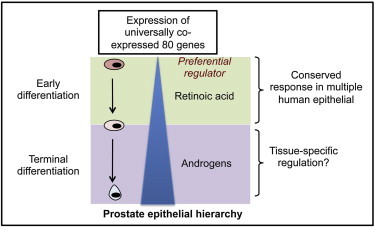
Summary
Human epithelia are organized in a hierarchical structure, where stem cells generate terminally differentiated cells via intermediate progenitors. This two-step differentiation process is conserved in all tissues, but it is not known whether a common gene set contributes to its regulation. Here, we show that retinoic acid (RA) regulates early human prostate epithelial differentiation by activating a tightly coexpressed set of 80 genes (e.g., TMPRSS2). Response kinetics suggested that some of these genes could be direct RA targets, whereas others are probably responding indirectly to RA stimulation. Comparative bioinformatic analyses of published tissue-specific microarrays and a large-scale transcriptomic data set revealed that these 80 genes are not only RA responsive but also significantly coexpressed in many human cell systems. The same gene set preferentially responds to androgens during terminal prostate epithelial differentiation, implying a cell-type-dependent interplay between RA and tissue-specific transcription factor-mediated signaling in regulating the two steps of epithelial differentiation.
Graphical Abstract
Highlights
-
•
Four sets of coexpressed genes mark primary human prostate stem cell differentiation
-
•
These gene sets are also tightly coexpressed in >150 human cell types
-
•
Retinoic acid induces early differentiation while upregulating one of the gene sets
-
•
Androgens preferentially regulate the same gene set during terminal differentiation
Maitland and colleagues demonstrate that a set of 80 genes could be important for the regulation of adult human epithelial differentiation, irrespective of tissue of origin. They also show that the identified gene set responds to retinoic acid during early differentiation while preferentially responding to a tissue-specific transcription factor during terminal differentiation.
Introduction
Differentiation of self-renewing adult human epithelial stem cells into rapidly proliferating progenitor cells is a conserved early event in human tissue development and homeostasis. Systematic and coordinated regulation by master transcription factors, noncoding RNA-mediated networks, or the establishment of progressive epigenetic marks have been proposed as likely regulatory mechanisms (Hanna et al., 2010). Identification of control mechanisms will enable the basic understanding of epithelial dynamics and could identify common perturbations leading to the disruption of epithelial homeostasis in cancers. We have investigated the nature of stem cell regulation during early human epithelial differentiation using patient-derived prostate epithelium as our primary experimental tool. In human prostate, stem-like cells (SCs) of a basal phenotype and their early differentiated progeny (transit amplifying [TA] and committed basal cells [CB]) can be reproducibly enriched from patient-derived benign and malignant tissues by selecting for differential CD133, CD44, and α2β1 integrin expression (Collins et al., 2005; Richardson et al., 2004). Transcriptional profiling of prostate SCs and CBs identified consistent gene expression changes associated with differentiation and carcinogenesis (Birnie et al., 2008). We have now investigated the expression pattern and the regulation of genes that are differentially expressed in the SC and CB populations, supplemented by a large-scale analysis of published transcriptomic experiments, to identify shared signaling pathways that regulate general epithelial differentiation.
Results and Discussion
Identification of Genes Coregulated during Prostate Stem Cell Differentiation
Coexpression analysis was performed on differentially expressed Affymetrix probes between SC and CB from the Birnie et al. (2008) data set (Figure 1A) to identify functionally related and coregulated genes without introducing observer or selection bias (Lee et al., 2004). Probes with high pairwise correlation (Pearson’s correlation coefficient, R ≥ 0.8 or R ≤ −0.8) were clustered into four distinct groups (A–D) (Figures 1B and 1C). All the correlations (above the threshold) were positive and no significant connections between probes within the four groups were seen. Analysis of independent published microarrays (Shepherd et al., 2008) showed similar changes in the expression of group A–D genes during prostate stem cell differentiation (data not shown). Gene ontology (GO) analysis further revealed that the genes in each group are functionally related. Groups B, C, and D genes were principally enriched for differentiation-associated processes (Figure 1B). Group C genes exhibited a distinct enrichment for genes constituting the “epidermal differentiation complex” (SPRR and S100 families) (Benitah, 2012; Mischke et al., 1996). Interactive Pathway Analysis (IPA) (Kececioglu and Kim, 2006) of group C further revealed a significant enrichment for genes having critical functions in generic “tissue developmental events” (p < 0.01) and genes known to be coregulated by strong differentiation-associated regulators such as retinoic acid (RA) (p = 2.94 × 10−13) and ROCK2 (p = 9.89 × 10−8). IPA analysis did not provide convincing evidence for differentiation-associated coregulation of group B. These results framed the hypothesis that group C genes could be coexpressed in a variety of human tissues and could play a broader functional role in differentiation.
Figure 1.
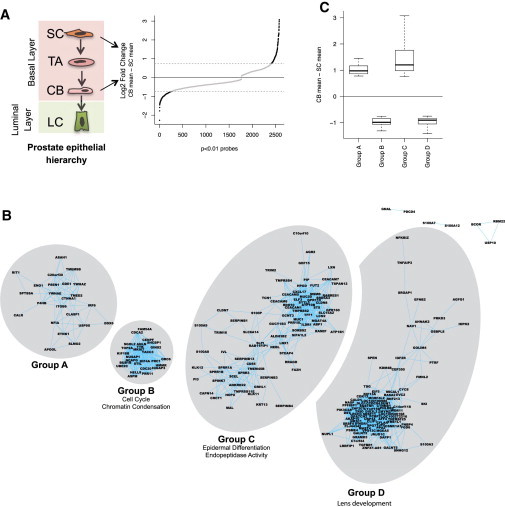
Identification of Coregulated Genes
(A) Selection of differentially expressed probes between SC and CB (p < 0.01; log2 fold change ≥ 0.75) are represented as black dots in the Birnie et al. (2008) microarray data.
(B) Differentially expressed probes were clustered in distinct A–D groups on coexpression analysis. Principal GO terms enriched for groups B and C are shown below the group name. Group A was not enriched for any GO terminology, whereas group C was enriched for genes involved in (lens) development.
(C) Normalized expression of group A–D probes. Probes overexpressed in committed basal cells (CB) are above the horizontal line at 0 (groups A and C) and probes overexpressed in stem-like cells (SC) (groups B and D) are below the line. Boxplot shows minimum, 25%, median, 75%, and maximum.
Gene Sets Coexpressed during Prostate Stem Cell Differentiation Are Coexpressed in the Majority of Human Cell Types
A compendium data set of 24,536 human Affymetrix microarrays from 806 experiments, representing at least 150 distinct human cell types, was generated to test whether group A–D genes are coexpressed across multiple cell types, pathologies, and treatments (Figures 2A and 2B). This data set is the largest singly normalized human microarray data set we are aware of. Unlike most others (Rhodes et al., 2007; Seita et al., 2012), it is not restricted to any particular sample type, tissue type, or pathology. Assessment of the raw microarray data for technical and experimental accuracies resulted in the removal of 507 corrupted or experimentally faulty experiments (Supplemental Experimental Procedures available online). Overall, the final data set yielded about 1.2 billion expression correlations between genes. We confirmed the quality and utility of this data set for gene variation analysis by performing GO analysis on the 100 least variable probes, which identified the most invariable genes (by coefficient of variation after normalization) in human cells, e.g., ribosomal genes and control genes for quantitative RT-PCR (qRT-PCR)/western blot normalization (Table S1). The data set can also be used to construct background interaction modeling to identify interactions representing normal physiology or common occurrences. Overlaying this background network on a transcriptomic data set of interest facilitates removal of background interactions, leaving behind experiment-specific biological behavior. However, the more restricted analysis in this study clearly demonstrated that group A–D genes were significantly coexpressed (Figure 2C), confirming that genes coexpressed during prostate epithelial differentiation show similar expression patterns (and thus could share common functions and regulation) in multiple human cell types.
Figure 2.
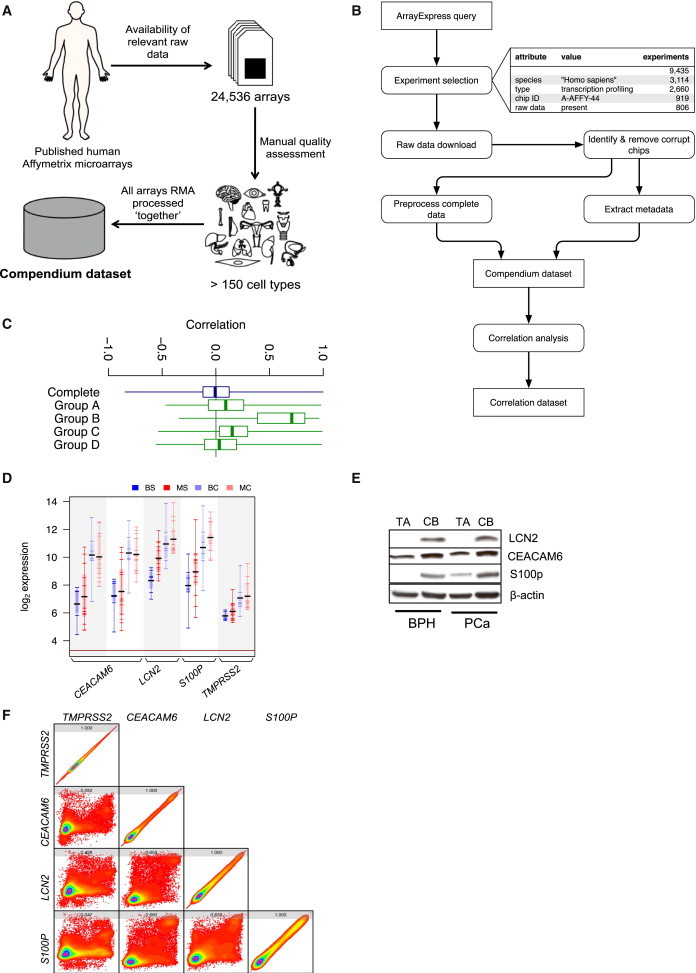
Genes in Groups A–D Are Coexpressed in the Majority of Human Cell Types
(A) Overview of the composition of compendium data set.
(B) Schematic representation of the methodology for the compendium data set.
(C) Correlation of coexpression of genes in groups A–D compared to all genes in the compendium data set. All the groups are significantly different where the p value is computationally indistinguishable from zero.
(D) Expression of candidate group C genes in the Birnie et al. (2008) microarray data. BS, benign stem-like cells; MS, malignant stem-like cells; BC, benign committed basal cells; MC, malignant committed basal cells.
(E) Western blot analysis of LCN2, CEACAM6, and S100P expression. Representative image, n = 6 each for benign prostatic hyperplasia (BPH) and treatment-naïve prostate cancer (PCa) (biological replicates).
(F) Rainbow contour graphs for the correlation of coexpression for TMPRSS2, CEACAM6, LCN2, and S100P with each other in compendium data set. Values at the top of each square indicate correlation coefficient. All values are significant with p < 0.05.
All boxplots show minimum, 25%, median, 75%, and maximum. See also Figure S1 and Table S1.
Group C Genes Are Likely Markers or Mediators of Stem Cell Differentiation
While group B genes showed stronger coexpression in the compendium data set and exhibited interesting GO term enrichment, group C genes provided more compelling evidence for their coregulatory and differentiation-associated properties. IPA and literature analyses (62 out of 80 genes in group C had at least one PubMed publication linking them to differentiation) strongly implied differentiation-specific coregulation of group C. We selected LCN2, CEACAM6, S100P, and TMPRSS2 as group C representative genes to assess group C coregulation, on the basis of strong evidence for their role in SC differentiation or prostate epithelial dynamics and their tight coexpression pattern. Microarray expression of the candidate genes (Figure 2D) was confirmed by both qRT-PCR (Figure S1) and western blot analysis (Figure 2E). In a wide variety of mammalian tissues, low expression of these genes has been shown to denote SC properties (He et al., 2009; Polson et al., 2013; Zheng et al., 2009). Furthermore, TMPRSS2 and S100P are considered to be classical androgen responsive genes in terminally differentiated cells, which can promote oncogenesis in prostate epithelium (Averboukh et al., 1996; Lin et al., 1999), and S100P has also been shown to be responsible for PC3 cell differentiation into luminal-like cells (Floryk and Thompson, 2008). Similar functions therefore translate into tight coexpression of the genes within the compendium data set, where S100P had a more similar expression pattern to that of LCN2 and CEACAM6 than any other gene in the human genome (Figure 2F). TMPRSS2 was also significantly coexpressed with these three genes (Figure 2F). Such functional attributes and bioinformatic analyses both indicated that LCN2, CEACAM6, S100P, and TMPRSS2 (and group C) are coregulated during prostate SC differentiation.
Retinoic Acid Regulates Group C Genes Inducing Early Prostate Stem Cell Differentiation
Multiple bioinformatic tools and a published literature evaluation were utilized to identify potential regulators of group C genes (Figure 3A). The analyses identified RA as the most likely regulator of gene expression because (1) IPA analysis suggested that RA can regulate group C; (2) ENCODE chromatin immunoprecipitation (ChIP) analysis of HepG2 cells also showed a significant enrichment for retinoid X receptor α (RXRA) binding sites on the promoters of group C; (3) LCN2, CEACAM6, S100P, and TMPRSS2 have putative RAR and RXR binding sites within their promoters (data not shown); (4) our previous work has demonstrated that two other genes in group C (RARRES1 and LXN) are also directly regulated by RA (Oldridge et al., 2013); and (5) RA-related transcription factors (RXRa, RARg, and RARa) are actively expressed in prostate epithelial cells (Figures 3B and S2). RA acts as an important regulator of both embryonic and adult SC differentiation. However, in spite of strong historical evidence for a role in the developing mouse prostate (Lohnes et al., 1995), the outcome of RA signaling manipulation in adult human prostate epithelial differentiation is unknown. The case for transcription-factor-mediated regulation of group C is also enhanced by the lack of evidence for a common regulatory mechanism in terms of microRNA, histone acetylation, and DNA methylation even for the group C candidate genes (data not shown).
Figure 3.
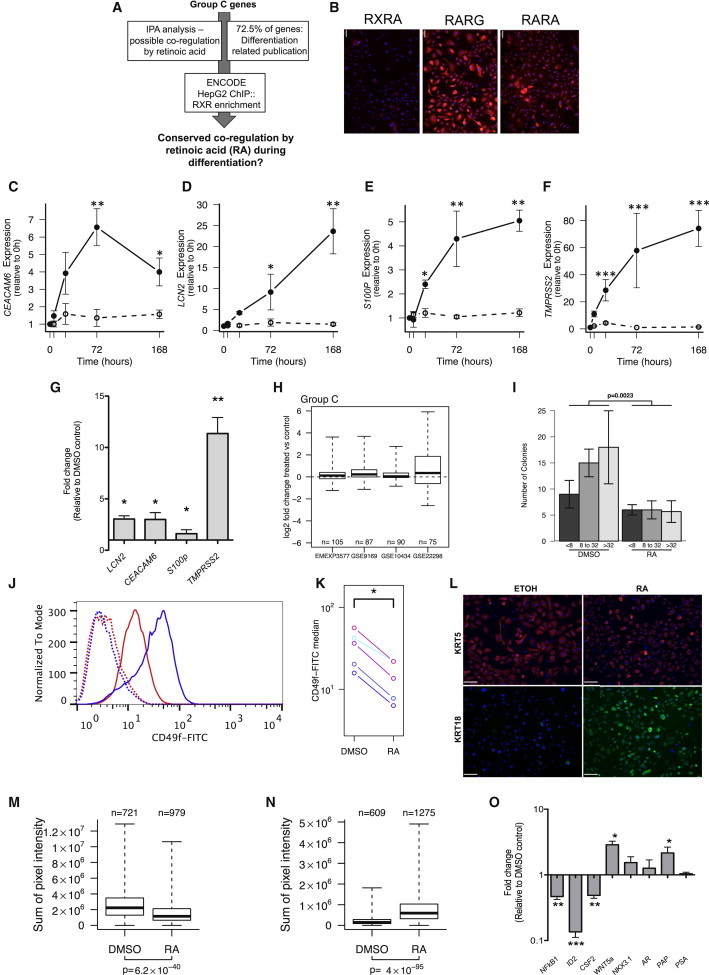
Retinoic Acid Can Regulate Group C while Inducing Early Prostate Stem Cell Differentiation
(A) Schematic representation for selection process of retinoic acid (RA) as a group C regulator.
(B) Expression of RA receptors in primary prostate epithelial cultures (PPECs) by immunofluorescence (representative image); n = 3 each for BPH and PCa (biological replicates). Scale bar: 62 μm.
(C–F) qRT-PCR analysis of CEACAM6, LCN2, S100P, and TMPRSS2 expression after 100 nM at-RA treatment of PPECs; n = 5 for BPH (3) and PCa (2) (biological replicates).
(G) qRT-PCR analysis of CEACAM6, LCN2, S100P, and TMPRSS2 expression after 100 nM at-RA treatment of SCs; mean values shown for BPH (five) and PCa (four) (biological replicates).
(H) Changes in the expression of group C genes after RA treatment in four published microarray experiments.
(I) Changes in colony-forming efficiency of PPECs after 72 hr 100 nM at-RA treatment; n = 4 for BPH and PCa.
(J and K) Fluorescence-activated cell sorting analysis of CD49f expression after 72 hr treatment of PPECs with 100 nM at-RA; n = 4 each for BPH and PCa (biological replicates). In (J), the blue line indicates DMSO control, whereas the red line indicates RA-treated samples. Dotted lines represent unlabeled controls.
(L) Immunofluorescence (IF) analysis of KRT5 and KRT18 after 72 hr treatment of PPECs with 100 nM at-RA. Scale bar, 100 μm.
(M and N) Quantification of changes in KRT5 (M) and KRT18 (N) protein expression by IF after 72 hr treatment of PPECs with 100 nM at-RA using velocity software.
(O) qRT-PCR expression analysis of markers of prostate epithelial differentiation after 72 hr treatment of PPECs with 100 nM at-RA; n = 4 each for BPH and PCa (biological replicates).
Error bars represent mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Box plots in (H), (M), and (N) show minimum, 25%, median, 75%, and maximum. See also Figure S2.
To test whether RA is indeed a principal regulator of prostate epithelial SC differentiation, benign and malignant patient-derived prostate epithelial cells (PPECs) were treated with 100 nM all-trans retinoic acid (at-RA). The group C candidate genes showed a 4- to 60-fold increase in expression within 72 hr (Figures 3C–3F), accompanied by a modest but statistically insignificant decrease in cell number (not shown). The rapid increase in TMPRSS2 expression levels (6 hr posttreatment) suggested that this gene could be a direct target of RA, whereas the other three genes responded more slowly and are more likely, on a kinetic basis, to be RA-“responsive” genes. A similar trend was also noted in at-RA-treated prostate SCs (Figure 3G). The RA responsiveness of these genes has been shown in other experimental models; for example, LCN2, S100P, and CEACAM6 were upregulated in sebaceous cells after 13-cis-RA treatment (Nelson et al., 2008). Analysis of publically available microarray data (Figure 3H) confirmed that group C genes are also upregulated in response to RA treatment in four different cell types, including human embryonic SCs. Taken together, these findings demonstrate that the majority of group C genes are RA-responsive genes.
Next, we assessed whether RA treatment alone, while modulating group C expression, could also induce early prostate SC differentiation. Treatment of PPECs with 100 nM at-RA for 72 hr induced (1) suppression of colony-forming efficiency by approximately 60% (Figure 3I), indicating a decrease in self-renewing cells; (2) a >50% decrease in the expression of the progenitor cell marker CD49f (Figures 3J and 3K) and a 12% decrease in CD49b expression (data not shown); (3) a decrease in CK5 (basal CK) expression with a concomitant increase in luminal CK18 expression (Figures 3L–3N); (4) a decrease in NF-κB1 and ID2 expression levels, which promote a SC phenotype; and (5) increased expression of the differentiation-associated genes WNT5a and PAP (Figure 3O) (Birnie et al., 2008). Classical prostate luminal differentiation markers, such as AR, PSA, and NKX3.1, remained unaltered after at-RA treatment (Figure 3O). A similar increase in endogenous RA levels in prostate cancer patients after treatment with liarozole also resulted in a minimal increase in luminal differentiation (Denis et al., 1998). Two human clinical trials also showed only a modest increase in the expression of PSA and PAP (in only about 30% of patients). These findings indicated that at-RA treatment can induce differentiation of prostate SCs to CBs, which are primed for luminal differentiation (as suggested by the increase in PAP and CK18 levels) but confirm that RA alone cannot force luminal differentiation. In this scenario, overexpression of all the candidate group C genes in luminal cells (Oudes et al., 2006) prompted us to assess the existence of additional regulators of group C genes in prostate epithelial differentiation.
Androgens Can Regulate Group C during Terminal Human Prostate Epithelial Differentiation
The majority of group C genes were indeed overexpressed in patient-derived luminal compared to basal cells (Oudes et al., 2006) (Figure 4A and S3), implying that another transcription factor, distinct from RA, may regulate group C to sustain and promote gene expression during luminal differentiation. Because systemic and niche-driven AR-mediated signaling is the main regulator of luminal differentiation in prostate and is capable of inducing luminal differentiation, even in 3D culture, with an obligatory contribution from stroma (Lang et al., 2006), we assessed the responsiveness of group C to AR stimulation in publically available microarray data. Group C was consistently upregulated upon AR stimulation in multiple prostate cell line models (Figure 4B), confirming that group C genes can respond to a tissue-specific transcription factor (like AR) in cells primed for terminal differentiation. However, this presented a conundrum about the “principal” regulator of group C genes during prostate epithelial differentiation. We therefore treated prostate cells, possessing a distinct basal or luminal phenotype, with at-RA or R1881 (an AR agonist). In cells with a basal phenotype, at-RA, but not R1881, upregulated TMPRSS2 expression (up to 80-fold), whereas TMPRSS2 responded preferentially to R1881 (up to a 10-fold increase) in cells with a luminal phenotype (Figures 4C and S4A). Similar observations were also made for CEACAM6 and S100P (Figures S4B and S4C). Inhibition of AR signaling has also been shown to inhibit the expression of these genes in LNCaP cells (Figure 4D; Averboukh et al., 1996). This experimental evidence indicated that RA and AR act in a cell-type-dependent manner to coregulate group C, providing a fuller explanation for the observed expression changes in TMPRSS2 (strongly considered solely as an androgen-regulated gene) in androgen-independent basal cells (Polson et al., 2013).
Figure 4.
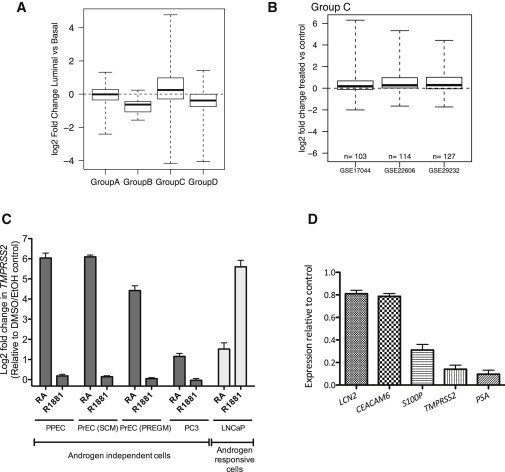
Androgens Regulate Group C Gene Expression during Terminal Differentiation
(A) Expression of group A–D genes in prostate luminal cells relative to basal cells.
(B) Changes in expression of group C genes after androgen stimulation in three published microarrays.
(C) qRT-PCR analysis of TMPRSS2 expression after 48 hr treatment with at-RA (RA) and R1881 (n = 6 for PPECs [3 BPH and 3 PCa, biological replicates] and n = 3 for cell lines [experimental replicates]).
(D) qRT-PCR for the expression of LCN2, CEACAM6, S100P, TMPRSS2, and PSA in LNCaP cells after 1 nM bicalutamide treatment for 48 hr. The data are plotted considering the untreated gene expression value as 1 (n = 3, experimental replicates).
Error bars represent mean ± SD. Box plots show minimum, 25%, median, 75%, and maximum. See also Figures S3 and S4.
This study provides direct evidence for the mechanistic role of RA during adult prostate SC differentiation, where ALDHhi cells with SC properties in prostate (van den Hoogen et al., 2010) can serve as a potential source of RA. Prostate epithelial SCs with no detectable AR expression respond to RA stimulation through a set of 80 genes, transforming SCs into basal cells committed to terminal differentiation (luminal). The genes can then preferentially respond to androgens during terminal differentiation. The change in regulatory preference could be due to multiple factors, such as weakening of a RA morphogenetic gradient from the basal to the luminal layer (Shimozono et al., 2013), loss of contact with the basement membrane and niche (Lang et al., 2006), chromatin modifications induced by RA (Kashyap et al., 2013), or changes in the expression of independent differentiation-associated chromatin modifying genes (e.g., group B genes). When the distinction of a basal-luminal cell type is not clear, as in intermediate prostate epithelial cells (van Leenders et al., 2000) and cell lines, competition between RA- and AR-mediated signaling may result in competitive inhibition of one of the two response pathways (Young et al., 1994). The reduced RA content of prostate cancer tissue compared to normal tissue (Pasquali et al., 1996), where AR-responsive terminally differentiated cells predominate (∼99% of total cells), suggests that the tissue strives to minimize AR-RA antagonism by downregulating the less efficient signaling mechanism. Cooperation/interference between RA and AR signaling (Rivera-Gonzalez et al., 2012) may therefore regulate prostate epithelial differentiation and dynamics through a similar set of genes. In mammary epithelial development, RA also regulates early differentiation, whereas a tissue-specific transcription factor (estrogen receptor) can then promote terminal epithelial differentiation (Förster et al., 2002; Seewaldt et al., 1997). Our data now reveal the existence of similar two-step regulation of differentiation in other epithelial tissues.
In terms of prostate cancer management, our data strongly suggest that RA treatment should lock prostate cancer stem cells into an AR-independent committed basal cell phenotype, which will not be affected by prevailing androgen-blocking therapy (Rane et al., 2012). Our data link two of the most conserved prostate epithelial differentiation-regulatory pathways to a shared gene set, providing a mechanistic insight into adult human epithelial dynamics.
Experimental Procedures
Establishment of Primary Cultures
Patient tissue material was obtained following written consent and full ethical approval (NHS Research Governance Framework: NRES reference number 07/H1304/121). Cultures were established from patient-derived cancer (all Gleason 7) and benign prostatic hyperplasia biopsy specimens and SC, TA, and CB cells were selected as described previously (Collins et al., 2005) at passage 2.
Construction and Analysis of Compendium Data Set
Transcriptomic data were normalized by RMA and annotated. Row-wise correlation analysis was used to yield a matrix of correlations between all probes. See the Supplemental Experimental Procedures for additional details.
Expression Analysis
qRT-PCR analysis was performed as described previously (Oldridge et al., 2013). Details of TaqMan probes and antibodies used for western blot, fluorescence-activated cell sorting, and immunofluorescence analyses are in the Supplemental Experimental Procedures.
Colony-Forming Efficiency
A total of 200 cells were plated in each well of a six-well collagen-coated plate with 500 μl of irradiated mouse fibroblasts. Colonies generated by individual cells were counted after 12–18 days.
Statistical Analysis
Experiments were carried out with at least three different samples (each in triplicate) representing at least two different experiments. Error bars shown are the SD. The significance was determined by Student’s two-tailed t test. Wilcoxon rank sum test was used to calculate significance of cytokeratin immunofluorescence.
Acknowledgments
We would like to thank all the patients and urology surgeons (L. Coombes, G. Cooksey, and J. Hetherington) at Castle Hill Hospital, Cottingham. The work was funded by PRO-NEST Marie-Curie grant (J.K.R.), Yorkshire Cancer Research (A.P.D., D.P., A.T.C., and N.J.M.), and The Freemasons’ Grand Charity (D.P.).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Averboukh L., Liang P., Kantoff P.W., Pardee A.B. Regulation of S100P expression by androgen. Prostate. 1996;29:350–355. doi: 10.1002/(SICI)1097-0045(199612)29:6<350::AID-PROS2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Benitah S.A. Defining an epidermal stem cell epigenetic network. Nat. Cell Biol. 2012;14:652–653. doi: 10.1038/ncb2538. [DOI] [PubMed] [Google Scholar]
- Birnie R., Bryce S.D., Roome C., Dussupt V., Droop A., Lang S.H., Berry P.A., Hyde C.F., Lewis J.L., Stower M.J. Gene expression profiling of human prostate cancer stem cells reveals a pro-inflammatory phenotype and the importance of extracellular matrix interactions. Genome Biol. 2008;9:R83. doi: 10.1186/gb-2008-9-5-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.T., Berry P.A., Hyde C., Stower M.J., Maitland N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Denis L., Debruyne F., De Porre P., Bruynseels J. Early clinical experience with liarozole (Liazal) in patients with progressive prostate cancer. Eur. J. Cancer. 1998;34:469–475. doi: 10.1016/s0959-8049(97)10120-4. [DOI] [PubMed] [Google Scholar]
- Floryk D., Thompson T.C. Perifosine induces differentiation and cell death in prostate cancer cells. Cancer Lett. 2008;266:216–226. doi: 10.1016/j.canlet.2008.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster C., Mäkela S., Wärri A., Kietz S., Becker D., Hultenby K., Warner M., Gustafsson J.A. Involvement of estrogen receptor beta in terminal differentiation of mammary gland epithelium. Proc. Natl. Acad. Sci. USA. 2002;99:15578–15583. doi: 10.1073/pnas.192561299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Marchionni L., Hansel D.E., Yu W., Sood A., Yang J., Parmigiani G., Matsui W., Berman D.M. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem Cells. 2009;27:1487–1495. doi: 10.1002/stem.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V., Laursen K.B., Brenet F., Viale A.J., Scandura J.M., Gudas L.J. RARγ is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells. J. Cell Sci. 2013;126:999–1008. doi: 10.1242/jcs.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kececioglu J., Kim E. Simple and fast inverse alignment. In: Apostolico A., Guerra C., Istrail S., Pevzner P.A., Waterman M., editors. Research in Computational Molecular Biology. Springer-Verlag; Berlin: 2006. pp. 441–455. [Google Scholar]
- Lang S.H., Smith J., Hyde C., Macintosh C., Stower M., Maitland N.J. Differentiation of prostate epithelial cell cultures by matrigel/ stromal cell glandular reconstruction. In Vitro Cell. Dev. Biol. Anim. 2006;42:273–280. doi: 10.1290/0511080.1. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Hsu A.K., Sajdak J., Qin J., Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Ferguson C., White J.T., Wang S., Vessella R., True L.D., Hood L., Nelson P.S. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- Lohnes D., Mark M., Mendelsohn C., Dollé P., Decimo D., LeMeur M., Dierich A., Gorry P., Chambon P. Developmental roles of the retinoic acid receptors. J. Steroid Biochem. Mol. Biol. 1995;53:475–486. doi: 10.1016/0960-0760(95)00094-g. [DOI] [PubMed] [Google Scholar]
- Mischke D., Korge B.P., Marenholz I., Volz A., Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J. Invest. Dermatol. 1996;106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- Nelson A.M., Zhao W., Gilliland K.L., Zaenglein A.L., Liu W., Thiboutot D.M. Neutrophil gelatinase-associated lipocalin mediates 13-cis retinoic acid-induced apoptosis of human sebaceous gland cells. J. Clin. Invest. 2008;118:1468–1478. doi: 10.1172/JCI33869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldridge E.E., Walker H.F., Stower M.J., Simms M.S., Mann V.M., Collins A.T., Pellacani D., Maitland N.J. Retinoic acid represses invasion and stem cell phenotype by induction of the metastasis suppressors RARRES1 and LXN. Oncogenesis. 2013;2:e45. doi: 10.1038/oncsis.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudes A.J., Campbell D.S., Sorensen C.M., Walashek L.S., True L.D., Liu A.Y. Transcriptomes of human prostate cells. BMC Genomics. 2006;7:92. doi: 10.1186/1471-2164-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali D., Thaller C., Eichele G. Abnormal level of retinoic acid in prostate cancer tissues. J. Clin. Endocrinol. Metab. 1996;81:2186–2191. doi: 10.1210/jcem.81.6.8964849. [DOI] [PubMed] [Google Scholar]
- Polson E.S., Lewis J.L., Celik H., Mann V.M., Stower M.J., Simms M.S., Rodrigues G., Collins A.T., Maitland N.J. Monoallelic expression of TMPRSS2/ERG in prostate cancer stem cells. Nat. Commun. 2013;4:1623. doi: 10.1038/ncomms2627. [DOI] [PubMed] [Google Scholar]
- Rane J.K., Pellacani D., Maitland N.J. Advanced prostate cancer—a case for adjuvant differentiation therapy. Nat. Rev. Urol. 2012;9:595–602. doi: 10.1038/nrurol.2012.157. [DOI] [PubMed] [Google Scholar]
- Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson G.D., Robson C.N., Lang S.H., Neal D.E., Maitland N.J., Collins A.T. CD133, a novel marker for human prostatic epithelial stem cells. J. Cell Sci. 2004;117:3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- Rivera-Gonzalez G.C., Droop A.P., Rippon H.J., Tiemann K., Pellacani D., Georgopoulos L.J., Maitland N.J. Retinoic acid and androgen receptors combine to achieve tissue specific control of human prostatic transglutaminase expression: a novel regulatory network with broader significance. Nucleic Acids Res. 2012;40:4825–4840. doi: 10.1093/nar/gks143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seewaldt V.L., Caldwell L.E., Johnson B.S., Swisshelm K., Collins S.J., Tsai S. Inhibition of retinoic acid receptor function in normal human mammary epithelial cells results in increased cellular proliferation and inhibits the formation of a polarized epithelium in vitro. Exp. Cell Res. 1997;236:16–28. doi: 10.1006/excr.1997.3694. [DOI] [PubMed] [Google Scholar]
- Seita J., Sahoo D., Rossi D.J., Bhattacharya D., Serwold T., Inlay M.A., Ehrlich L.I., Fathman J.W., Dill D.L., Weissman I.L. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS ONE. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd C.J., Rizzo S., Ledaki I., Davies M., Brewer D., Attard G., de Bono J., Hudson D.L. Expression profiling of CD133+ and CD133- epithelial cells from human prostate. Prostate. 2008;68:1007–1024. doi: 10.1002/pros.20765. [DOI] [PubMed] [Google Scholar]
- Shimozono S., Iimura T., Kitaguchi T., Higashijima S., Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496:363–366. doi: 10.1038/nature12037. [DOI] [PubMed] [Google Scholar]
- van den Hoogen C., van der Horst G., Cheung H., Buijs J.T., Lippitt J.M., Guzmán-Ramírez N., Hamdy F.C., Eaton C.L., Thalmann G.N., Cecchini M.G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- van Leenders G., Dijkman H., Hulsbergen-van de Kaa C., Ruiter D., Schalken J. Demonstration of intermediate cells during human prostate epithelial differentiation in situ and in vitro using triple-staining confocal scanning microscopy. Lab. Invest. 2000;80:1251–1258. doi: 10.1038/labinvest.3780133. [DOI] [PubMed] [Google Scholar]
- Young C.Y., Murtha P.E., Andrews P.E., Lindzey J.K., Tindall D.J. Antagonism of androgen action in prostate tumor cells by retinoic acid. Prostate. 1994;25:39–45. doi: 10.1002/pros.2990250106. [DOI] [PubMed] [Google Scholar]
- Zheng L.T., Lee S., Yin G.N., Mori K., Suk K. Down-regulation of lipocalin 2 contributes to chemoresistance in glioblastoma cells. J. Neurochem. 2009;111:1238–1251. doi: 10.1111/j.1471-4159.2009.06410.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


