Abstract
Background
The complexity of protein glycosylation makes it difficult to characterize glycosylation patterns on a proteomic scale. In this study, we developed an integrated strategy for comparatively analyzing N-glycosylation/glycoproteins quantitatively from complex biological samples in a high-throughput manner. This strategy entailed separating and enriching glycopeptides/glycoproteins using lectin affinity chromatography, and then tandem labeling them with 18O/16O to generate a mass shift of 6 Da between the paired glycopeptides, and finally analyzing them with liquid chromatography-mass spectrometry (LC-MS) and the automatic quantitative method we developed based on Mascot Distiller.
Results
The accuracy and repeatability of this strategy were first verified using standard glycoproteins; linearity was maintained within a range of 1:10–10:1. The peptide concentration ratios obtained by the self-build quantitative method were similar to both the manually calculated and theoretical values, with a standard deviation (SD) of 0.023–0.186 for glycopeptides. The feasibility of the strategy was further confirmed with serum from hepatocellular carcinoma (HCC) patients and healthy individuals; the expression of 44 glycopeptides and 30 glycoproteins were significantly different between HCC patient and control serum.
Conclusions
This strategy is accurate, repeatable, and efficient, and may be a useful tool for identification of disease-related N-glycosylation/glycoprotein changes.
Keywords: 18O labeling, Hepatocellular carcinoma, Mass spectrometry, N-glycoproteome
Background
The glycosylation of proteins is a common post-translational modification. The occupancy of the glycosylation site and the glycan structure in the glycoproteins have a profound effect on their biological functions [1]. Alteration of this glycosylation influences growth, differentiation, transformation, adhesion, metastasis, and immune surveillance of cancers [2-6]. Glycans are classified as either O-linked (through Ser or Thr) or N-linked (through Asn on the Asn-X-Thr/Ser recognition sequence, X ≠ P) depending on their polypeptide attachment site. In particular, N-linked glycosylation is prevalent in secreted proteins found in body fluids (such as blood and urine) [7] and plays a significant role in cellular recognition and signal transduction and can therefore be considered a potential therapeutic target or biomarker for diseases, including cancers [8,9].
Currently, the most effective and accurate method of quantitatively analyzing glycopeptides and glycoproteins is mass spectrometry (MS). MS is usually combined with other techniques such as various protein/peptide enrichment, labeling, and data analysis techniques to obtain a complete understanding of protein glycosylation patterns (including glycosylation sites), site occupancy, and glycan structures. However, accurate, quantitative, high-throughput techniques for comprehensive analyses of protein glycosylation in complex biological samples have only rarely been established [8,10].
N-glycosylated sites in a glycopeptide are usually labeled and identified with 18O. Kuster et al. reported a method in which N-linked glycans were enzymatically removed from glycopeptides by peptide N-glycosidase F (PNGase-F), and the glycosylated Asn residues labeled with 18O. They demonstrated that the process generated a mass shift of 2 Da and the glycosylated sites were subsequently identified accurately via MS [11]. Kaji et al. further modified this method, performing quantitative comparative analyses using 16O-labeled residues as a control. However, the partial overlap in isotopic distribution of the 16O- and 18O-labeled peptides affects the accuracy of this quantitative method [12]. Recently, Liu et al. established the tandem 18O stable isotope labeling technique, which includes enriching glycopeptides via hydrophilic affinity extraction and labeling three 18O or 16O tandems at their C-terminus and N-glycosylation sites. The mass shift between paired 18O- and 16O-labeled glycopeptides is 6 Da [13]. This method overcomes isotope distribution overlap and enhances the accuracy of quantification. However, these 18O labeling-related techniques are time-consuming and low-throughput due to a lack of software for automatic quantitative analysis [13], particularly when analyzing a large number of complex biological samples. Moreover, these methods are not able to identify glycan structure alterations in specific glycoproteins, which are important for understanding the effects of glycan changes in glycoproteins on pathological processes.
Lectin affinity chromatography is an accurate glycan separation technology, and is extensively used to expound upon the glycan structure in glycoproteins [14,15]. For example, using this technique, Lens culinaris (LCH)-affinitive alpha-fetoprotein (AFP-L3) is separated, which is more accurate in diagnosing liver cancer than total AFP [16-19]. In the present study, we combined lectin affinity chromatography, tandem 18O/16O labeling, MS, and a self-build automatic quantitative method based on Mascot Distiller software to develop an integrated strategy for high-throughput quantitative analysis of N-glycosylation changes in complex biological samples. The accuracy and repeatability of this strategy were verified using glycoprotein standards. We also utilized this strategy to analyze the serum of healthy individuals and hepatocellular carcinoma (HCC) patients to confirm its feasibility in complex biological samples.
Results and discussion
An integrated strategy for glycoproteomic study
It is well known that protein glycosylation varies widely between different glycoproteins; indeed, a single glycoprotein can be glycosylated at multiple sites with various glycans. Such complexity makes it difficult to characterize protein glycosylation patterns on a proteomic scale. The major limitations of the current glycoproteomic studies include: (1) Searching for changes in glycan structure or glycosylation site occupancy of a single glycoprotein, rather than in a high-throughput manner on a large proteomic scale [16,20]; (2) Investigating glycosyltransferase or the general changing trends of glycan structure in biological samples, regardless of the glycan structure of each specific glycoprotein [21-23]; and (3) Analyzing the expression levels of glycoproteins with specific glycan structure, but not describing the glycosylation sites and glycosylation site occupancy [24-27]. In order to overcome these limitations, we developed an integrated strategy which can be used to quantitatively analyze the abundance of glycoproteins/glycopeptides in a high-throughput manner, as well as the glycan structure and sites of altered N-glycosylation.
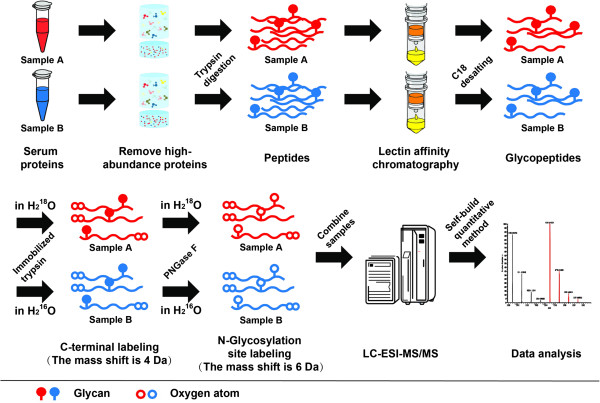
Our overall experimental strategy is shown in Figure 1. The glycopeptides with specific glycan structure were enriched via digestion and lectin affinity chromatography, and then treated with immobilized trypsin and PNGase-F in 16O or 18O water. During this process, the C terminus of the peptide was labeled by two 18O or 16O, and the N-glycosylation site was labeled by one 18O or 16O. Thus, when they were mixed at the same ratio, a mass shift of 6 Da was generated between the paired 18O- and 16O-labeled glycopeptides, while only a mass shift of 4 Da was present between the paired non-glycopeptides, which could subsequently be identified by MS. A detailed description of 18O labeling is supplied in Additional file 1. The relative concentration ratio could be directly quantitated through the relative signal strength of the peptide ion pair in the precursor scan because corrections for the overlapping distributions of monoisotopic peaks were built into the software.
Figure 1.
Flow chart of the integrated strategy. Serum samples were digested with trypsin after removal of high-abundance proteins. Glycopeptides were enriched with lectin chromatography from two different samples, desalted, and then catalyzed by immobilized trypsin and PNGase-F in 18O or 16O water, as indicated. Equal amounts of 18O- and 16O-labeled glycopeptides were mixed, and the 6 Da mass shifts were generated between paired, labeled glycopeptides, which could be identified by subsequent LC-ESI-MS/MS. The data were quantitatively analyzed automatically with the self-build quantitative method.
The glycan structure of glycoproteins is specifically recognized by lectin and the glycan structure changes of glycoproteins distinguished by LCH, WGA, or ConA are associated with cancer development, and therefore have potential diagnostic and prognostic values [28-31]. As such, in this study, we used LCH, WGA, and ConA lectin chromatography to separate and enrich glycopeptides with a specific glycan structure. Using this method, we were able to obtain information regarding changes in the abundance of glycoproteins with different types of glycan, as well as the glycosylation site and glycosylation site occupancy in each altered glycoprotein, all with one experiment. Our strategy pinpoints the glycosylation changes to each glycoprotein on a large glycoproteomics scale, providing a valuable supplement to techniques currently used in glycoproteomics.
However, lectin affinity chromatography is far from ideal [32-34], as the enrichment efficiency of the method is unsatisfactory, is easily affected by buffer conditions, and non-specifically recognizes glycans [35,36]. In this study, we attempted to stabilize the binding and elution buffers, including adjustment of pH, concentration, and binding/eluting time, in different experiments, to overcome these disadvantages.
Incomplete 18O labeling generates negative results, primarily due to the reversible labeling reaction at the C-terminus [37]. A number of factors may affect the efficiency of 18O labeling at the C-terminus, such as the catalytic activity of trypsin, the purity of H218O, H216O, and other reagents, the back-exchange caused by incomplete trypsin quenching, and the relative positions of Lys and Asp/Glu at the C-terminus [38-40]. In order to remove these interference factors, we made the following modifications to our experiments based on previous studies: (1) Immobilized trypsin was applied to increase the mole ratio of protease to substrate and improve labeling efficiency [41]. The immobilized enzymes could be completely removed physically after the reaction and the carboxyl oxygen exchange nearly ceased and reduced back-exchange; (2) Acidic conditions were adopted to facilitate catalysis of the carboxyl oxygen reaction and obtain better efficiency of the immobilized trypsin labeling [40,42]; and (3) After digestion with trypsin, samples were boiled for 10 min followed by freezing for 5 min, and methanoic acid was added prior to PNGase-F labeling to fully quench the trypsin and avoid back-exchange [39].
Validation of the feasibility and accuracy of the integrative strategy
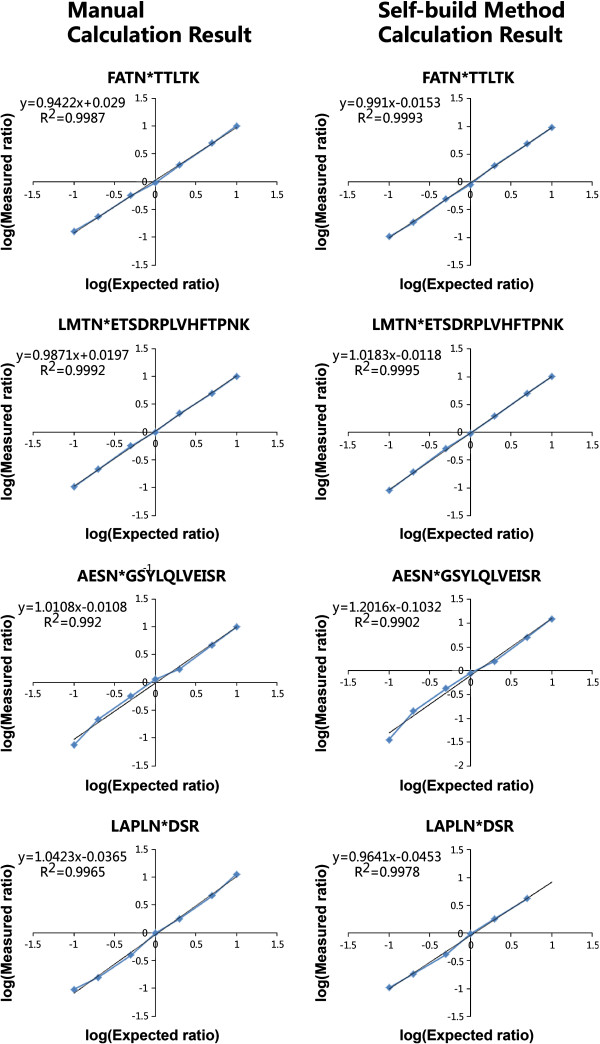
The glycoproteins invertase and Fetuin were used as standards to evaluate the accuracy and feasibility of this integrated strategy for quantitation of N-glycoproteins. The glycopeptides were enriched from the glycoprotein standards with ConA lectin chromatography, and labeled with 18O and 16O. The 18O- and 16O-labeled glycopeptides were mixed in ratios of 1:1, 1:2, 2:1, 1:5, 5:1, 1:10, and 10:1, then analyzed by LC-MS. The relative concentration ratios of the glycopeptides (18O3/16O3) were calculated using Formula 1, and the relative concentration ratios of the non-glycopeptides (18O2/16O2) were calculated according to Formula 2 (see details in the methods section). In the mass spectrum of the mixed glycopeptides, the mass shift of 6 Da was easily identified for paired glycopeptides, and the mass shift of 4 Da was identified for paired non-glycopeptides (Additional file 2), accurately distinguishing between the two. Four glycopeptides and four non-glycopeptides in invertase and Fetuin were selected to further manually calculate relative concentration ratios. We found that peptide concentration ratios from manual calculation were similar to theoretical values (Table 1), and correlation coefficients (R2) were all >0.99 (Figure 2). These results indicated that our strategy had good linearity and accuracy in a 100-fold dynamic range.
Table 1.
Relative abundance ratios of the 18 O/ 16 O-labeled glycopeptides and non-glycopeptides in glycoprotein standards
| Protein | Peptide sequence | Expected ratio (Sample A:B) | Manually calculated ratio | Self-build quantitative method ratio |
|---|---|---|---|---|
| Invertase |
FATN*TTLTK |
1 (1:1) |
0.961 |
0.897 |
| |
|
2 (2:1) |
1.997 |
1.982 |
| |
|
5 (5:1) |
4.880 |
4.856 |
| |
|
0.5 (1:2) |
0.563 |
0.491 |
| |
|
0.2 (1:5) |
0.236 |
0.188 |
| |
|
0.1 (1:10) |
0.129 |
0.104 |
| |
|
10 (10:1) |
9.989 |
9.456 |
| |
LMTN*ETSDRPLVHFTPNK |
1 (1:1) |
0.992 |
0.953 |
| |
|
2 (2:1) |
2.169 |
1.939 |
| |
|
5 (5:1) |
4.980 |
5.033 |
| |
|
0.5 (1:2) |
0.567 |
0.517 |
| |
|
0.2 (1:5) |
0.216 |
0.192 |
| |
|
0.1 (1:10) |
0.103 |
0.089 |
| |
|
10 (10:1) |
10.110 |
10.050 |
| |
ENPYFTNR |
1 (1:1) |
0.937 |
0.997 |
| |
|
2 (2:1) |
2.040 |
2.144 |
| |
|
5 (5:1) |
5.217 |
5.462 |
| |
|
0.5 (1:2) |
0.474 |
0.539 |
| |
|
0.2 (1:5) |
0.177 |
0.208 |
| |
|
0.1 (1:10) |
0.085 |
0.111 |
| |
|
10 (10:1) |
10.558 |
11.160 |
| |
GLEDPEEYLR |
1 (1:1) |
0.769 |
0.837 |
| |
|
2 (2:1) |
1.714 |
1.788 |
| |
|
5 (5:1) |
5.014 |
4.437 |
| |
|
0.5 (1:2) |
0.472 |
0.458 |
| |
|
0.2 (1:5) |
0.184 |
0.183 |
| |
|
0.1 (1:10) |
0.084 |
0.086 |
| |
|
10 (10:1) |
9.636 |
11.110 |
| Fetuin |
AESN*GSYLQLVEISR |
1 (1:1) |
1.138 |
0.886 |
| |
|
2 (2:1) |
1.735 |
1.588 |
| |
|
5 (5:1) |
4.664 |
5.070 |
| |
|
0.5 (1:2) |
0.562 |
0.424 |
| |
|
0.2 (1:5) |
0.219 |
0.143 |
| |
|
0.1 (1:10) |
0.075 |
0.036 |
| |
|
10 (10:1) |
9.849 |
12.330 |
| |
LAPLN*DSR |
1 (1:1) |
0.980 |
0.965 |
| |
|
2 (2:1) |
1.797 |
1.824 |
| |
|
5 (5:1) |
4.627 |
4.174 |
| |
|
0.5 (1:2) |
0.397 |
0.415 |
| |
|
0.2 (1:5) |
0.158 |
0.184 |
| |
|
0.1 (1:10) |
0.096 |
0.103 |
| |
|
10 (10:1) |
11.289 |
N
a
|
| |
ALGGEDVR |
1 (1:1) |
0.904 |
0.969 |
| |
|
2 (2:1) |
2.116 |
1.913 |
| |
|
5 (5:1) |
5.748 |
4.589 |
| |
|
0.5 (1:2) |
0.469 |
0.471 |
| |
|
0.2 (1:5) |
0.175 |
0.170 |
| |
|
0.1 (1:10) |
0.088 |
0.080 |
| |
|
10 (10:1) |
12.237 |
11.540 |
| |
TPIVGQPSIPGGPVR |
1 (1:1) |
1.067 |
1.042 |
| |
|
2 (2:1) |
2.102 |
1.986 |
| |
|
5 (5:1) |
5.823 |
5.559 |
| |
|
0.5 (1:2) |
0.460 |
0.479 |
| |
|
0.2 (1:5) |
0.183 |
0.163 |
| |
|
0.1 (1:10) |
0.063 |
0.100 |
| 10 (10:1) | 10.414 | 10.120 |
*Denotes the N-glycosylation site.
a self-build quantitative method did not give a ratio.
Sample A was treated in 18O water and Sample B in 16O water. N = self-build quantitative method did not give a ratio.
Figure 2.
Linearity and accuracy of the self-build quantitative method in glycopeptides from glycoprotein standards. Four glycopeptides, FATN*TTLTK and LMTN*ETSDRPLVHFTPNK from invertase [Swiss-Prot:P00724] and AESN*GSYLQLVEISR and LAPLN*DSR from Fetuin [Swiss-Prot:P12763], were serially diluted to determine the linearity and accuracy of the self-build quantitative method. On the left are the results from manual calculation and on the right are the results from the self-build quantitative method. Glycopeptide concentration ratios calculated manually and those from the self-build method were all significantly associated with the theoretical values, with both correlation coefficients (R2) being >0.99. *denotes the N-glycosylation site.
Confirmation of the precision of the self-build method
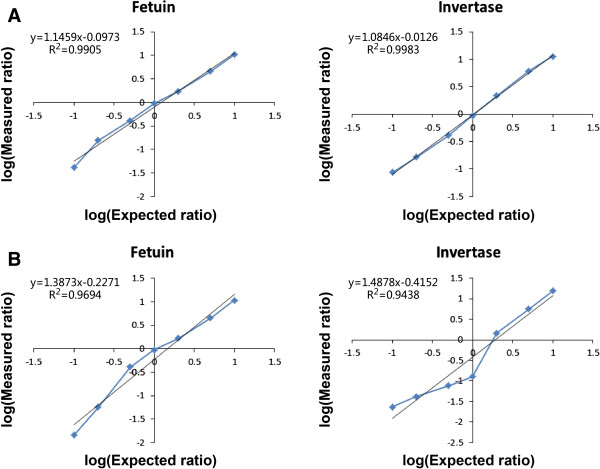
Although data analysis of some 18O labeling methods can be supported by some automatic software, the tandem 18O3 labeling technique lacks matched software, and the data obtained has so far been analyzed with time-consuming manual calculation. Managing data from complex biological samples using manual calculation is difficult, necessitating an accurate, reliable, and user-friendly automated analysis method for data generated with 18O3 labeling [13]. In our study, two customized software packages, XPRESS and ASAPRatio, including the Trans-Proteomic Pipeline Ver. 4.5 (TPP, Seattle Proteome Center), were first used to quantitatively analyze data generated from the 18O3-labeled glycoprotein standards. The results were disappointing; XPRESS gave linear results to non-glycopeptides labeled with 18O2, rather than to the 18O3-labeled glycopeptides, and the quantitative results generated by ASAPRatio were even less satisfactory than those of XPRESS (Additional file 3). We established an automatic quantitative method for the 18O3 labeling technique based on Mascot Distiller and applied it to analyze the glycoprotein standard data obtained from LC-MS. As shown in Table 1, the peptide concentration ratios calculated by this quantitative method were similar to both the theoretical values and the manually calculated results, and had good linearity and accuracy within the ratio range of 1:10–10:1 (Table 1 and Figure 2). Similar results were found with the protein concentration ratios (Figure 3). These data indicate that this quantitative method is reliable for calculating concentration ratios of both peptides and proteins labeled with 18O3, and may replace the time-consuming manual calculation in time.
Figure 3.
Linearity and accuracy of the self-build quantitative method for glycoprotein standards. The glycoprotein standards Fetuin and invertase were serially diluted to determine the linearity and accuracy of the self-build quantitative method. (A) The ratio of (Group A + Group B)/Group C. Glycopeptide concentration ratios calculated by the self-build method were significantly associated with the theoretical values, with both correlation coefficients (R2) being >0.99. (B) The ratio of Group A/Group C was calculated without considering back-labeling and incomplete labeling in glycoprotein standards; a difference was found between the result of the relative quantitation ratio of 18O/16O in (B) and the theoretical ratio.
As mentioned in the methods section, three modification groups were defined in the self-build quantitative method, and the quantitative ratio of 18O/16O was finally generated via Formula 3 [(Group A + Group B)/Group C] to avoid the influences of back-exchange and incomplete C-terminal labeling on the final result. The ratio of Group A to Group C was used to evaluate the influences of back-exchange and incomplete C-terminal labeling. We found that if these influencing factors had not been excluded, the final quantitative ratios of 18O3/16O3 differed from the theoretical ones (Figure 3), demonstrating that even the labeling efficiency was improved in this study. These data indicate that the quantitative setting in our study is correct and can minimize the influence of incomplete labeling and back-exchange of 18O.
Establishment of the quantitative criteria for this integrative strategy
In order to establish the measurement criteria for the relative quantification of glycoproteins and glycopeptides, the 16O/18O-labeled glycoprotein standard mixture at a ratio of 1:1 was repeatedly analyzed by LC-MS and quantitatively calculated seven times. The SD was detected in the spectrum of the glycoprotein standard, with a SD range of 0.023–0.186 for the glycopeptide and 0.075–0.216 for the glycoprotein. The relative quantitative ratios generated are listed in Table 2. An 16O/18O-labeled glycopeptide or glycoprotein ratio >3 times the SD value was considered a significant change; in contrast, when the ratio was within 1–3 times the SD, it was considered a minor change [13,43]. Thus, the quantitative criteria were defined as follows: Significant changes were determined when the ratio was smaller than 0.63 or greater than 1.57 for glycopeptides and less than 0.60 or over 1.65 for glycoproteins, whereas minor changes were assumed when the ratio was 0.63–0.84 or 1.19–1.57 for glycopeptides and 0.60–0.82 or 1.22–1.65 for glycoproteins.
Table 2.
The SD for glycoproteins and four glycopeptides from ConA-enriched Fetuin and invertase in a 1:1 18O/16O ratio calculated by the self-build quantitative method
| Protein and peptide sequence | SD ratio |
|---|---|
|
Invertase glycoprotein |
0.216 |
| FATN*TTLTK |
0.186 |
| NPVLAAN*STQFR |
0.156 |
|
Fetuin glycoprotein |
0.075 |
| LAPLN*DSR |
0.054 |
| AESN*GSYLQLVEISR | 0.023 |
*denotes the N-glycosylation site.
Validation of the feasibility of the integrative strategy in complex biological samples
Serum samples from three HCC patients and three healthy individuals were used to determine the feasibility of this strategy in complex clinical samples. Considering that gender and age may partially affect serum glycan distributions and a number of environmental variables (such as smoking) may also be associated with serum glycome components [44-46], we matched the HCC patients and healthy individuals as much as possible to decrease bias caused by individual differences.
The glycopeptides in the serum samples were separated and enriched with ConA, LCH, or WGA lectin chromatography, generating three subgroups of glycopeptides specifically recognized by ConA, LCH, and WGA, respectively. The glycopeptides were then labeled with 18O or 16O, followed by mixing the glycopeptides in a ratio of 1:1 from each subgroup of glycopeptides. Each mixture was repeatedly analyzed by LC-MS and quantitatively calculated seven times. We found that 44 unique glycopeptides and 30 glycoproteins with a specific glycan structure were differently expressed between HCC patients and healthy individuals (Additional file 4). Among these differentially expressed glycopeptides and glycoproteins, 14 and 13 changed in more than one lectin subgroup, respectively (see detailed data in Additional files 5 and 6). There were 67 unchanged glycopeptides in serum samples (see detailed data in Additional file 7). All N-linked glycopeptides had a consensus motif of Asn-X-Thr/Ser (X ≠ P). However, there were very low amounts of the differentially expressed glycopeptides/glycoproteins identified in our study, partially due to the limited volume of the serum samples and the multi-step processing of samples. All detailed data of detected glycopeptides in HCC patient and health control serum is shown in Additional file 8.
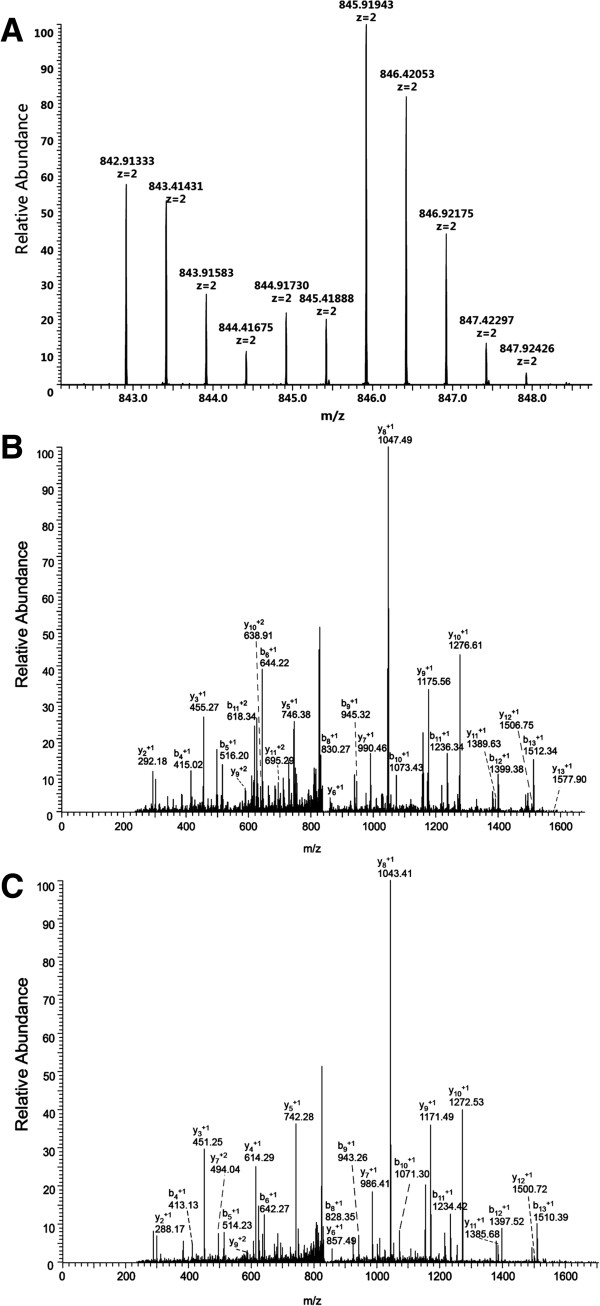
A representative Nano LC-ESI-MS/MS spectrum of a clusterin (CLUS) protein glycopeptide, LAN*LTQGEDQYYLR, in the ConA subgroup is shown in Figure 4; Figure 4A shows a magnified MS spectrum with a monoisotopic peak of double-charged peptide at m/z 845.91943 (18O) and 842.91333 (16O), representing a 6 Da mass shift. The MS spectrum indicated that there were three 18O atom labels and a mono-glycosylation site on this glycopeptide. The fragmented ion MS/MS spectrum had a mass shift of 117 Da between the y11 and y12 ions, equal to the mass shift generated by aspartic acid after being labeled by one 18O atom, and characteristically verified the deamidation of Asn in this position. The mass shift of 4 Da was displayed in all singly charged y ions (Figure 4B and C), indicating that the C-terminus was labeled by two 18O. A 2-Da mass shift was displayed in the b-ion series of monocharges, confirming that one 18O was present at the monoglycosylation site (Asn residue).
Figure 4.
MS and quantitation of a deglycosylated peptide, LAN*LTQGEDQYYLR, from ConA-enriched CLUS [Swiss-Prot: P10909] glycoprotein in human serum. (A) Double-charged peptide labeled with 18O and 16O. The m/z 845.91943 (18O) and 842.91333 (16O) displays a 6 Da mass shift in the monoisotopic peaks; the intensity ratio of the 18O/16O isotope cluster was 1.812. (B) Mass spectrum of the glycopeptide labeled with 18O in m/z 845.91943. (C) Mass spectrum of the glycopeptide labeled with 16O in m/z 842.91333. *denotes the N-glycosylation site.
The quantitative results of the serum samples were verified again by manual calculation of the four selected glycopeptides. There was no significant difference between the automatically quantitated ratios and the manually calculated ones (Additional file 9), suggesting that this automatic quantitative method is reliable for analysis of complex biological samples.
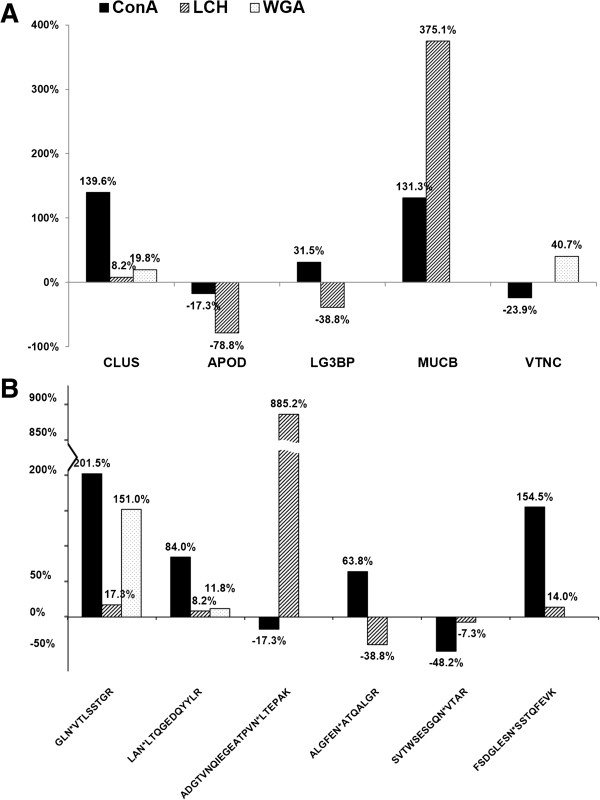
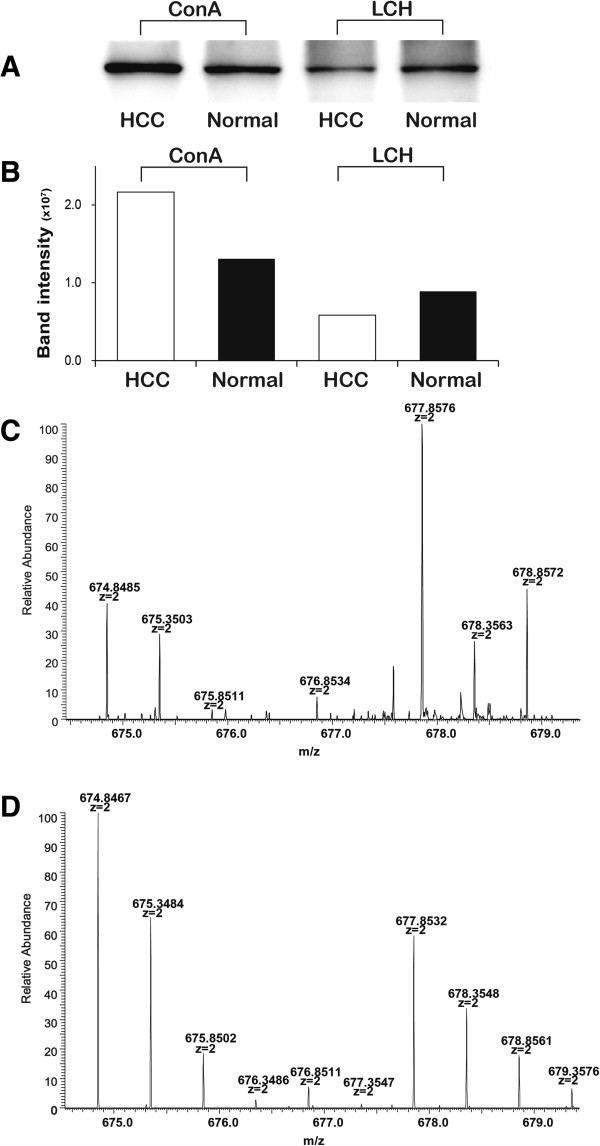
Compared with the healthy individuals, the alterations of some glycopeptide/glycoprotein levels in HCC patients were inconsistent, even converse, among the three subgroups of glycopeptides (Figure 5). These data suggest that the glycan structure on specific glycosylation sites may also be altered in these glycoproteins. Compared with the studies using total serum or tissue glycoproteome, glycoprotein subgroups separated by lectin chromatography could reduce the complexity of tested samples and improve the detection of low-abundance proteins. Therefore, these glycan changes on specific glycoproteins may be sensitive potential biomarkers for disease diagnosis, which are worthy of further investigation. Among these proteins, apolipoprotein D (APOD) was down-regulated in HCC patient serum in all three lectin subgroups, and CLUS was up-regulated in all three lectin subgroups, consistent with previous data [47,48]. To further validate the quantitative results obtained by our strategy, we determined the expression levels of glycoprotein LG3BP by western blot in the ConA and LCH lectin subgroups from HCC patient and healthy individual serum. As shown in Figure 6, the band intensity ratio of HCC patients versus healthy individuals was 1.66 in the ConA subgroup and 0.66 in the LCH subgroup. These were very similar to the ratios of glycoproteins (1.32 in the ConA subgroup and 0.61 in the LCH subgroup) and glycopeptide ratios of the proteins (1.64 in the ConA subgroup and 0.61 in the LCH subgroup) obtained by our integrated strategy. The quantity changes observed in the integrated strategy were independently confirmed by western blot. These differentially expressed glycoproteins might play an important role in screening for sporadic HCC in the general population. All of the above results indicate that the present labeling strategy is feasible and reliable.
Figure 5.
Glycopeptides/glycoproteins with inconsistent changes in the ConA, LCH, and WGA subgroups. (A) When comparing HCC patients with healthy individuals, five glycoproteins were altered inconsistently among the three lectin subgroups (ConA, LCH, and WGA); (B) When comparing the serum of HCC patients with that of healthy individuals, six glycopeptides changed discordantly among the three lectin subgroups (ConA, LCH, and WGA). Each column represents the percentage of the amount of glycopeptide/glycoprotein (HCC patients versus healthy individuals) in each lectin subgroup. Positive percentages denote upregulation, whereas negative percentages denote downregulation. Glycopeptide GLN*VTLSSTGR was from CO4A_HUMAN [Swiss-Prot:P0C0L4], LAN*LTQGEDQYYLR from CLUS_HUMAN [Swiss-Prot:P10909], ADGTVNQIEGEATPVN*LTEPAK from APOD_HUMAN [Swiss-Prot:P05090], ALGFEN*ATQALGR from LG3BP_HUMAN [Swiss-Prot:Q08380], SVTWSESGQN*VTAR from IGHA2_HUMAN [Swiss-Prot:P01877], and FSDGLESN*SSTQFEVK from CO4B_HUMAN [Swiss-Prot:P0C0L5]. Other glycoproteins were MUCB_HUMAN [Swiss-Prot:P04220] and VTNC_HUMAN [Swiss-Prot:P04004]. *denotes the N-glycosylation site.
Figure 6.
Validation of glycoprotein LG3BP in different lectin subgroups by western blot. (A) The expression levels of LG3BP in ConA and LCH lectin subgroups from the serum of HCC patients and healthy individuals were detected by western blot. (B) The target band intensities were quantified by ImageQuantTL software. The band intensity ratio of HCC patients versus healthy individuals was 1.66 in the ConA subgroup and 0.66 in the LCH subgroup. (C) The mass spectrum of a glycopeptide, ALGFEN*ATQALGR, from glycoprotein LG3BP in the ConA subgroups. (D) The mass spectrum of the glycopeptide, ALGFEN*ATQALGR, from glycoprotein LG3BP in the LCH subgroups. The 18O/16O ratios (HCC patients versus healthy individuals) of these glycopeptides were 1.64 in the ConA subgroup and 0.61 in the LCH subgroup. *denotes the N-glycosylation site.
Conclusions
In this study, we established an integrated research strategy for the high-throughput, quantitative analysis of N-linked glycoproteomics. This strategy integrated lectin chromatography and tandem 18O/16O labeling with LC-MS analysis and our novel automatic data analysis method. We also made modifications to the techniques used to avoid various interferences and enhance the labeling efficiency of 18O3. We demonstrated this strategy to be accurate and reliable using glycoprotein standards, and then identified a number of N-glycoproteins with specific glycan structures that were differently expressed between HCC patients and healthy individuals, as well as N-glycoproteins with modified glycosylation site occupancy. Western blot analysis further confirmed these results. This integrated strategy provides a useful tool for identifying disease-related N-glycosylation changes and glyco-biomarkers for diagnosis and prognosis of diseases.
Methods
Chemicals and materials
The ProteoMiner Protein Enrichment Kit was purchased from Bio-Rad (Hercules, CA), the PNGase-F from New England BioLabs (Ipswich, MA), the C18 cartridge from Waters (Milford, MA), the 3-kDa spin column from Millipore (Billerica, MA), and the immobilized trypsin beads from Applied Biosystems (Framingham, MA). The concanavalin A (ConA)-based and wheat germ agglutinin (WGA)-based glycoprotein isolation kit, the bicinchoninic acid (BCA) assay kit and MicroSpin column were obtained from Pierce (Rockford, IL). The LCH-based isolation kit was from GALAB (Germany). The glycoprotein standards (bovine Fetuin and yeast invertase), 18O water (97%), and other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Preparation of serum samples
The archived serum samples of patients with HCC were obtained from Zhongshan Hospital, Fudan University (Shanghai, China). Healthy individuals served as normal controls. Physiological conditions such as age, etc., were matched to decrease bias caused by individual differences. Detailed information regarding the HCC patients and controls were summarized in Additional file 10. This study was approved by the Research Ethics Committee of Zhongshan Hospital, and informed consent was obtained from all subjects.
The serum samples were stored at -80°C before processing. Equal volumes of serum from three HCC patients or three healthy individuals were pooled together to generate two sample pools, which were used in subsequent experiments. The most abundant serum proteins were removed by the ProteoMiner Protein Enrichment Kit according to the manufacturer's instruction. The protein concentrations were determined using the BCA assay kit.
Digestion of glycoprotein standards and serum samples
The paired serum samples and the glycoprotein standards, Fetuin and yeast invertase, in solution were denatured at 100°C for 10 min. The samples were reduced with 10 mM dithiothreitol (DTT) at 57°C for 30 min and alkylated with 30 mM iodoacetamide at room temperature for 1 h in the dark. After desalting by spin column, the samples were digested with trypsin at an enzyme-to-substrate ratio of 1:50 (w/w) at 37°C for 16 h. To quench the trypsin and prevent back-exchange of 18O, the digested samples were boiled in a water bath for 10 min and then placed on ice for 5 min, as previously described [39].
Lectin affinity chromatography
Lectin affinity chromatography was performed using ConA-, LCH-, and WGA-based isolation kits to separate out glycopeptides with specific glycan structure. Briefly, the digested serum samples or glycoprotein standards were diluted with binding/wash buffer and then added to the resin bed and incubated for 10 min at room temperature. The resin was then washed and the bound glycopeptides eluted and collected.
Isotope labeling with 18O or 16O water
After lectin affinity chromatography, the peptides obtained from the samples and glycoprotein standards were desalted using SepPak C18 cartridges, and then dried in a vacuum centrifuge. The peptides were then mixed with immobilized trypsin (20% slurry v/w) for 20 min with gentle shaking, and then lyophilized. The lyophilized peptides were dissolved in 100 μL acetonitrile in 50 mM NH4HCO3 (pH 6.8) (ACN/NH4HCO3, 20% v/v) prepared with H216O or H218O in advance, then incubated at 37°C for 24 h to catalyze the labeling of tryptic peptides at the C-terminus. The immobilized trypsin beads were then removed by MicroSpin columns. A total of 5 μL formic acid was added to further inhibit any possible residual trypsin activity. The peptides were lyophilized and then dissolved in 100 mM NH4HCO3 buffer prepared in H216O or H218O. PNGase F was added at a concentration of 1 μL PNGase-F/mg of crude protein, and the labeling was conducted at 37°C overnight. Finally, the 16O- and 18O-labeled peptides were mixed at designated ratios (1:1, 2:1, 1:2, 5:1, 1:5, 10:1, and 1:10 for glycoprotein standards; 1:1 for samples) and lyophilized.
Nano LC-electrospray ionization (ESI)-MS/MS
The lyophilized peptides were resuspended with 2% ACN in 0.1% formic acid, separated by nano LC, and then analyzed by online electrospray tandem mass spectrometry. The experiments were performed on a Nano Aquity UPLC system (Waters) connected to an LTQ Orbitrap XL mass spectrometer (Thermo Electron Corp., Bremen, Germany) interfaced with an online nano electrospray ion source (Michrom Bioresources, Auburn, CA). The peptide separation was performed in a Michrom CAPTRAP (500 μm i.d. × 2 mm trap column) and a Michrom C18 (3.5 μm, 100 μm i.d. × 15 cm reverse phase column) (Michrom Bioresources). The model glycoprotein digests (0.5 μg) were loaded onto the trap column and leached at a flow rate of 20 μL/min for 3 min. The mobile phases included 2% ACN in 0.1% formic acid (phase A and the loading phase) and 95% ACN in 0.1% formic acid (phase B). To achieve sufficient separation, a 60-min (for glycoprotein standards) or 90-min (for serum samples) linear gradient from 5% to 45% at phase B was employed. The flow rate of the mobile phase was set at 500 nL/min, and the electrospray voltage used was 1.6 kV. The linear gradient was adjusted to 90 min for serum samples analyses, while all other parameters remained unchanged. The LTQ Orbitrap XL mass spectrometer was operated in the data-dependent mode with an automatic switch between MS and MS/MS acquisition. The survey full-scan MS spectra with two microscans (m/z 350–1800) was acquired in Orbitrap at a resolution of 100,000 (at m/z 400) followed by eight MS/MS scans in LTQ trap. Dynamic exclusion was set to initiate a 60 s exclusion for ions analyzed twice within a 10 s interval.
Manual calculation of relative concentration ratios
The mass spectra acquired by Nano LC-ESI-MS/MS of the samples were searched against the human International Protein Index (IPI) database (IPI human v3.45 FASTA with 71,983 entries, with bovine Fetuin and yeast invertase manually added), using the SEQUEST algorithm integrated into the Bioworks package (Version 3.3.1; Thermo Electron). The parameters for the SEQUEST search included: enzyme, partial trypsin; missed cleavages allowed, two; fixed modification, carboxyamidomethylation (Cys); variable modifications, deamidation (Asn +0.98 Da), deamidation plus 18O (Asn +2.98 Da), C-term (+4.01 Da), and oxidation (Met +15.99 Da); peptide tolerance, 10 ppm; and MS/MS tolerance, 1.00 Da. The statistical significance of the database search results was evaluated with the aid of PeptideProphet [49]. A minimum PeptideProphet probability score (P) filter of 0.9 was selected as a threshold to remove low-probability peptides.
The relative concentration ratios of the peptides were then manually calculated. Formula 1 was used to calculate the ratio (16O/18O) of glycopeptides as described previously [13]:
Formula 2 was used to calculate the ratio (16O/18O) of the non-glycopeptides as described previously [50]:
M0, M2, M4, and M6 are the corresponding theoretical relative intensities of the isotopic envelope of the peptide, calculated using MS-Isotope (http://prospector.ucsf.edu).
Calculation of relative concentration ratios with the self-build quantitative method
The raw data acquired by Nano LC-ESI-MS/MS were searched against the Swiss-Prot database using the Mascot Distiller software (Version 2.3.2.0; Matrix Science) and user-defined search criteria. The search parameters were set according to the preceding settings of Bioworks. The relative concentration ratios were generated by the Mascot Distiller software with a self-build quantitative method. In this method, the quantitative protocol is the precursor. Taking into account the incomplete label or back-exchange on the C-terminus, three exclusive modification groups were used for calculating the ratios: Group A was comprised of two 18O labels on the C-terminus and one 18O label on each N-glycosylated Asn residue; Group B included one 18O label on the C-terminus and one 18O label on each N-glycosylated Asn residue; and Group C were labeled by 16O on both of these sites. As one given peptide may only carry one or another set of modifications, but never have a mixture of both sets, the “exclusive” modification group was used to avoid interference from the too-complex resultant data and too many variable modifications derived from the pooled samples. The isotope and impurity correction factors were set to 97% 18O based on actual use. The relative concentration ratios were calculated by Formula 3:
The glycoprotein ratios were calculated according to the median of the glycopeptide ratios with the self-built quantitation software. Additional file 11 is self-build quantitation setting file and Additional file 12 is a modified unimod file.
Western blot
The expression level of glycoprotein galectin-3-binding protein (LG3BP) was evaluated by western blot to validate the results of the integrated research strategy. The glycoproteins in the depleted pooled serum from three HCC patients and three healthy individuals were enriched by lectin affinity chromatography, and then separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer onto polyvinylidene fluoride membranes. Anti-LG3BP (Santa Cruz Biotechnology, Dallas, TX) was used as the primary antibody. The quantitative signals were acquired and quantified via a LAS-4000 imager and ImageQuantTL software (Version 7.0; GE Healthcare, Piscataway, NJ).
Statistical evaluation
Based on the statistical three-sigma rule which states that nearly all values lie within 3 standard deviations (SD) of the mean for a normal distribution [51], we first established a set of criteria for statistical evaluation of glycopeptide/glycoprotein differences using the glycoprotein standards. Compared with the controls, a change of more than 3-fold of SD at an abundance ratio of 1.0 was considered statistically significant at a 99% confidence level [52,53].
Abbreviations
AFP-L3: Lens culinaris affinitive alpha-fetoprotein; APOD: Apolipoprotein D; CLUS: Clusterin; ConA: Concanavalin A; DTT: Dithiothreitol; HCC: Hepatocellular carcinoma; LCH: Lens culinaris; LG3BP: Galectin-3-binding protein; WGA: Wheat germ agglutintin.
Competing interests
The authors declare no conflict of interests with any company or financial organization.
Authors’ contributions
JW carried out the experimental steps and wrote the paper; JW and CZ were involved in serum peptide purification; JW, WZ, JY and HJL performed the mass spectrometric analysis of proteins; CZ and QZD were involved in serum samples and clinical data collection; HJZ and LXQ designed the experiments and supervised the research manuscript. All authors read and approved the manuscript.
Supplementary Material
The reaction of three 18O atoms labeling happened in the glycopeptides by catalysis with Trypsin and PNGase F. The reaction is reversible at the 18O labeling of the C-terminal catalyzed with trypsin, indicating that the back-exchange and C-terminal single 18O labeling in the C-terminal cannot be completely avoided in the reaction product. This feature is a problem identified in the experimental operation, result analysis, and quantitative method design, but do not need to consider in the labeling process of PNGase-F catalysis.
The mass shifts identified by mass spectrum between paired 18 O and 16 O labeled glycopeptides/non-glycopeptides from standard glycoprotein Invertase. (A) For the non-glycopeptides VFWYEPSQK, the mass shift of 4 Da was generated in mass spectrometry. (B) For the glycopeptide FATN*TTLTK, the mass shift of 6 Da was generated in mass spectrometry. *denotes the N-glycosylation site.
ASAP and XPRESS ratios of glycopeptides and non-glycopeptides in the dynamic range of 1:10–10:1 derived by Trans-Proteomic Pipeline Ver. 4.5.
The number of differently expressed glycoproteins/glycopeptides between HCC patients and healthy individuals in the three lectin subgroups. The data calculated by self-build quantitative method.
The table of changed glycopeptides in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
The table of changed glycoproteins in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
The table of unchanged glycopeptides in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
The detailed data of detected glycopeptides in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
Calculation of abundance ratios of four glycopeptides between HCC patients and healthy individuals. The ratios calculated manually were similar to the ratios obtained by self-build quantitative method, which indicated the reliability of the quantitative results of our integrated research strategy.
Physiological/pathological characteristics of patients and healthy individuals enrolled in this study.
Our self-build quantitation setting file.
A Modified unimod file. You can download Additional files 9 and 10, copy them into the directory of /mascot/config/, please backup the original files before copying.
Contributor Information
Ji Wang, Email: wangji123456@gmail.com.
Chuang Zhou, Email: zhouchuang126@126.com.
Wei Zhang, Email: weizhf@gmail.com.
Jun Yao, Email: yaojun123@fudan.edu.cn.
Haojie Lu, Email: luhaojie@fudan.edu.cn.
Qiongzhu Dong, Email: qzhdong@fudan.edu.cn.
Haijun Zhou, Email: zhou1997@gmail.com.
Lunxiu Qin, Email: qin_lx@yahoo.com.
Acknowledgments
This research was supported by China National Key Projects for Infectious Disease (2012ZX10002-012), National Major Scientific Research Project (2013CB910500), the State Key Basic Research Program of China (2009CB521701), the National Natural Science Foundation of China (81272733), and the National Science and Technology Key Project of China (2012CB910602).
References
- Dove A. The bittersweet promise of glycobiology. Nat Biotechnol. 2001;19:913–917. doi: 10.1038/nbt1001-913. [DOI] [PubMed] [Google Scholar]
- Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053:79–88. doi: 10.1016/j.chroma.2004.08.150. [DOI] [PubMed] [Google Scholar]
- Alper J. Glycobiology. Turning sweet on cancer. Science. 2003;301:159–160. doi: 10.1126/science.301.5630.159. [DOI] [PubMed] [Google Scholar]
- Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation–potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol Cell Biol. 2005;83:429–439. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102:285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- An HJ, Kronewitter SR, de Leoz ML, Lebrilla CB. Glycomics and disease markers. Curr Opin Chem Biol. 2009;13:601–607. doi: 10.1016/j.cbpa.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracaula R, Barrabes S, Sarrats A, Rudd PM, de Llorens R. Altered glycosylation in tumours focused to cancer diagnosis. Dis Markers. 2008;25:207–218. doi: 10.1155/2008/797629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M, Quelhas D, Critchley AJ, Carchon H, Hebestreit HF, Hibbert RG, Vilarinho L, Teles E, Matthijs G, Schollen E. et al. Detailed glycan analysis of serum glycoproteins of patients with congenital disorders of glycosylation indicates the specific defective glycan processing step and provides an insight into pathogenesis. Glycobiology. 2003;13:601–622. doi: 10.1093/glycob/cwg079. [DOI] [PubMed] [Google Scholar]
- Kuster B, Mann M. 18O-labeling of N-glycosylation sites to improve the identification of gel-separated glycoproteins using peptide mass mapping and database searching. Anal Chem. 1999;71:1431–1440. doi: 10.1021/ac981012u. [DOI] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- Liu Z, Cao J, He Y, Qiao L, Xu C, Lu H, Yang P. Tandem 18O stable isotope labeling for quantification of N-glycoproteome. J Proteome Res. 2010;9:227–236. doi: 10.1021/pr900528j. [DOI] [PubMed] [Google Scholar]
- Kaji H, Yamauchi Y, Takahashi N, Isobe T. Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site-specific stable isotope tagging. Nat Protoc. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- Kubota K, Sato Y, Suzuki Y, Goto-Inoue N, Toda T, Suzuki M, Hisanaga S, Suzuki A, Endo T. Analysis of glycopeptides using lectin affinity chromatography with MALDI-TOF mass spectrometry. Anal Chem. 2008;80:3693–3698. doi: 10.1021/ac800070d. [DOI] [PubMed] [Google Scholar]
- Li D, Mallory T, Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta. 2001;313:15–19. doi: 10.1016/S0009-8981(01)00644-1. [DOI] [PubMed] [Google Scholar]
- Taketa K, Sekiya C, Namiki M, Akamatsu K, Ohta Y, Endo Y, Kosaka K. Lectin-reactive profiles of alpha-fetoprotein characterizing hepatocellular carcinoma and related conditions. Gastroenterology. 1990;99:508–518. doi: 10.1016/0016-5085(90)91034-4. [DOI] [PubMed] [Google Scholar]
- Sato Y, Nakata K, Kato Y, Shima M, Ishii N, Koji T, Taketa K, Endo Y, Nagataki S. Early recognition of hepatocellular carcinoma based on altered profiles of alpha-fetoprotein. N Engl J Med. 1993;328:1802–1806. doi: 10.1056/NEJM199306243282502. [DOI] [PubMed] [Google Scholar]
- Shiraki K, Takase K, Tameda Y, Hamada M, Kosaka Y, Nakano T. A clinical study of lectin-reactive alpha-fetoprotein as an early indicator of hepatocellular carcinoma in the follow-up of cirrhotic patients. Hepatology. 1995;22:802–807. doi: 10.1002/hep.1840220317. [DOI] [PubMed] [Google Scholar]
- Zhang S, Shu H, Luo K, Kang X, Zhang Y, Lu H, Liu Y. N-linked glycan changes of serum haptoglobin beta chain in liver disease patients. Mol Biosyst. 2011;7:1621–1628. doi: 10.1039/c1mb05020f. [DOI] [PubMed] [Google Scholar]
- Lei Z, Beuerman RW, Chew AP, Koh SK, Cafaro TA, Urrets-Zavalia EA, Urrets-Zavalia JA, Li SF, Serra HM. Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J Proteome Res. 2009;8:1992–2003. doi: 10.1021/pr800962q. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Na K, Choi EY, Kim KS, Kim H, Paik YK. Simple method for quantitative analysis of N-linked glycoproteins in hepatocellular carcinoma specimens. J Proteome Res. 2010;9:308–318. doi: 10.1021/pr900649b. [DOI] [PubMed] [Google Scholar]
- Saravanan C, Cao Z, Head SR, Panjwani N. Analysis of differential expression of glycosyltransferases in healing corneas by glycogene microarrays. Glycobiology. 2010;20:13–23. doi: 10.1093/glycob/cwp133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, Wang L, Zhuang H, Callewaert N, Libert C. et al. N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus. Hepatology. 2007;46:1426–1435. doi: 10.1002/hep.21855. [DOI] [PubMed] [Google Scholar]
- Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Noninvasive diagnosis of liver cirrhosis using DNA sequencer-based total serum protein glycomics. Nat Med. 2004;10:429–434. doi: 10.1038/nm1006. [DOI] [PubMed] [Google Scholar]
- Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, Abdel-Hamid M, Gouda I, Ezzat S, Kyselova Z. et al. Detection of hepatocellular carcinoma using glycomic analysis. Clin Cancer Res. 2009;15:1808–1813. doi: 10.1158/1078-0432.CCR-07-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Varghese RS, Bekesova S, Loffredo CA, Hamid MA, Kyselova Z, Mechref Y, Novotny MV, Goldman R, Ressom HW. Identification of N-glycan serum markers associated with hepatocellular carcinoma from mass spectrometry data. J Proteome Res. 2010;9:104–112. doi: 10.1021/pr900397n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RR, Schwegler EE, Malik G, Diaz J, Block T, Mehta A, Semmes OJ. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- Li C, Zolotarevsky E, Thompson I, Anderson MA, Simeone DM, Casper JM, Mullenix MC, Lubman DM. A multiplexed bead assay for profiling glycosylation patterns on serum protein biomarkers of pancreatic cancer. Electrophoresis. 2011;32:2028–2035. doi: 10.1002/elps.201000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongsachart P, Huang-Liu R, Sinchaikul S, Pan FM, Phutrakul S, Chuang YM, Yu CJ, Chen ST. Glycoproteomic analysis of WGA-bound glycoprotein biomarkers in sera from patients with lung adenocarcinoma. Electrophoresis. 2009;30:1206–1220. doi: 10.1002/elps.200800405. [DOI] [PubMed] [Google Scholar]
- Shetty V, Nickens Z, Shah P, Sinnathamby G, Semmes OJ, Philip R. Investigation of sialylation aberration in N-linked glycopeptides by lectin and tandem labeling (LTL) quantitative proteomics. Anal Chem. 2010;82:9201–9210. doi: 10.1021/ac101486d. [DOI] [PubMed] [Google Scholar]
- Lazar IM, Lazar AC, Cortes DF, Kabulski JL. Recent advances in the MS analysis of glycoproteins: Theoretical considerations. Electrophoresis. 2011;32:3–13. doi: 10.1002/elps.201000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Nakano M, Hincapie M, Kolarich D, Baker MS, Hancock WS, Packer NH. The lectin riddle: glycoproteins fractionated from complex mixtures have similar glycomic profiles. Omics : J Integr Biol. 2010;14:487–499. doi: 10.1089/omi.2010.0075. [DOI] [PubMed] [Google Scholar]
- Dai Z, Zhou J, Qiu SJ, Liu YK, Fan J. Lectin-based glycoproteomics to explore and analyze hepatocellular carcinoma-related glycoprotein markers. Electrophoresis. 2009;30:2957–2966. doi: 10.1002/elps.200900064. [DOI] [PubMed] [Google Scholar]
- Jung K, Cho W, Regnier FE. Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J Proteome Res. 2009;8:643–650. doi: 10.1021/pr8007495. [DOI] [PubMed] [Google Scholar]
- Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52:1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- Capelo JL, Carreira RJ, Fernandes L, Lodeiro C, Santos HM, Simal-Gandara J. Latest developments in sample treatment for 18O-isotopic labeling for proteomics mass spectrometry-based approaches: a critical review. Talanta. 2010;80:1476–1486. doi: 10.1016/j.talanta.2009.04.053. [DOI] [PubMed] [Google Scholar]
- Shakey Q, Bates B, Wu J. An approach to quantifying N-linked glycoproteins by enzyme-catalyzed 18O3-labeling of solid-phase enriched glycopeptides. Anal Chem. 2010;82:7722–7728. doi: 10.1021/ac101564t. [DOI] [PubMed] [Google Scholar]
- Petritis BO, Qian WJ, Camp DG 2nd, Smith RD. A simple procedure for effective quenching of trypsin activity and prevention of 18O-labeling back-exchange. J Proteome Res. 2009;8:2157–2163. doi: 10.1021/pr800971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang L, Palmer Toy D, Hancock WS, Sgroi DC, Karger BL. Proteomic analysis of ductal carcinoma of the breast using laser capture microdissection, LC-MS, and 16O/18O isotopic labeling. J Proteome Res. 2004;3:604–612. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- Mirza SP, Greene AS, Olivier M. 18O labeling over a coffee break: a rapid strategy for quantitative proteomics. J Proteome Res. 2008;7:3042–3048. doi: 10.1021/pr800018g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova D, Rao KC, Miyagi M. pH dependency of the carboxyl oxygen exchange reaction catalyzed by lysyl endopeptidase and trypsin. J Proteome Res. 2006;5:1667–1673. doi: 10.1021/pr060033z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Kojima S, Yanagi K, Kanaoka M. 18O-labeling quantitative proteomics using an ion trap mass spectrometer. Proteomics. 2005;5:16–23. doi: 10.1002/pmic.200300885. [DOI] [PubMed] [Google Scholar]
- Gornik O, Wagner J, Pucic M, Knezevic A, Redzic I, Lauc G. Stability of N-glycan profiles in human plasma. Glycobiology. 2009;19:1547–1553. doi: 10.1093/glycob/cwp134. [DOI] [PubMed] [Google Scholar]
- KnezevićParekh R, Roitt I, Isenberg D, Dwek R, Rademacher T. Age-related galactosylation of the N-linked oligosaccharides of human serum IgG. J Exp Med. 1988;167:1731–1736. doi: 10.1084/jem.167.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic A, Polasek O, Gornik O, Rudan I, Campbell H, Hayward C, Wright A, Kolcic I, O'Donoghue N, Bones J. et al. Variability, heritability and environmental determinants of human plasma N-glycome. J Proteome Res. 2009;8:694–701. doi: 10.1021/pr800737u. [DOI] [PubMed] [Google Scholar]
- Utsunomiya T, Ogawa K, Yoshinaga K, Ohta M, Yamashita K, Mimori K, Inoue H, Ezaki T, Yoshikawa Y, Mori M. Clinicopathologic and prognostic values of apolipoprotein D alterations in hepatocellular carcinoma. Int J Cancer. 2005;116:105–109. doi: 10.1002/ijc.20986. [DOI] [PubMed] [Google Scholar]
- Lau SH, Sham JS, Xie D, Tzang CH, Tang D, Ma N, Hu L, Wang Y, Wen JM, Xiao G. et al. Clusterin plays an important role in hepatocellular carcinoma metastasis. Oncogene. 2006;25:1242–1250. doi: 10.1038/sj.onc.1209141. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- Ruan D, Chen G, Kerre EE, Wets G. Intelligent Data Mining Techniques and Applications. Berlin Heidelberg: Springer-Verlag GmbH; 2005. [Google Scholar]
- Grubbs F. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. doi: 10.1080/00401706.1969.10490657. [DOI] [Google Scholar]
- Barnett V, Lewis T. Outliers in statistical data. 3rd edn. Chichester. New York: Wiley & Sons; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The reaction of three 18O atoms labeling happened in the glycopeptides by catalysis with Trypsin and PNGase F. The reaction is reversible at the 18O labeling of the C-terminal catalyzed with trypsin, indicating that the back-exchange and C-terminal single 18O labeling in the C-terminal cannot be completely avoided in the reaction product. This feature is a problem identified in the experimental operation, result analysis, and quantitative method design, but do not need to consider in the labeling process of PNGase-F catalysis.
The mass shifts identified by mass spectrum between paired 18 O and 16 O labeled glycopeptides/non-glycopeptides from standard glycoprotein Invertase. (A) For the non-glycopeptides VFWYEPSQK, the mass shift of 4 Da was generated in mass spectrometry. (B) For the glycopeptide FATN*TTLTK, the mass shift of 6 Da was generated in mass spectrometry. *denotes the N-glycosylation site.
ASAP and XPRESS ratios of glycopeptides and non-glycopeptides in the dynamic range of 1:10–10:1 derived by Trans-Proteomic Pipeline Ver. 4.5.
The number of differently expressed glycoproteins/glycopeptides between HCC patients and healthy individuals in the three lectin subgroups. The data calculated by self-build quantitative method.
The table of changed glycopeptides in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
The table of changed glycoproteins in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
The table of unchanged glycopeptides in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
The detailed data of detected glycopeptides in HCC patient serum (18O Labeling) compared to health control (16O Labeling).
Calculation of abundance ratios of four glycopeptides between HCC patients and healthy individuals. The ratios calculated manually were similar to the ratios obtained by self-build quantitative method, which indicated the reliability of the quantitative results of our integrated research strategy.
Physiological/pathological characteristics of patients and healthy individuals enrolled in this study.
Our self-build quantitation setting file.
A Modified unimod file. You can download Additional files 9 and 10, copy them into the directory of /mascot/config/, please backup the original files before copying.