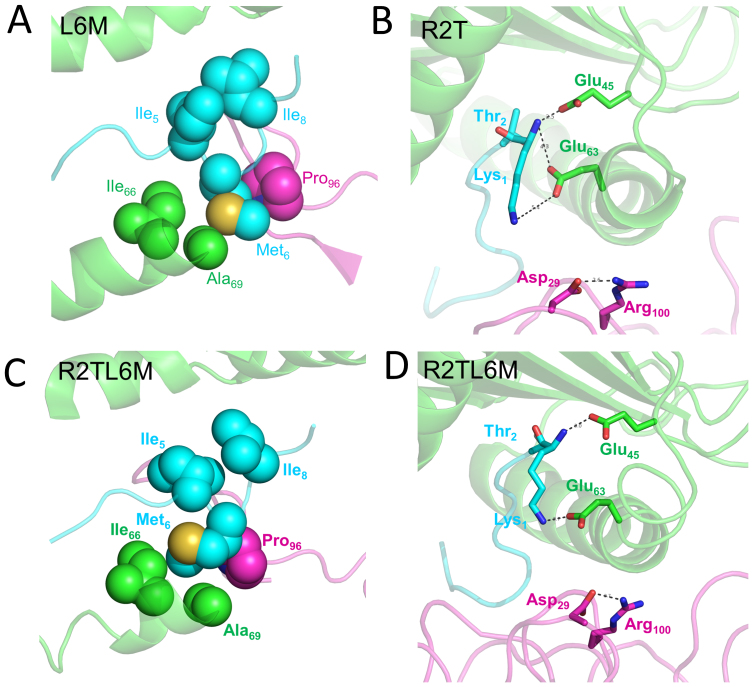
In order to further assess the sensitivity to viral peptide mutants of clonotype B3, which was effective against WT virus but less so against mutants, we calculated binding affinity changes due to mutations frequently occurring in vivo (i.e., L6M, R2T and R2TL6M) in KK10 (Fig. 1B). Overall, our in silico mutagenesis studies show excellent agreement with those in vivo data. Our rigorous FEP calculations show a dramatic weakening in the binding affinities for both R2T (ΔΔG = 7.88 ± 1.80 kcal/mol) and R2TL6M (ΔΔG = 13.17 ± 0.83 kcal/mol) mutants, albeit a somewhat milder reduction in the single L6M mutation (ΔΔG = 0.81 ± 0.24 kcal/mol, which is equivalent to a ~4-fold increase of the dissociation constant Kd) (see Table S3), consistent with the specific lysis assay data (Fig. 1A). For more details, the structural comparison reveals that the L6M variant introduces a relatively less hydrophobic side chain to position 6 of KK10, which slightly weakens the well-packed hydrophobic core formed in the WT peptide (Fig. 4A), while the R2T mutant directly destroys the highly conserved salt-bridge between Arg2 in KK10 and Glu45 in HLA. Moreover, the R2T mutation also indirectly loosens another important salt-bridge between Lys1 in KK10 and Asp29 at the CDR1α of the TCR through conformational distortion (Fig. 4B). The even larger decrease in binding affinity for the R2TL6M double mutant is found to be related to larger conformational distortions and serious damage in conserved “signature salt-bridges” (Fig. 4C and 4D).
Figure 4. Structural comparison of HLA B*2705-KK10-TCR complexes due to in vivo occurring mutants.
The following mutations of KK10 peptide for FW56 clone B3 is shown (green, HLA; cyan, the HIV KK10 peptide; magenta, TCR): (A) L6M mutant. (B) R2T mutant. (C) and (D) R2TL6M double mutant. The overall complex is represented as cartoon and the residues at the binding site are rendered with spheres (non-polar interactions) or sticks (polar interactions).
The binding between the clonotype B5, which is potent against WT virus and variants (see Fig. 1A), and the central residues of KK10 is more complicated due to combined effects of both specific side-chain and non-specific backbone interactions (Fig. 3C). In WT KK10, Leu6 makes hydrophobic contacts with Tyr31 in the CDR1β and Val50 in the CDR2β of the TCR. Meanwhile, Gln98 in the CDR3β forms three hydrogen bonds with nitrogen in the indole ring of Trp3, and backbone carbonyl groups of Leu6 and Gly7 of KK10, respectively. These hydrogen bonds are considered as energetically favored, which could enhance the binding affinity between TCR and KK10. We note that two of the hydrogen bonds are formed with the backbone of KK10 (Gly7 with B5 TCR's Gln98), indicating a lesser sensitivity to the side-chain variations of KK10 (they only interact with the backbone amide and carbonyl groups) (Fig. 3C), which is different from the very specific hydrophobic interaction patterns in B3 clonotype.
These structural observations explain why the clonotype B3 has higher sensitivity to viral peptide mutants against HIV-1 in vivo mutants than the clonotype B5. Our FEP study of the clonotype B5 further confirms this (Fig. 1B), in that there is no apparent binding affinity change in the mutant peptide L6M (ΔΔG = 0.07 ± 0.30 kcal/mol), and slightly unfavorable changes in R2T (ΔΔG = 0.78 ± 1.21 kcal/mol) and R2TL6M (ΔΔG = 0.92 ± 0.95 kcal/mol) mutants (Table S3), again in excellent agreement with the in vitro experiment. The structural comparisons between these mutants also reveal that the key interactions observed in the WT KK10 peptide are well preserved with no observable binding mode change (Fig. S2), providing a structural basis for the superior efficacy of the clonotype B5 over the broad viral mutations.
The interaction between the least effective clonotype, B6, and WT KK10 peptide is less complex. Only two hydrogen bonds are available between the CDR3β of the TCR (Arg99 and Arg102) and KK10 peptide (the backbone of Gly7, and the side chain of Asn9, see Fig. 3C). The relatively weak antiviral efficacy, shown in the functional lysis assay (Fig. 1A), is consistent with a lack of favorable hydrophobic interaction, as compared to the clonotypes B3 and B5 (Fig. 3C; also see Fig. S3 and Note 1 in supplementary data).
Alanine substitutions further confirm the structural basis for antiviral efficacy
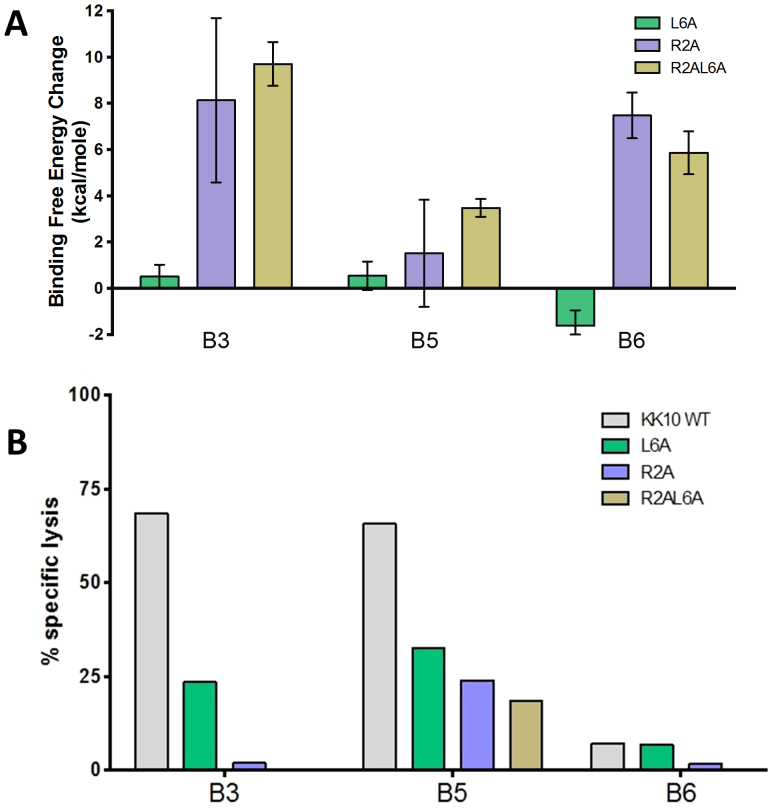
These in silico data suggest specific molecular interactions that modulate antiviral efficacy of specific clonotypes. We next extended the FEP calculations with alanine substitutions (R2A, L6A and R2AL6A) in KK10 peptide, to further investigate the structural basis and intrinsic roles of these important residues in the binding specificity and functional attributes of these clonotypes. Overall, the alanine mutants display a similar trend in binding affinity changes as those naturally occurring mutants (Table S4). For the clonotype B3, the binding affinity is slightly reduced with the L6A mutant (ΔΔG = 0.50 ± 0.51 kcal/mol) as compared to the L6M mutant (Fig. 5A). Although alanine is a smaller and less hydrophobic residue than the original leucine, it is observed that other hydrophobic residues Ile66 and Ala69 in the HLA can adjust themselves to closely pack with Ile4 of KK10 (Fig. S4A). Similar to the R2T variant, a dramatic decrease in the binding affinity (ΔΔG = 8.13 ± 3.55 kcal/mol) is observed in the R2A mutation, again due to the loss of conserved signature salt-bridges at N-terminus (Fig. S4B). For the clonotype B5, all alanine mutants result in relatively small binding affinity losses, but slightly larger than those in vivo mutants L6M, R2T, and R2TL6M (Fig. 5A and Fig. S5), meaning that the low sensitivity to viral peptide mutants is still largely maintained in the alanine mutants in the clonotype B5, as found experimentally. Similarly, the binding affinity changes of alanine mutants in the clonotype B6 are very close to those in vivo mutants (Fig. S6, also see Note 2 in supplementary data). These FEP predictions are then confirmed by our experimental data from the specific lysis assay (Fig. 5B), indicating our combined approach can quantitatively measure even small differences in binding affinities, thus revealing a direct relationship between peptide-HLA-TCR structure and function.
Figure 5. Impact of alanine substitutions on FEP and antiviral efficacy.
(A). The FEP simulation results for the predicted TCR-KK10 binding free energy change due to L6A and R2A single mutations, and R2AL6A double mutation in viral peptide KK10. (B). The ability of KK10-specific clonotypes to recognize KK10 WT peptide and alanine mutants was tested in the standard 4-h chromium release assay with peptide-loaded HLA-B*2705-expressing GXR cells at an effector/target cell ratio of 1:1.
A second co-crystal structure template further confirms our homology models
It should be noted that a new co-crystal structure with the same HLA-KK10 but a different TCR clonotype was recently reported11. We repeated the structural modeling process for clonotypes B3, B5 and B6 using this new co-crystal structure as a second template, and found the newly built homology models shared very similar structures, with comparable binding modes for KK10 as those from the previous template24,25 (Fig. S7). For example, we found similar salt-bridges and polar interactions between TCR and KK10 peptide in different clonotypes, and similar hydrophobic interactions in B3 and B5 clonotypes. We also repeated the FEP calculations for two representative in vivo mutants, L6M and R2T in KK10, which again displayed very similar binding affinity changes with the previous case for all the three clonotypes (see data in Table S5). These findings further support our conclusion that specific structural characteristics of epitope-specific clonotypic TCRs modulate TCR engagement and CTL antiviral efficacy.
Discussion
Multiple mechanisms (e.g. the induced-fit model) have been proposed for the TCR and pMHC recognition and binding25,26,27. The TCR-peptide-MHC ternary complex cannot be easily extrapolated from the free peptide-MHC structure, since TCR-peptide recognition is mainly determined by the highly flexible and variable CDR loops. Using a panel of TCR clonotypes specific for the same protective pMHC but differing in antiviral function, we show that conserved polar interactions, including salt-bridges and hydrogen bonds, are presented in both effective and ineffective clonotypes, despite TCR usage of different Vα and Vβ genes. Meanwhile, significantly different packing modes are observed among the clonotypes, especially with the mostly hydrophobic central residues of the viral peptide, which suggests a key role in antiviral efficacy as well as in cross-reactivity.
These studies also allowed us to examine structure and function relationships in the relative ability of different TCR clonotypes to recognize naturally occurring variants within the targeted epitope. CTL-mediated control in rapidly mutating viral infections results from specific TCR-pMHC interactions that trigger antiviral efficacy as well as non-specific interactions that provide a degree of tolerance to the sequence variation of the viral peptide26,28,29. Our results using functionally distinct clonotypes from an HIV-1 controller show the impact of the interplay of both specific hydrophobic interactions and non-specific hydrogen bonds in the TCR-viral peptide interaction. In the case examined here, the strong antiviral efficacy of the clonotype B5 is mainly derived from the favorable hydrophobic interactions between the central viral peptide residues and the corresponding hydrophobic residues in the CDR loops. Additionally, the low sensitivity to viral peptide mutants of the clonotype B5 is largely maintained by the strong non-specific hydrogen bonds (also known as dehydrons, which are buried or partially buried hydrogen bonds30) between the viral peptide backbone and Gln98 at the CDR3β of the TCR. These dehydrons are energetically very favored, which enhance the binding affinity between KK10 and TCR, resulting in a less sensitivity to mutations. Compared to the clonotype B5, B3's TCR predominantly uses highly specific hydrophobic interactions for contacts with the central residues Leu6 and Leu8 of KK10. This may explain why the high antiviral efficacy is achieved in the clonotype B3, but with a rather high sensitivity to viral peptide variants. There are larger conformational distortions in the clonotype B3 upon mutations. For example, in the R2T mutant, serious damages to the conserved signature salt-bridge (KK10 Lys1 – TCR Asp29) were seen (Figure S8).
These structural analyses were further validated by calculating the binding affinity changes due to point mutations in the viral peptide with the rigorous FEP method. Intrapatient analyses in subject FW56 for clonotypes B3, B5 and B6 showed a clear hierarchy between binding affinity and antiviral efficacy and cross-reactivity. Specific structural alterations accounting for much larger binding affinity losses were found for the KK10 in vivo mutants for clonotype B3, indicating a high sensitivity to viral peptide mutants, such as R2T, L6M and R2TL6M, which was confirmed by in silico FEP calculations. Additional FEP predictions for all the corresponding alanine substitutions were then further validated by in silico mutagenesis studies. The excellent agreements between in silico and in vivo mutagenesis studies indicate our current structural models and analyses might be reasonable. In particular, our FEP calculations successfully predicted a larger binding affinity change for L6A and R2A mutants than those naturally occurring L6M and R2T ones in the KK10 peptide with the clonotype B5, which were confirmed by our experimental assays. These findings are consistent with recent studies showing that TCR composition and associated signaling potential may modulate effector cell patterning10.
We conclude that by applying rigorous binding affinity prediction tools with in vivo mutagenesis studies, we not only capture structural and energetic details of particular substitutions, but also reveal the underlying molecular mechanism for differences among TCR clonotypes in their potency and cross-reactivity. This study provides structural and mechanistic insights into T cell-mediated antiviral immunity in a chronic human viral infection.
Methods
Chromium release assay
HLA-B*2705-expressing green fluorescent protein (GFP) reporter CEM-derived GXR cells (which contain a plasmid encoding GFP driven by the long terminal repeat of HIV-1) were constructed as described31 and infected with the NL4-3 virus or viral variants expressing one or more mutations in the gene encoding Gag p24 KK10 peptide (KRWIILGLNK, Gag aa 263–272) by site-directed mutagenesis32 at the specified multiplicity of infection (MOI). On day 5 after infection, viable virally infected cells were sorted on a FACS Aria cell-sorting instrument (BD Biosciences) and labeled with chromium for 1 hr at 37°C. CTL clones were then added at the indicated effector-target ratios, and a standard 4-h chromium release assay was performed as previously described33. Percent specific lysis was calculated as [(mean experimental cpm − mean spontaneous cpm)/(mean maximum cpm − mean spontaneous cpm)] × 100. Spontaneous and maximum releases were determined by incubating the labeled target cells with medium alone or 2% Triton X-100, respectively.
Structural modeling of HLA B*2705-KK10-TCR complexes
Two independent templates were used in our homology structure modeling for the viral peptide-HLA-TCR complexes. For the first template, the binary structure of HLA B*2705 and KK10 peptide (KRWIILGLNK) complex was taken directly from the co-crystallized x-ray structure (PDB entry 2BSS)34. The structures of three different TCR clonotypes, TCRBV4-3 (B3), TRBV6-5 (B5), and TRBV20-1 (B6) of a controller FW56, were built from their closest homologs, with MODELLER software package35,36, based on TCR sequence details from the respective clonotypes B3, B5, and B6 (Table S1). After that, the entire complex was aligned to an available ternary complex (PDB entry 3H9S)25. The binding interface between complementarity determining regions (CDRs) of TCR and the KK10 peptide was optimized globally at atomic level by a Potential Smoothing and Search method in TINKER package37. The second template followed a similar procedure, but with a newly released co-crystal structure (PDB entry: 4G8G) as the input (see main text).
Molecular dynamics simulations
Each HLA B*2705-KK10-TCR complex was solvated in a 77.5 Å × 128.0 Å × 76.5 Å water box, where the system was firstly neutralized with counter ions, and then further ionized in a 150 mM NaCl in order to mimic in vivo physiological environment. The solvated system was minimized by 20,000 steps, then followed by a ~1 ns equilibration (with a 0.5-fs timestep) in 1 atm and 310 K. In each system, five snapshots at the second half of the equilibration were randomly picked as starting structures for up to 50 ns long molecular dynamics simulations and independent 60 + ns free energy perturbation (FEP) calculations for each system, with an aggregate of 1.7 microseconds MD simulation time for all clonotypes and their variants. The unbound (free) state of the FEP calculation was modeled with the binary complex of only HLA-KK10, which was prepared with a similar protocol employed for the bound system of HLA B*2705-KK10-TCR complex. The particle-mesh Ewald (PME) method was used for the long-range electrostatic interactions38, while the van der Waals interactions were handled with usual smooth cutoff with a cutoff distance of 12 Å. All molecular dynamics simulations have been performed with a specially optimized NAMD2 molecular modeling package for Blue Gene39,40, with a 1.5-fs timestep in NPT ensemble at 1 atom and 310 K. Molecular dynamics simulations have been widely used in modeling biological systems to complement experiments, which can provide atomic details that are often inaccessible in experiments due to resolution limits even with the current most sophisticated experimental techniques22,41,42,43,44,45,46,47,48,49,50. The CHARMM22 force field51 and TIP3P water model52 are used for proteins and solvents, respectively.
Free energy perturbation protocol
The binding affinity changes, due to antigenic variations, between the TCR and HLA-KK10 complex were estimated by the free energy perturbation (FEP) method16,17,53,54,55,56,57,58,59,60. We calculate the free energy changes for the same mutation(s) in both the bound state (HLA-KK10-TCR 3-way binding complex) and the free state (HLA-KK10 binary complex). For each mutation, at least five independent runs starting from different initial configurations (taken from the molecular dynamic simulations) are performed for better sampling. The simulation time for each run is 6.0 ns, thus, at least 60 ns (6.0-ns × 5-runs × 2-states) simulation time was generated for each mutation. Larger window sizes and longer simulation durations have also been tested in our previous studies, and we found that the current protocol gives us a reasonable convergence in the final binding affinities16,18. Please see Note 3 in supporting data for more detailed description of the FEP protocol.
Author Contributions
R.Z. and B.D.W. conceived and designed the research. Z.X., H.C., S.K., T.H., B.D.W. and R.Z. co-wrote the manuscript. Z.X. and T.H. carried out the computational modeling, molecular dynamics simulations, and free energy calculations. H.C., J.W.F. and P.A.L., carried the chromium release assay. Z.X., H.C., T.H., S.G. and R.Z. analyzed the data. All authors discussed the results and commented on the manuscript.
Supplementary Material
Srep04087 Supplementary Information
Acknowledgments
This work was supported by the IBM Blue Gene Science Program (R.Z.), the Harvard University Center for AIDS Research (5 P30 AI060354-04), grants from the Bill and Melinda Gates Foundation (B.D.W.), the Doris Duke Charitable Foundation (B.D.W.), the NIH (B.D.W. AI030914), the Howard Hughes Medical Institute (B.D.W.), and the Mark and Lisa Schwartz Foundation (B.D.W.). We also thank IBM Watson Blue Gene Supercomputer Center for computational resources.
References
- McMichael A. J., Borrow P., Tomaras G. D., Goonetilleke N. & Haynes B. F. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10, 11–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo M. M. et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77, 2081–92 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts M. R. et al. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75, 11983–91 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P. et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13, 46–53 (2007). [DOI] [PubMed] [Google Scholar]
- Harari A. et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc. Natl. Acad. Sci. U. S. A. 104, 16233–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes. J. Virol. 83, 3138–49 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat. Immunol. 13, 691–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M. C. et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood 118, 2138–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T. M. et al. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 79, 12952–60 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo Noah J. et al. Single Naive CD4+ T Cells from a Diverse Repertoire Produce Different Effector Cell Types during Infection. Cell 153, 785–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladell K. et al. A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells. Immunity 38, 425–36 (2013). [DOI] [PubMed] [Google Scholar]
- Mendoza D. et al. HLA B*5701-positive long-term nonprogressors/elite controllers are not distinguished from progressors by the clonal composition of HIV-specific CD8+ T cells. J. Virol. 86, 4014–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbazian L. et al. Clonotype and repertoire changes drive the functional improvement of HIV-specific CD8 T cell populations under conditions of limited antigenic stimulation. J. Immunol. 188, 1156–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzetto D. et al. Evaluation of template-based models in CASP8 with standard measures. Proteins 77, 18–28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani V., Kiefer F., Schmidt T., Haas J. & Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins 79, 37–58 (2011). [DOI] [PubMed] [Google Scholar]
- Zhou R., Das P. & Royyuru A. K. Single Mutation Induced H3N2 Hemagglutinin Antibody Neutralization: A Free Energy Perturbation Study. J. Phys. Chem. B 112, 15813–15820 (2008). [DOI] [PubMed] [Google Scholar]
- Das P., Li J., Royyuru A. K. & Zhou R. Free energy simulations reveal a double mutant avian H5N1 virus hemagglutinin with altered receptor binding specificity. J. Comput. Chem. 30, 1654–1663 (2009). [DOI] [PubMed] [Google Scholar]
- Xia Z., Huynh T., Kang S. G. & Zhou R. Free-energy simulations reveal that both hydrophobic and polar interactions are important for influenza hemagglutinin antibody binding. Biophys J 102, 1453–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F. P. et al. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science 330, 1551–1557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren P. J. et al. Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum. Mol. Genet. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R. H., Huang X. H., Margulis C. J. & Berne B. J. Hydrophobic collapse in multidomain protein folding. Science 305, 1605–1609 (2004). [DOI] [PubMed] [Google Scholar]
- Liu P., Huang X., Zhou R. & Berne B. J. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature 437, 159–62 (2005). [DOI] [PubMed] [Google Scholar]
- Wu Y., Vadrevu R., Kathuria S., Yang X. & Matthews C. R. A tightly packed hydrophobic cluster directs the formation of an off-pathway sub-millisecond folding intermediate in the alpha subunit of tryptophan synthase, a TIM barrel protein. J. Mol. Biol. 366, 1624–38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L. et al. A structural basis for the selection of dominant alpha beta T cell receptors in antiviral immunity. Immunity 18, 53–64 (2003). [DOI] [PubMed] [Google Scholar]
- Borbulevych O. Y. et al. T Cell Receptor Cross-reactivity Directed by Antigen-Dependent Tuning of Peptide-MHC Molecular Flexibility. Immunity 31, 885–896 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald W. A. et al. T Cell Allorecognition via Molecular Mimicry. Immunity 31, 897–908 (2009). [DOI] [PubMed] [Google Scholar]
- Yin Y. Y. & Mariuzza R. A. The Multiple Mechanisms of T Cell Receptor Cross-reactivity. Immunity 31, 849–851 (2009). [DOI] [PubMed] [Google Scholar]
- Dunn S. M. et al. Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity. Protein Sci. 15, 710–721 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L. et al. The structure of HLA-B8 complexed to an immunodominant viral determinant: Peptide-induced conformational changes and a mode of MHC class I dimerization. J. Immunol. 169, 5153–5160 (2002). [DOI] [PubMed] [Google Scholar]
- Fernandez A. & Scheraga H. A. Insufficiently dehydrated hydrogen bonds as determinants of protein interactions. Proc. Natl. Acad. Sci. U. S. A. 100, 113–118 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman M. A., Tanzi G. O., Walker B. D. & Allen T. M. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J. Virol. Methods 131, 134–42 (2006). [DOI] [PubMed] [Google Scholar]
- Schneidewind A. et al. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82, 5594–605 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang O. O. et al. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J. Immunol. 171, 3718–24 (2003). [DOI] [PubMed] [Google Scholar]
- Stewart-Jones G. B. et al. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 35, 341–51 (2005). [DOI] [PubMed] [Google Scholar]
- Sali A. & Blundell T. L. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- Fiser A., Do R. K. G. & Sali A. Modeling of loops in protein structures. Protein Sci. 9, 1753–1773 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu R. V., Hart R. K. & Ponder J. W. Analysis and application of potential energy smoothing and search methods for global optimization. J. Phys. Chem. B 102, 9725–9742 (1998). [Google Scholar]
- Darden T. A., York D. M. & Pedersen L. G. Particle mesh Ewald: An NlogN method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- Kumar S. et al. Scalable Molecular Dynamics with NAMD on Blue Gene/L. IBM J. Res. Dev. 52, 177–188 (2008). [Google Scholar]
- Morrone J. A., Zhou R. H. & Berne B. J. Molecular Dynamics with Multiple Time Scales: How to Avoid Pitfalls. J. Chem. Theo. Comp. 6, 1798–1804 (2010). [DOI] [PubMed] [Google Scholar]
- Eleftheriou M., Germain R. S., Royyuru A. K. & Zhou R. Thermal denaturing of mutant lysozyme with both the OPLSAA and the CHARMM force fields. J. Am. Chem. Soc. 128, 13388–95 (2006). [DOI] [PubMed] [Google Scholar]
- Zhou R., Berne B. J. & Germain R. The free energy landscape for beta hairpin folding in explicit water. Proc Natl Acad Sci U S A 98, 14931–6 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Eleftheriou M., Royyuru A. K. & Berne B. J. Destruction of long-range interactions by a single mutation in lysozyme. Proc Natl Acad Sci U S A 104, 5824–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. Q., Yang W. & Karplus M. A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase. Cell 123, 195–205 (2005). [DOI] [PubMed] [Google Scholar]
- Hummer G., Rasaiah J. C. & Noworyta J. P. Water conduction through the hydrophobic channel of a carbon nanotube. Nature 414, 188–190 (2001). [DOI] [PubMed] [Google Scholar]
- Kamberaj H. & van der Vaart A. An optimized replica exchange molecular dynamics method. J. Chem. Phys. 130, 074906 (2009). [DOI] [PubMed] [Google Scholar]
- Karplus M., Gao Y. Q., Ma J., van der Vaart A. & Yang W. Protein structural transitions and their functional role. Philos. Transact. A Math. Phys. Eng. Sci. 363, 331–355; discussion 355–356 (2005). [DOI] [PubMed] [Google Scholar]
- Zheng L., Chen M. & Yang W. Random walk in orthogonal space to achieve efficient free-energy simulation of complex systems. Proc. Natl. Acad. Sci. U. S. A. 105, 20227–20232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch B. G. et al. Blue Matter: Strong scaling of molecular dynamics on Blue Gene/L, (Springe rBerlin Heidelberg, 2006). [Google Scholar]
- Kaminski G. A., Friesner R. A. & Zhou R. A computationally inexpensive modification of the point dipole electrostatic polarization model for molecular simulations. J Comput Chem 24, 267–76 (2003). [DOI] [PubMed] [Google Scholar]
- MacKerell A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- Deng Y. & Roux B. Calculation of standard binding free energies: aromatic molecules in the T4 lysozyme L99A mutant. J. Chem. Theo. Comp. 2, 1255–1273 (2006). [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L. Free-energy calculations - a breakthrough for modeling organic-chemistry in solution. Acc. Chem. Res. 22, 184–189 (1989). [Google Scholar]
- Kollman P. Free-energy calculations - applications to chemical and biochemical phenomena. Chem. Rev. 93, 2395–2417 (1993). [Google Scholar]
- Simonson T., Archontis G. & Karplus M. Free energy simulations come of age: Protein-ligand recognition. Acc. Chem. Res. 35, 430–437 (2002). [DOI] [PubMed] [Google Scholar]
- Tembe B. L. & McCammon J. A. Ligand receptor interactions. Computers & Chemistry 8, 281–283 (1984). [Google Scholar]
- Warshel A. Simulating the Energetics and Dynamics of Enzymatic Reactions. Specificity in Biological Interactions 55, 59–81 (1984). [Google Scholar]
- Warshel A., Sharma P. K., Kato M. & Parson W. W. Modeling Electrostatic Effects in Proteins. Biochim. Biophys. Acta 1764, 1647–1676 (2006). [DOI] [PubMed] [Google Scholar]
- Xia Z., Das P., Huynh T., Royyuru A. K. & Zhou R. Modeling mutations of influenza virus with IBM Blue Gene. IBM J. Res. Dev. 55 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Srep04087 Supplementary Information