Abstract
To identify genes that promote facial skin youthfulness (SY), a genome-wide association study on an Ashkenazi Jewish discovery group (n=428) was performed using Affymetrix 6.0 Single-Nucleotide Polymorphism (SNP) Array. After SNP quality controls, 901,470 SNPs remained for analysis. The eigenstrat method showed no stratification. Cases and controls were identified by global facial skin aging severity including intrinsic and extrinsic parameters. Linear regression adjusted for age and gender, with no significant differences in smoking history, body mass index, menopausal status, or personal or family history of centenarians. Six SNPs met the Bonferroni threshold with Pallele<10−8; two of these six had Pgenotype<10−8. Quantitative trait loci mapping confirmed linkage disequilibrium. The six SNPs were interrogated by MassARRAY in a replication group (n=436) with confirmation of rs6975107, an intronic region of KCND2 (potassium voltage-gated channel, Shal-related family member 2) (Pgenotype=0.023). A second replication group (n=371) confirmed rs318125, downstream of DIAPH2 (diaphanous homolog 2 (Drosophila)) (Pallele=0.010, Pgenotype=0.002) and rs7616661, downstream of EDEM1 (ER degradation enhancer, mannosidase α-like 1) (Pgenotype=0.042). DIAPH2 has been associated with premature ovarian insufficiency, an aging phenotype in humans. EDEM1 associates with lifespan in animal models, although not humans. KCND2 is expressed in human skin, but has not been associated with aging. These genes represent new candidate genes to study the molecular basis of healthy skin aging.
Introduction
Genes promoting overall youthfulness in the form of delayed aging have been reported in the centenarian population (Perls et al., 2002; Barzilai et al., 2003; Atzmon et al., 2009b; Tazearslan et al., 2011), but whether these genes also promote skin youthfulness (SY) is unclear. In fact, genes that promote SY have, to date, not been identified. Multiple sources have estimated that over US$140 billion yearly is spent on cosmetics worldwide (Oh, 2006), testifying to the intense and broad interest in promoting SY. Anecdotally, some centenarians possess facial skin that appears decades younger than their chronologic age. This dissociation between phenotypic and chronologic age may be a unique window to examine whether particular genotypes associate with SY.
Prior skin aging studies have categorized skin aging parameters into intrinsic versus extrinsic characteristics, believed to reflect genetic versus environmental factors contributing to skin aging features, respectively (e.g., Fisher et al., 2002; Vierkötter and Krutmann, 2012). However, data suggest that extrinsic skin aging parameters can have a strong genetic basis. For instance, MC1R gene mutations can promote severe photoaging (Elfakir et al., 2010). Heritability analyses in twins have shown that extrinsic skin aging parameters such “sun damage” exhibit 60% heritability (Gunn et al., 2009). Recently, extrinsic skin aging parameters of facial photoaging have been linked to a genetic change, a single-nucleotide polymorphism (SNP) in the intronic area of the STXBP5L gene, in a genome-wide association study of French women (Le Clerc et al., 2013). Hence, this study aims to examine skin aging in a global sense, and includes both intrinsic and extrinsic skin aging parameters, both of which have genetic underpinnings.
The primary objective of this study was to assess whether there may be genes that confer SY in a unique centenarian population, by identifying individuals that lack global skin aging features (both intrinsic and extrinsic as both have a genetic basis) compared with individuals of similar chronologic age. The secondary objective of this study was to determine whether SY genes overlap with longevity genes in this centenarian population.
Results
Cases and controls for facial SY phenotype were identified using a global skin aging score (SAS) incorporating intrinsic and extrinsic skin aging parameters (Guinot et al., 2002). The average difference between chronologic age and SAS (i.e., CA minus SAS), or average “delta,” for the entire discovery group was 15.5. To avoid bias in assigning SY cases and controls for the genome-wide association study (GWAS), individuals whose “deltas” were greater than the average “delta” were defined as cases (individuals with SY), and those whose “deltas” were less than or equal to the group average were defined as controls (individuals without SY). There were no individuals whose “delta” equaled the group average.
The global SASs were determined by a board-certified dermatologist experienced in assessing skin aging parameters (see Materials and Methods section). Bland–Altman analysis showed the differences in skin aging ratings at two time points (Supplementary Figure S1 online). The rating differences at two time points that fell outside the 95th percentile were distributed across the range of average skin ages, indicating lack of bias in rating for any particular skin age (Supplementary Figure S1 online).
Clinical characteristics of the case and control groups are reported in Table 1. Linear regression adjusted for age, gender, and personal or family history of centenarians. There were no significant differences in smoking history, body mass index, or personal history of skin cancer between cases and controls. All female subjects in the discovery group were postmenopausal.
Table 1. Characteristics of cases (SY group) and controls (no-SY group) in the discovery group (n=428).
| Characteristic | Cases: SY Delta>avg. delta | Controls: no-SY Delta⩽avg. delta | P-value |
|---|---|---|---|
| N | 218 | 210 | |
| Chronologic age, mean in years±SD | 79.3±15.4 | 93.3±10.4 | <0.0001 |
| Delta SY, mean±SD | 24.6±6.6 | 6.7±6.7 | <0.0001 |
| Female (%) | 67% | 70% | 0.43 |
| Body mass index (kg m−2), mean±SD | 24.4±4.03 | 23.6±4.34 | 0.48 |
| % With no family history of centenarians | 65% | 16% | <0.0001 |
| Positive smoking history (%) | 25% | 37% | 0.85 |
| Positive personal history of skin cancer (%) | 13% | 18% | 0.32 |
Abbreviations: GWAS, genome-wide association study; SAS, skin aging score; SY, skin youthfulness.
Cases and controls were identified using a global SAS incorporating intrinsic and extrinsic skin aging parameters. The average difference between CA and SAS (that is, CA minus SAS), or average “delta,” for the entire discovery group was 15.5. To avoid bias in assigning SY cases and SY controls for the GWAS, individuals whose “deltas” were greater than the average “delta” were defined cases (individuals with SY), and those whose “deltas” were less than or equal to the group average were defined as cases (individuals without SY). There were no individuals whose “delta” equaled the group average. P-values are calculated by Fisher's exact test or t-test to assess for significant differences overall between cases and controls. Examples of SY cases and SY controls are shown in Supplementary Figure S8 online.
A conventional genome-wide association study on an Ashkenazi Jewish discovery group (n=428) was performed using Affymetrix 6.0 SNP Array. SNP quality controls included removal of 22,118 SNPs that were not in Hardy–Weinberg equilibrium, with 901,470 SNPs remaining for analysis. Eigenstrat method showed no stratification, with good homogeneity between cases and controls (Supplementary Figure S2 online). Isolation-by-distance studies in this Jewish diaspora were reported previously (Atzmon et al., 2010; Kenny et al., 2012). Identity-by-descent analysis showed that all individuals in the discovery group were unrelated (Supplementary Figure S3 online). Quantile–quantile plots of the P-values are shown in Supplementary Figure S4 online.
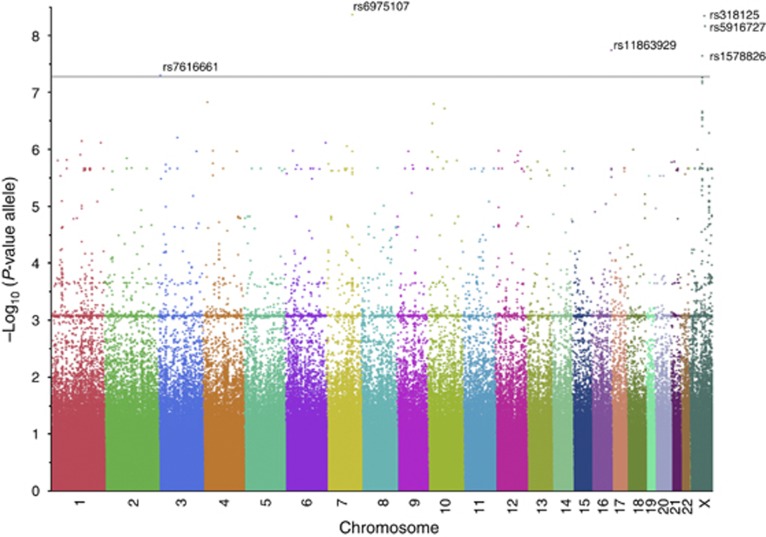
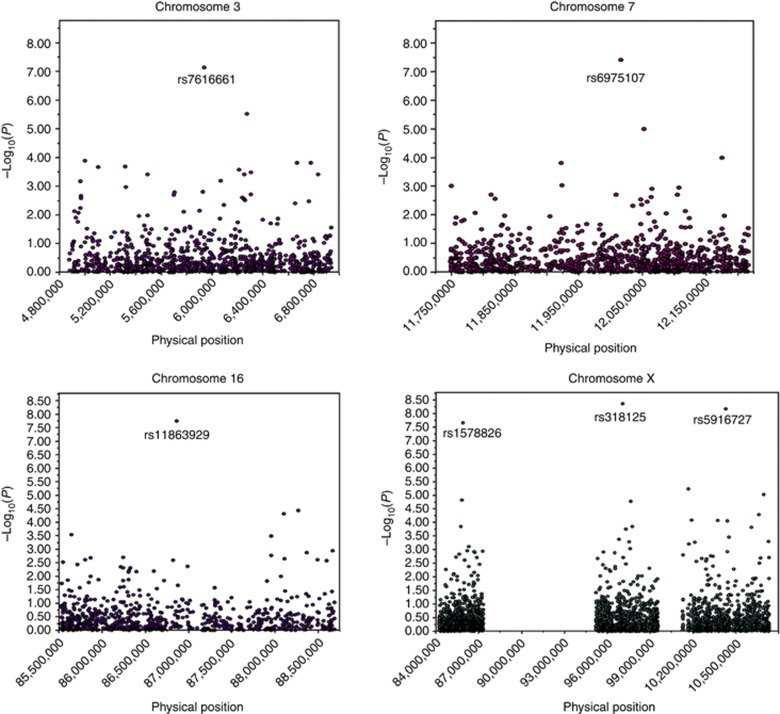
Six SNPs met the Bonferroni threshold with Pallele<10−8; two of these six had Pgenotype<10−8 (Table 2a). A Manhattan plot of the discovery group is shown in Figure 1. Quantitative trait loci mapping confirmed linkage disequilibrium (Figure 2).
Table 2. Significant SNPs identified in the discovery group, and results of the first and second replication groups.
| dbSNP | Chromosome | Physical position | Gene relationship | Gene distance | Gene | BP change | Prob allele | Prob genotype |
|---|---|---|---|---|---|---|---|---|
| (A) Discovery group | ||||||||
| Rs6975107 | 7 | 120,168,143 | Intron | 0 | KCND2 | C−> T | 4.15 × 10−9 | 3.74 × 10−9 |
| Rs318125 | X | 97,090,551 | Downstream | 348,298 | DIAPH2 | C−> T | 4.35 × 10−9 | 1.25 × 10−5 |
| Rs5916727 | X | 104,294,270 | Intron | 0 | IL1RAPL2 | C−> T | 6.66 × 10−9 | 1.34 × 10−5 |
| Rs11863929 | 16 | 86,861,934 | Upstream | 159,445 | ZNF469 | C−> G | 1.77 × 10−8 | 5.48 × 10−7 |
| Rs1578826 | X | 85,889,112 | Intron | 0 | DACH2 | T−> C | 2.20 × 10−8 | 5.46 × 10−6 |
| Rs7616661 | 3 | 5,940,543 | Downstream | 703,894 | EDEM1 | T−> G | 4.82 × 10−8 | 3.37 × 10−8 |
| (B) First replication group | ||||||||
| Rs6975107 | 7 | 120,168,143 | Intron | 0 | KCND2 | C−> T | 0.133 | 0.023 |
| Rs318125 | X | 97,090,551 | Downstream | 348,298 | DIAPH2 | C−> T | 0.332 | 0.332 |
| Rs5916727 | X | 104,294,270 | Intron | 0 | IL1RAPL2 | C−> T | 0.623 | 0.456 |
| Rs11863929 | 16 | 86,861,934 | Upstream | 159,445 | ZNF469 | C−> G | 0.370 | 0.371 |
| Rs1578826 | X | 85,889,112 | Intron | 0 | DACH2 | T−> C | 0.576 | 0.245 |
| Rs7616661 | 3 | 5,940,543 | Downstream | 703,894 | EDEM1 | T−> G | 0.390 | 0.180 |
| (C) Second replication group | ||||||||
| Rs6975107 | 7 | 120,168,143 | Intron | 0 | KCND2 | C−> T | 0.709 | 0.494 |
| Rs318125 | X | 97,090,551 | Downstream | 348,298 | DIAPH2 | C−> T | 0.010 | 0.002 |
| Rs5916727 | X | 104,294,270 | Intron | 0 | IL1RAPL2 | C−> T | 0.686 | 0.347 |
| Rs11863929 | 16 | 86,861,934 | Upstream | 159,445 | ZNF469 | C−> G | 0.344 | 0.604 |
| Rs1578826 | X | 85,889,112 | Intron | 0 | DACH2 | T−> C | 0.341 | 0.117 |
| Rs7616661 | 3 | 5,940,543 | Downstream | 703,894 | EDEM1 | T−> G | 0.216 | 0.042 |
Abbreviations: dbSNP, single-nucleotide polymorphism database; GWAS, genome-wide association study.
(A) Results of GWAS on discovery group showing SNPs with a significance level of P<10−8, in bold. (B) Results of targeted interrogation of candidate SNPs in first replication group. Significance level is P<0.05, in bold. (C) Results of targeted interrogation of candidate SNPs in a second replication group. Significance level is P<0.05, in bold. Prob allele denotes P-value of allele (Pallele) frequency differences between case and control groups; Prob genotype denotes P-value of genotype (Pgenotype) frequency differences between case and control groups. NA indicates that the SNP was monomorphic (i.e., only one genotype).
Figure 1.
Manhattan plot shows that single-nucleotide polymorphisms (SNPs) are distributed across all chromosomes in the discovery group with significant SNPs labeled. Results are plotted as negative log10-transformed P-values from a genotypic association test using allele mode. Significant SNPs are shown in Table 2.
Figure 2.
Quantitative trait loci analysis for candidate skin youthfulness (SY) genes identified in discovery group. y-Axis shows negative transformed log10 of significant Palleles.
To explore the effects of the asymmetry in percent of centenarians between SY cases and SY controls in the discovery group, we corrected for this and report the effect on the Palleles for each of the six SNPs Supplementary Table S1 online. These P-values remain significant when the asymmetry is corrected for.
A gene network to assess coexpression of the six significant SNPs identified in the discovery group is shown in Supplementary Figure S5 online. The six SNPs form four clusters of coexpression but no physical interaction.
To confirm these findings of the discovery group, the six candidate SNPs were interrogated by MassARRAY in a replication group (n=436). Clinical characteristics of the case and control groups of the replication group are reported in Supplementary Table S2a online. After covariate correction for age and gender, the significance of rs6975107 was confirmed. This SNP is located in the intronic region of KCND2 (potassium voltage-gated channel, Shal-related family member 2) (Pgenotype=0.023).
The six candidate SNPs were interrogated by MassARRAY in a second replication group (n=371). Clinical characteristics of the case and control groups of the second replication group are reported in Supplementary Table S2B online. After covariate correction for age and gender, the significance of rs318125 and rs7616661 were confirmed (Tables 2b and c). The SNP rs318125 is downstream of DIAPH2 (diaphanous homolog 2 (Drosophila)) (Pallele=0.010, Pgenotype=0.002). The SNP rs7616661 is downstream of EDEM1 (ER degradation enhancer, mannosidase α-like 1) (Pgenotype=0.042).
An alternative method to analyze the X-chromosome SNPs identified from the discovery group would be to assume a dominant mode instead of correcting for gender. Using the dominant mode, only one out of three of the X-linked SNPs, rs5916727, was significant (Pdominant=4.70 × 10−8) in the discovery group. None of the three X-linked SNPs were confirmed in the two replication groups using this dominant mode analysis.
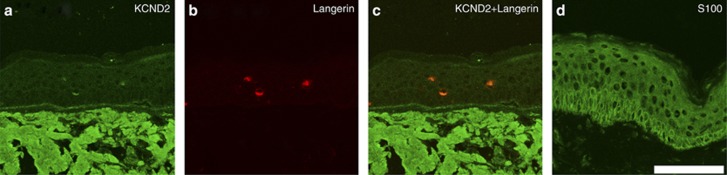
As it is not known whether KCND2 is expressed in human skin, we performed immunohistochemistry for KCND2 in older adult human facial skin. Double immunofluorescence staining with anti-KCND2 and an anti-Langerin antibody confirmed that KCND2 is located on Langerhans cells (LCs) in human facial epidermis (Figure 3). DIAPH2 has been associated with human premature ovarian (Bione et al., 1998; Marozzi et al., 2000a, 2000b), although it is not reported to be significantly expressed in human skin. EDEM1 is expressed in human skin and is associated with endoplasmic reticulum–associated degradation of glycoproteins (Shenkman et al., 2013).
Figure 3.
KCND2 stains Langerhans cells (LCs) in facial skin of an older adult individual. (a) Confocal microscopy of skin immunostained with anti-KCND2 antibody (green) shows positivity in three cells in the epidermis (original magnification, × 63). (b) Same section of skin with positive staining of the same three cells in the epidermis using anti-Langerin antibody (red). (c) Red and green fluorescence in the same three cells in epidermis, confirming KCND2 stains LCs. (d) S100-positive control. Bar=100 μm.
In case X-chromosome–associated SNPs could be missed using the genotypic mode for association analysis, the P-values for genotypic mode when females alone are included in the groups are reported in Supplementary Table S3 online. There were no new significant signals from the X-chromosome SNPs in genotypic mode for either the first or second replication group. However, rs5916727 located in an intronic region of IL1RAPL2 on the X chromosome (Pallele=0.017) and rs1578826 located in an intronic region of DACH2 on the X chromosome (Pallele=0.003) did become significant in the second replication group when females alone were included. IL1RAPL2 has been associated with autism (Piton et al., 2008), a phenotype located in the head region. Gene expression of DACH2 in humans is unknown, but does appear in the dermatomyotome of embryonic mice head region (Supplementary Figure S6 online).
Discussion
This study supports the hypothesis that facial SY may be associated with specific genes. In particular, we identify and replicate three genes that may promote human facial SY in an Ashkenazi Jewish population: KCND2, DIAPH1, and EDEM1. Although functional studies may directly confirm the biologic relevance of these three genes in facial SY, the informed consent process in this study did not include the provision for skin biopsies.
Nevertheless, KCND2, a voltage-gated potassium channel expressed in LCs, suggests an intriguing potential link of SY to skin immune response. Physiologically, the number of LCs in human skin is known to decrease with age (Thiers et al., 1984). Functionally, voltage-gated potassium channels can modulate phagocytic capacity of LCs in mice (Matzner et al., 2008) Recently, dendritic cells have been linked to inflammation and age-related diseases (Cavanagh et al., 2012; Jonsson et al., 2013). A direct link between KCND2 gene regulation, altered LC function, and subsequent potential effect on skin phenotype remains to be determined.
The possible role of DIAPH2 in SY is plausible, as defects in DIAPH2 underlie premature ovarian insufficiency in humans (Bione et al., 1998; Marozzi et al., 2000a). The association of menopause and skin aging has been widely reported (Pontius and Smith, 2011; Emmerson et al., 2012), although the precise mechanism by which the SNP downstream of DIAPH2 identified in this study exerts its effect on the SY phenotype is unclear.
EDEM1 is an intriguing and logical candidate SY gene. EDEM1 associates with lifespan in Drosophila melanogaster and Caenorhabditis elegans (Liu et al., 2006). EDEM1 levels are reduced in fibroblasts taken from a dwarf mouse model of longevity, and these fibroblasts are more resistant to cell death from stressors such as UV light (Akha et al., 2011). Future studies on fibroblasts from individuals with and without SY may determine whether EDEM1 levels are different, and whether this difference confers resistance to cell death from UV light in humans. One could speculate that if the SY phenotype was due to resistance to cell death in fibroblasts, then SY individuals might have more collagen fibers in the dermis than those without SY.
In this initial study, we aimed to broadly control for UV exposures by only including individuals residing in the same location, the New York metropolitan area. Individuals who were immigrants to New York were all from Poland or Ukraine with similar average UV index as New York metropolitan area. Current studies are ongoing in which detailed UV exposure histories are being collected to more precisely account for this covariate. However, extrinsic factors such as UV exposures have been shown to account for only 10% of the discriminant ability of the skin aging scoring method that we utilized in this study (Guinot et al., 2002). Hence, the effect of UV exposures on the phenotypes in this GWAS is not likely to be large.
As this study included a large number of centenarians, it was unclear whether the SY genes would overlap with longevity genes, such as IGF1 (Barzilai et al., 2003; Atzmon et al., 2009a, 2009b; Tazearslan et al., 2011; Conneely et al., 2012). The candidate SY genes in this study do not clearly overlap with known longevity genes in humans. However, EDEM1 levels appear to be lower in a dwarf mouse model for longevity that also possesses lower levels of IGF1, suggesting a possible connection between IGF1 and EDEM1 that merits further study (Akha et al., 2011). Interestingly, a recent GWAS on facial photoaging suggested an association with an SNP in linkage disequilibrium with FBXO40, which can modulate the IGF1 pathway (Le Clerc et al., 2013), a regulator of human longevity and lifespan in animal models (van Heemst et al., 2005). It is possible that with larger sample size, we might be able to detect additional candidate SY genes, especially if more than one SNP contributes to the SY phenotype; SNPs that interact to lead to SY phenotype might not individually reach genome-wide significance with our current sample size.
Finally, studies are underway to examine whether genes that promote SY also protect against age-associated diseases in other organ systems such as diabetes, cancer, and neurologic disease, which can impact human longevity.
Materials and Methods
Study design and population
This study was performed in accordance with the Declaration of Helsinki Principles, including Institutional Review Board approval at both Albert Einstein College of Medicine and Stanford University School of Medicine. After written informed consent, women and men of Ashkenazi Jewish descent (defined as having four Ashkenazi Jewish grandparents) living in the New York metropolitan area who were are part of the LonGenity Project at Albert Einstein College of Medicine were included in the discovery group for this GWAS. The LonGenity database has been useful in identifying gene variants promoting longevity (Barzilai et al., 2003; Atzmon et al., 2009a, 2009b; Tazearslan et al., 2011). Individuals who had immigrated to New York were from Poland or Ukraine with an average UV index from March to September comparable to New York metropolitan area (5.74 vs. 6.34, respectively). Individuals with self-reported history of facial cosmetic procedures or use of topical antiaging medication such as tretinoin at time photographs were taken were excluded from the study. A first replication group was assembled after the discovery group. Several years after, a second replication group was accrued. The study flow is depicted in Supplementary Figure S7 online.
Statistical adjustment for covariates
Mean ages, gender distribution, centenarian status, mean body mass index, personal history of skin cancer, smoking history, and menopausal status were assessed in the discovery group. Significant differences between SY cases and SY controls were adjusted for by multivariate regression in the GWAS. These covariates were also assessed and adjusted if significantly different in the two replication groups.
Facial skin aging assessments
Frontal facial photographs were rated for skin aging features by a board-certified dermatologist experienced in assessing skin aging and who was blinded to the chronological ages of the study participants. Whenever possible, the same blue background and 12 megapixel digital camera was used to obtain photographs.
A global SAS to include both intrinsic and extrinsic skin aging parameters was adapted from Guinot et al. (2002), and applied to all participants in this study (Supplementary Table S4 online). To ensure reproducibility, this scale was tested using 300 randomly selected photographs from the study on the dermatologist who eventually rated all the photographs in this study. The same set of photographs was shown to the dermatologist rater after a 2-week interval. Intrarater reproducibility and potential bias across a range of SASs was assessed by Bland–Altman analysis (Supplementary Figure S1 online).
To assess whether a GWAS restricted to an intrinsic aging parameter might be informative, we analyzed the correlation between an intrinsic aging feature (fine wrinkling) and an extrinsic aging parameter (coarse wrinkling). Using a four-point severity scale, fine wrinkles and coarse wrinkles were rated on 300 individuals of various chronologic ages in this study by the dermatologist rater who rated all the participants in this study. The Spearman's rank-order correlation test showed a significant correlation between the two scores at 0.43, P<0.0001, 95% confidence interval: 0.33–0.52. Hence, patients with higher fine wrinkle scores also had higher coarse wrinkle scores, and a GWAS with fine wrinkling alone was not performed.
Definition of SY cases and SY controls were as described in Results section. Examples of individuals with and without SY are shown in Supplementary Figure S8 online.
Methods of genotyping and quality control
Genotypes for the discovery group were generated from peripheral blood DNA processed with Affymetrix 6.0 SNP Array (Affymetrix, Santa Clara, CA) to interrogate 906,000 SNPs. SNPs with more than 10 missing calls were removed from the analysis. SNPs that were not in Hardy–Weinberg equilibrium (using PLINK software) were also removed from the analysis. In total, 901,470 SNPs remained for analysis. GWAS was performed using JMP Genomics and SAS software (Cary, NC). Genome-wide statistical significance utilized the Bonferroni threshold at P<10−8. P-values were plotted as a quantile–quantile plot (Supplementary Figure S4 online). Manhattan plot was generated to examine the distribution of significant SNPs across all chromosomes (Figure 1). Quantitative trait locus analysis was performed to confirm linkage disequilibrium (Figure 2).
Targeted interrogation of candidate SNPs in the replication groups was performed using MassARRAY system (Sequenom, San Diego, CA). After adjustment for covariates as described above, χ2 analysis was performed on the candidate SNPs between cases and controls using a significance threshold P<0.05.
Assessment for population stratification
Principal component analysis plots of all the participants in the discovery group were used to assess for population stratification by Eigensoft package (version 2.0). (Supplementary Figure S2 online).
Additional quality controls
Isolation-by-distance studies have been previously reported for the subjects in this study (Atzmon et al., 2010; Kenny et al., 2012). The impact of isolation by distance does not apply to this study population as they are an Ashkenazi Jewish diaspora in the New York metropolitan area. This population has been shown to be genetically distinct but still more similar to other Jews such as European Jews and Sephardic Jews than non-Jews (Kenny et al., 2012).
Identity-by-descent analysis among all possible pairs of individuals in the discovery group was performed (Supplementary Figure S4 online). Values below 0.125 (second-degree cousins) were considered unrelated. All individuals in the discovery group were unrelated.
Gene associations, gene expression, and gene networks
The UCSC genome browser (http://genome.ucsc.edu), dbSNP (single-nucleotide polymorphism database) (http://www.ncbi.nlm.nih.gov/projects/SNP), and GeneCards version 3 were used to search the candidate SNPs for gene associations and gene expression in human skin. Gene Paint (http://www.genepaint.org) was used to search gene expression patterns in mice and humans. GeneMANIA (http://geneMANIA.org) was used to search for gene interactions in the most significant SNPs.
Immunohistochemistry
To assess the gene expression of KCND2 in human skin, immunohistochemistry was performed using 1:50 dilution of both rabbit anti-KCND2 (Sigma; HPA029068; St Louis, MO) and mouse anti-Langerin (Dendritics; clone 306G9; Waltham, MA) antibodies on methanol-fixed-frozen facial skin of an older individual. S100 (Dako; Z0311; Carpenteria, CA)-positive control staining was performed at 1:400 dilution. The tissue was visualized with confocal microscopy acquired on a Leica SP2 AOBS Laser Scanning Microscope with an HCX PL APO × 63 oil immersion objective. Images were arranged with ImageJ (Solms, Germany), Adobe Photoshop (San Jose, CA), and Adobe Illustrator (San Jose, CA).
In situ hybridization
Template for DACH2 mRNA for in situ hybridization was amplified from embryonic mouse cDNA by PCR using sequence-specific primers that included the promoter sites for T3 or T7 RNA polymerase. DACH2 antisense riboprobe was transcribed with T7 RNA polymerase in the presence of Dig-11-UTP (Roche, Indianapolis, IN). Section in situ hybridizations were performed by incubating tissue sections in hybridization buffer (Ambion Corporation, Austin, TX) at 70 °C for 12 hours, followed by the addition of riboprobe (approximate concentration of 0.2–0.3 μg ml probe per kilobase of probe complexity). Nonspecifically bound probe was removed with high stringency washes (0.1°—SSC, 65 °C). For color detection, embryos or slides were blocked with 10% sheep serum, 1% Boehringer–Mannheim Blocking Reagent (Roche) and levamisole, and developed using nitro blue tetrazolium chloride (Roche) and 5-bromo-4-chloro-3-indolyl phosphate (Roche). After developing, the slides were coverslipped with aqueous mounting medium (Supplementary Figure S6 online).
Acknowledgments
We would like to acknowledge Zakia Rahman, MD, Brent Kirkland, MD, and Katherine Arefiev, MD for assistance confirming reproducibility of the global facial skin aging assessment scale used in this study. We are grateful to Lisa Zaba, MD PhD, for expertise in skin immunology and assistance with analyzing skin histology. We thank Wanda Guzman-Delgado, William Greiner, RN, Janet Schein, and Omar Amir, MS, for assistance with data collection and management. We are indebted to Bharathi Lingala, PhD, for statistical consultation and analysis. We also thank Olena Mykhaylichenko for administrative support. This study was funded in part by a Dermatology Foundation Career Development Award (ASC), and by grants from the National Institutes of Health (P01AG021654) (NB), The Nathan Shock Center of Excellence for the Biology of Aging (P30AG038072) (NB), the Glenn Center for the Biology of Human Aging (Paul Glenn Foundation Grant) (NB), and Diabetes Center Grant (DK-20541) (NB). HYC was supported by the Ellison Medical Foundation, Glenn Foundation, and the Howard Hughes Medical Institute Early Career Scientist Award.
AUTHOR CONTRIBUTIONS
ASC and GA are co-first authors; ASC, GA, HYC, AB, and NB contributed to the design of the study; ASC and GA contributed the majority of the data in this manuscript; SB contributed data; SA provided technical support; GA was responsible for biostatistical analysis; ASC, GA, HYC, AB, and NB helped to interpret results; and ASC drafted the manuscript. All authors contributed to the final paper with ASC, GA, HYC, AB, and NB playing key roles.
Glossary
- GWAS
genome-wide association study
- LC
Langerhans cell
- SAS
skin aging score
- SNP
single-nucleotide polymorphism
- SY
skin youthfulness
The authors state no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Akha AAS, Harper JM, Salmon AB, et al. Heightened induction of proapoptotic signals in response to endoplasmic reticulum stress in primary fibroblasts from a mouse model of longevity. J Biol Chem. 2011;286:30344–30351. doi: 10.1074/jbc.M111.220541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Barzilai N, Hollowell JG, et al. Extreme longevity is associated with increased serum thyrotropin. J Clin Endocr Metab. 2009;94:1251–1254. doi: 10.1210/jc.2008-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Barzilai N, Surks MI, et al. Genetic predisposition to elevated serum thyrotropin is associated with exceptional longevity. J Clin Endocr Metab. 2009;94:4768–4775. doi: 10.1210/jc.2009-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Hao L, Pe'er I, et al. Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet. 2010;86:850–859. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Schechter C, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- Bione S, Sala C, Manzini C, et al. A human homologue of the Drosophila melanogaster diaphanous gene is disrupted in a patient with premature ovarian failure: evidence for conserved function in oogenesis and implications for human sterility. Am J Hum Genet. 1998;62:533–541. doi: 10.1086/301761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh MM, Weyand CM, Goronzy JJ. Chronic inflammation and aging: DNA damage tips the balance. Curr Opin Immunol. 2012;24:488–493. doi: 10.1016/j.coi.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely KN, Capell BC, Erdos MR, et al. Human longevity and common variations in the LMNA gene: a meta-analysis. Aging Cell. 2012;11:475–481. doi: 10.1111/j.1474-9726.2012.00808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakir A, Ezzedine K, Latreille J, et al. Functional MC1R-gene variants are associated with increased risk for severe photoaging of facial skin. J Invest Dermatol. 2010;130:1107–1115. doi: 10.1038/jid.2009.366. [DOI] [PubMed] [Google Scholar]
- Emmerson E, Campbell L, Davies FC, et al. Insulin-like growth factor-1 promotes wound healing in estrogen-deprived mice: new insights into cutaneous IGF-1R/ERalpha cross talk. J Invest Dermatol. 2012;132:2838–2848. doi: 10.1038/jid.2012.228. [DOI] [PubMed] [Google Scholar]
- Fisher GJ, Kang S, Varani J, et al. Mechanisms of photoaging and chronological skin aging. Arch Dermatol. 2002;138:1462–1470. doi: 10.1001/archderm.138.11.1462. [DOI] [PubMed] [Google Scholar]
- Guinot C, Malvy DJ, Ambroisine L, et al. Relative contribution of intrinsic vs extrinsic factors to skin aging as determined by a validated skin age score. Arch Dermatol. 2002;138:1454–1460. doi: 10.1001/archderm.138.11.1454. [DOI] [PubMed] [Google Scholar]
- Gunn DA, Rexbye H, Griffiths CE, et al. Why some women look young for their age. PLoS One. 2009;4:e8021. doi: 10.1371/journal.pone.0008021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EE, Pe'er I, Karban A, et al. A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci. PLoS Genet. 2012;8:e1002559. doi: 10.1371/journal.pgen.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clerc S, Taing L, Ezzedine K, et al. A genome-wide association study in Caucasian women points out a putative role of the STXBP5L gene in facial photoaging. J Invest Dermatol. 2013;133:929–935. doi: 10.1038/jid.2012.458. [DOI] [PubMed] [Google Scholar]
- Liu YL, Lu WC, Brummel TJ, et al. Reduced expression of alpha-1, 2-mannosidase I extends lifespan in Drosophila melanogaster and Caenorhabditis elegans. Aging Cell. 2006;8:370–379. doi: 10.1111/j.1474-9726.2009.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozzi A, Manfredini E, Tibiletti MG, et al. Molecular definition of Xq common-deleted region in patients affected by premature ovarian failure. Hum Genet. 2000;107:304–311. doi: 10.1007/s004390000364. [DOI] [PubMed] [Google Scholar]
- Marozzi A, Vegetti W, Manfredini E, et al. Association between idiopathic premature ovarian failure and fragile X premutation. Hum Reprod. 2000;15:197–202. doi: 10.1093/humrep/15.1.197. [DOI] [PubMed] [Google Scholar]
- Matzner N, Zemtsova IM, Nguyen TX, et al. Ion channels modulating mouse dendritic cell functions. J Immunol. 2008;181:6803–6809. doi: 10.4049/jimmunol.181.10.6803. [DOI] [PubMed] [Google Scholar]
- Oh CHRAM. Regional sales of multinationals in the world cosmetics industry. Eurn Manage J. 2006;24:163–173. [Google Scholar]
- Perls T, Kunkel LM, Puca AA. The genetics of exceptional human longevity. J Mol Neurosci. 2002;19:233–238. doi: 10.1007/s12031-002-0039-x. [DOI] [PubMed] [Google Scholar]
- Piton A, Michaud JL, Peng H, et al. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum Mol Genet. 2008;17:3965–3974. doi: 10.1093/hmg/ddn300. [DOI] [PubMed] [Google Scholar]
- Pontius AT, Smith PW. An antiaging and regenerative medicine approach to optimal skin health. Facial Plastic Surg. 2011;27:29–34. doi: 10.1055/s-0030-1270416. [DOI] [PubMed] [Google Scholar]
- Purcell S, Noale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population based linkage analysis. Am J Human Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkman M, Groisman B, Ron E, et al. A shared endoplasmic reticulum-associated degradation pathway involving the EDEM1 protein for glycosylated and nonglycosylated proteins. J Biol Chem. 2013;288:2167–2178. doi: 10.1074/jbc.M112.438275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C, Huang J, Barzilai N, et al. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell. 2011;10:551–554. doi: 10.1111/j.1474-9726.2011.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiers BH, Maize JC, Spicer SS, et al. The effect of aging and chronic sun exposure on human Langerhans cell populations. J Invest Dermatol. 1984;82:223–226. doi: 10.1111/1523-1747.ep12260055. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, et al. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Vierkötter A, Krutmann J. Environmental influences on skin aging and ethnic-specific manifestations. Dermatoendocrinology. 2012;4:227–231. doi: 10.4161/derm.19858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.