Abstract
Exposure to pathogens in the periphery elicits effector T cell differentiation in local lymph nodes followed by migration of activated T cells to and within the infected site. However, the relationships among pathogen abundance, antigen display on MHC molecules, effector T cell dynamics, and functional responses at the infected sites are incompletely characterized. Here we compared CD4+ T cell effector dynamics and responses during pulmonary mycobacterial infection vs. acute influenza infection. Two-photon imaging together with in situ as well as ex vivo analysis of cytokine production revealed that the proportion of migration-arrested, cytokine-producing effector T cells was dramatically higher in the influenza-infected lungs due to substantial differences in antigen abundance in the two infectious states. Despite the marked inflammatory conditions associated with influenza infection, histocytometric analysis showed that cytokine production was focal, with a restriction to areas of significant antigen burden. Optimal effector function is thus constrained by the availability of TCR ligands, pointing to the value of increasing antigen stimulation rather than effector numbers in harnessing CD4+ T cells for therapeutic purposes in such conditions.
Introduction
Cellular adaptive immunity is initiated in secondary lymphoid organs, where naïve recirculating T cells encounter presenting cells (APC) bearing cognate antigen. These interactions lead to T cell receptor engagement, T cell activation, proliferation, and acquisition of an effector phenotype. The stimulated T cells are then poised to exit secondary lymphoid organs, migrate to inflamed/infected sites, and carry out their effector functions, which in the case of infectious agents, are aimed at eliminating the pathogen. Although lymphocyte dynamic behavior during the early stages of T cell activation within lymph nodes has been well-described (1-4), there are only limited quantitative data on the spatiotemporal aspects of T cell function in peripheral sites. Most but not all studies of effector T cell dynamics in tissues have found that these cells exhibit reduced migration and/or arrest upon recognizing their cognate ligand (pMHC) presented by tissue APCs (5-14). Unfortunately, only a few reports link the assessment of cell motility to antigen-induced activation and local effector responses such as cytokine production by the T cells at the infectious site (5, 14), events that are central to host defense. Indeed, the most commonly used method to measure effector responses is assessment of cytokine production following restimulation of isolated effector T cells ex vivo with antigen or chemical stimuli, an approach that prevents developing an understanding of the extent to which these same T cells are activated to a functional level in vivo, or where in the tissue such function occurs. In considering how adaptive immunity can be harnessed for more effective eradication of infection, especially in organs such as the lungs where the pathogen-induced disturbance of physiological homeostasis can have severe consequences, it is critical to better understand how effector function is delivered, how pathogens evade such responses, and what approaches might be used to improve anti-microbial activity.
We have previously shown that only a very small fraction of antigen-specific CD4+ effector T cells within hepatic granulomas of mice infected intravenously with Mycobacterium tuberculosis (Mtb) or Bacillus Calmette-Guerin (BCG) actively produced IFNγ or TNFα within the infected liver at a given time. Likewise, only a correspondingly small proportion of the antigen-specific T cells showed migration arrest (14). However, arrest of nearly all antigen-specific effector CD4+ T cells within granulomas could be seen when a substantial amount of mycobacteria-derived antigenic peptide was introduced systemically into the infected animal and this in turn was accompanied by a parallel increase in the frequency of cytokine-producing effector CD4+ T cells and the magnitude of per cell cytokine synthesis. This implies there is no intrinsic effector CD4+ T cell deficiency or insurmountable suppressive activity in this infectious setting, but rather that antigen presentation in mycobacterial lesions is limiting (14). Bold et al. used this method of providing extra synthetic specific antigen to examine the potential therapeutic benefits of increased antigen presentation and subsequent increased cytokine production by effector CD4+ T cells in Mtb-infected mice, documenting greater CD4+ T cell effector function and reduced bacterial burden with such treatment (15). Thus, for mycobacterial infections, low levels of antigen presentation constrain effector activity and providing additional antigen at the infection site can be used as a strategy for treatment in experimental animal settings.
There are many reasons to wonder whether this striking limitation in antigen-dependent tissue activation of anti-pathogen effector T cells is generally the case or characteristic of only a subset of infections or specific tissue sites. Aerosol mycobacterial infection leads to a protracted immune response culminating in the formation of lung granulomas, which are agglomerations of macrophages and other immune cells including effector lymphocytes. The formation of granulomas is dependent on MHCII and IFNγ, which is mainly produced by effector CD4+ T cells (16, 17). Mycobacteria-derived peptides are presented on MHCII molecules and these peptide-MHCII complexes can subsequently activate CD4+ T cells (16). The inflammatory cytokines IFNγ and TNFα produced by antigen-specific CD4+ T cells then augment the anti-microbial activity of infected macrophages (16, 18-20). It is therefore evident why mycobacteria have developed mechanisms to modulate MHCII presentation to limit such effector CD4+ T cell responses (21, 22). In addition, mycobacteria are slowly growing organisms, which might itself result in relatively low levels of Ag presentation. For these reasons, it is important to understand if the limited CD4+ T cell activation at the effector sites is a phenomenon restricted to liver mycobacterial granulomas, or whether it applies to other tissues and pathogens.
To address this issue, we have examined effector CD4+ T cell dynamic behavior and cytokine responses in mycobacteria-infected lung, which is more physiologically relevant than the liver in the case of Mtb, and compared these results to those seen for antigen-specific effector CD4+ T cells in the lungs of influenza-infected animals. Our experiments show that effector CD4+ T cells specific for an influenza-encoded antigen and transferred into an influenza-infected animal exhibit a significantly greater reduction in migration as compared to BCG-specific effector CD4+ T cells transferred into a BCG-infected host. These differences in cell dynamics were also reflected in quantitative measurements of effector cytokine production, with a higher proportion of antigen-specific CD4+ T cells recovered from influenza-infected lungs actively producing IFNγ as compared to antigen-specific CD4+ T cells recovered from BCG-infected lungs. Peptide antigen administration could markedly increase the very small fraction of cytokine-producing cells in BCG-infected lungs, indicating that this population has a large, under-utilized capacity for effector molecule production. Interestingly, despite the highly inflammatory environment, cytokine production in response to influenza infection was largely focal and restricted to areas of substantial antigen burden. Taken as a whole, this study reveals markedly different efficiencies of effector CD4+ T cell activation in the infected lung in the presence of these two divergent pathogens, driven by differences in the extent of antigen presentation. Our findings have implications for how one might manipulate the adaptive immune response to better deal with mycobacteria and for understanding the contribution of effector responses to immunopathology when antigen reaches high levels and leads to high rate contemporaneous cytokine production by a large population of recruited effector T cells.
Materials and Methods
Mice
C57BL/6 mice were from Jackson Laboratories. C57BL/6 OTII TCR transgenic (23) RAG1 deficient (24) mice and C57BL/6 OTII TCR transgenic mice were from Taconic Laboratories. C57BL/6 P25 TCR transgenic RAG1 deficient mice (25) were kindly provided by J. Ernst (New York University School of Medicine, New York). These animals were also crossed to C57BL/6 mice expressing EGFP under the control of the human ubiquitin promoter (UbEGFP) (26), from Jackson Laboratories. C57BL/6 LysM-EGFP knock-in animals (27) and C57BL/6 CD11c-EYFP (28) animals were obtained through the NIAID/Taconic exchange program. All animals were maintained in specific pathogen-free facilities that are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All animal procedures were approved by and carried out in accordance with NIAID Animal Care and Use Committee guidelines under approved animal study protocols.
Pathogens
PR8OVA323 (variant of influenza genetically engineered to express the MHCII restricted peptide from ovalbumin (OVA323-339) presented in H-2b mice) virus stock was kindly provided by Drs. P. Thomas and P. Doherty, St. Jude's Children's Hospital (29). To grow the PR8OVA323 virus stock, Madin-Darby Canine Kidney (MDCK) cells (ATCC) were cultured to confluence in Dulbecco's Modified Eagle Medium (DMEM, Gibco) supplemented with 10% non heat-inactivated fetal calf serum (FCS, Atlanta Biologicals), L-Glutamine, Penicillin-Streptomycin (Lonza), and non-essential amino acids (Gibco). Once confluent, virus stock was added to the MDCK cell layer at 1:100 dilution, in DMEM with 7.5% IgG-free bovine serum albumin (BSA, Sigma-Aldrich), further supplemented with 2.5μg/ml of recombinant bovine trypsin (TrypZean, Sigma-Aldrich). After about 48hrs, when about 75% of the cells displayed cytopathic changes, the supernatant was harvested, aliquoted, and stored at −80° Celsius until used. Mice were inoculated with 50μl of this supernatant intranasally after a brief period of anesthesia, achieved with inhalational isoflurane. Wild type BCG strain Pasteur was kindly provided by Dr. Alan Sher, NIAID, NIH. 5×103 (low dose) or 5×105 (high dose) colony forming units were used for intranasal infection of the animals.
In vitro T cell stimulation and adoptive transfer
To generate in vitro effectors, lymph nodes and spleens of donor TCR transgenic mice (OTII or P25) were harvested and homogenized through a 70μm cell strainer. Following red blood cell lysis, cells underwent CD4 negative T cell selection using magnetic beads (Miltenyi Biotec). Three to 4×106 cells were plated with 1×107 mitomycin C-treated (Sigma Aldrich) C57BL/6 splenocytes, in RPMI 1640 (Lonza) supplemented with 10% non heat-inactivated FCS (Atlanta Biologicals), L-Glutamine, Penicillin-Streptomycin (Lonza), non-essential amino acids (Gibco), and 50μM 2-mercaptoethanol in each well of a 24-well plate. The splenocytes were pulsed with 2μM OVA323-339 peptide (OTII) or 1.26μM Ag85b 240-254 peptide (P25). The cells were cultured in the presence of 10ng/ml IFNγ (eBioscience), 10μg/ml anti-mouse IL-4 (eBioscience), and 10ng/ml recombinant human IL-12 (R&D). On day 2, 10ng/ml recombinant human IL-2 (R&D) was also added to the culture medium. On day 6, cells were harvested and for FACS experiments 2-4×106 cells were transferred into recipient mice, whereas for imaging experiments, about 6-8×106 T cells were transferred. Transferred populations were either congenically marked, genetically labeled with GFP, or labeled with intracellular dyes (CMTMR, CMFDA, TAMRA, or CMF 2HR, all from Invitrogen) for further identification by flow cytometry or imaging.
Cell isolation
For isolation of lymphocytes from lung tissue, mice were sacrificed, and the lungs were perfused via the right ventricle. Following isolation of the tissue, each lobe was perfused further with a digestion mixture, containing Liberase TL (Roche) at a concentration of 1.66 Wünsch units/mL and DNAseI (Roche) at 10μg/mL. The tissue was then disrupted and processed further and single cell suspensions obtained. For in vitro stimulation, cells were plated at about 1-2×106 cells in the presence of leukocyte stimulation cocktail (BD) according to protocol and incubated for 4 hours. All steps were performed in the presence of Brefeldin A (Sigma) at a final concentration of 200μg/mL. For in vivo antigen delivery experiments in BCG-infected mice, animals received 200μg of Ag85b intravenously and 200μg intranasally. In experiments involving influenza-infected mice, animals received 200μg of OVA323 intravenously per mouse.
Flow Cytometry
The following antibodies were used for flow cytometry: CD3e (145-2C11; eBioscience and 500A2; BD), CD4 (RM4-5; BD), CD45.2 (104; eBioscience), CD90.1 (HIS51; eBioscience). Intracellular cytokine staining was performed with anti-IFNγ (XMG1.2; BD) or anti-TNFα (MP6-XT22; BD), using the Cytofix/Cytoperm kit (BD Biosciences). Flow cytometric data were collected on an LSR II (BD Biosciences) and analyzed with FlowJo software (TreeStar).
Immunofluorescence Staining and Histochemistry
Lungs were harvested and fixed using PLP buffer (0.05 M phosphate buffer containing 0.1 M L-lysine [pH 7.4], 2 mg/mL NaIO4, and 10 mg/mL paraformaldehyde) for 12 hr, then dehydrated in 30% sucrose prior to embedding in OCT freezing media (Sakura Finetek). 30μm sections were cut on a CM3050S cryostat (Leica) and adhered to Superfrost Plus slides (VWR). Sections were then permeabilized and blocked in PBS containing 0.1% Triton X-100 (Sigma) and 10% normal mouse serum (Jackson Immunoresearch) followed by staining in the same blocking buffer. The following antibodies were used for staining: anti-IFNγ (XMG1.2; BD), anti-Ag85b (rabbit polyclonal, abcam), anti-PR8 (goat polyclonal serum; Virostat), anti-RSV (goat polyclonal serum; Virostat). Unconjugated primary antibodies were stained with AF-conjugated secondary antibodies (Invitrogen). For IFNγ staining, the signal was further amplified with a secondary goat anti-rat AF-conjugated antibody. Stained slides were mounted with Fluoromount-G (Southern Biotech) and images were acquired on a 710 confocal microscope (Carl Zeiss Microimaging). Data were processed and analyzed with Imaris (Bitplane). For hematoxylin and eosin stains (H&E) tissues were harvested in 4% paraformaldehyde and transferred into 70% ethanol 24h later. Tissue was then processed and stained by Histoserv, Inc. (Germantown, MD). Bright-field images were acquired on a Leica AF 6000 LX (Leica systems).
Detailed quantitative analysis methods are described elsewhere (30). Briefly, cellular surfaces were 3D rendered via the Imaris surface creation module. Influenza stained regions were also analyzed via the Imaris spots creation module. Mean voxel fluorescence intensity statistics and positional information for each surface were exported into Excel. Minimal distances were measured using 2D distance calculation formulas. Fluorescence intensity and distance data were then imported into FlowJo via the Text to FCS Java file converter.
Two-photon imaging of explanted lungs and image analysis
Mice were euthanized by cervical dislocation, following a brief period of inhalational anesthesia with isoflurane. Lungs were taken out en bloc, rinsed in warm PBS and the left lobe then placed on a custom-made imaging platform (Fig. S1). Continuous temperature monitoring of the setup was also performed to ensure a narrow range of 36-38o Celsius was maintained at all times. The images were then acquired on a Zeiss 510 system (Carl Zeiss Microimaging), equipped with a Chameleon laser (Coherent), tuned to 800nnm, 840nm, or 860nm, and a 25X air lens (NA 0.8), driven by LSM software. Z stacks of approximately 50μm with 3μm step size were acquired every 30 seconds. Data were processed and analyzed with Imaris (Bitplane). Through the surface and spot creation module in Imaris, spatial coordinates of objects of interest were obtained over time and data were further analyzed with MatLab.
Statistical analysis
Student's t test was used for statistical analysis of differences between groups.
Results
Establishment of a robust in vivo system designed to study CD4+ T cell effector responses in lung tissue
To compare the CD4+ T cell effector responses in chronic BCG vs. acute influenza infection we made use of two distinct TCR transgenic systems as the antigen-specific indicator populations, namely in vitro-generated effector CD4+ T cells derived from mycobacterial antigen Ag85b-specific (ovalbumin-unrelated) P25 TCR transgenic RAG1-deficient mice (P25) and from ovalbumin-specific (mycobacteria-unrelated) OTII TCR transgenic RAG1-deficient mice (OTII). Although it would be optimal to do the studies described here using exactly the same TCR transgenic T cell reporter population, this would require either mycobacterial peptide-expressing influenza recombinants or recombinant mycobacteria expressing an antigen shared with wild type or recombinant influenza. At present, such organisms with validated infectious behaviors have proved problematic to construct, as insertion of foreign peptides can markedly alter the infectivity of the virus and its virulence. We therefore obtained a PR8 strain expressing the MHCII-restricted peptide from ovalbumin, OVA323 (29). This strain has been shown to elicit a CD4+ T cell response and to cause a viral pneumonia similar to, if slightly milder, than the parental strain (29), permitting us to use this organism for analysis with a well-characterized T cell population (OTII) in the context of a near normal infectious process. Unfortunately, BCG expressing OVA has been very poorly immunogenic for OTII T cells in our hands (Egen JG and Rothfuchs AG, unpublished data). Given our expectation that influenza was likely not to limit MHCII antigen presentation in the same manner as BCG, we chose to use OTII T cells to analyze the response to influenza and the Ag85b-specific P25 transgenic T cells to assess the response to BCG infection, as we have done previously. This of course raises the possibility that observed differences in effector behavior might be attributable to intrinsic differences in sensitivity to cognate antigen rather than distinct biology associated with the two different pathogens.
To directly examine this critical issue of antigen sensitivity of the two transgenic T cell populations, wild type splenocytes were pulsed with titrated amounts of either cognate peptide (Ag85b or OVA323) and these presenting cells incubated for 5 hours with their respective antigen-specific CD4+ T effector cells: either P25 or OTII day 7 effector cells generated in vitro employing a Th1-polarizing protocol. Intracellular cytokine staining was performed, along with surface staining for congenic markers to discriminate the transgenic T cell populations. More P25 T cells produced IFNγ as compared to OTII T cells when exposed to APCs pulsed with the same molar concentration of cognate peptide (Fig. 1A). To examine this issue in vivo, we injected various equimolar amounts of OVA323 and Ag85b peptide intravenously after prior transfer of the respective effector cell populations. Two hours after peptide delivery, mice were sacrificed and spleens isolated. Single cell suspensions were then stained for intracellular IFNγ. Again, at each peptide dose, P25 T cells displayed a greater ability to produce cytokines, such as IFNγ (Fig. 1B), recapitulating the in vitro findings. In combination with other studies (31) that reveal the same response hierarchy using methods independent of the capacity of the two peptides to bind MHCII molecules (i.e., basal levels of partial TCR ζ chain tyrosine phosphorylation, levels of CD5 expression, and cognate tetramer binding), these data make a strong case that P25 T cells are intrinsically more sensitive to their cognate antigen as compared to OTIIs and that if at all, the present studies of effector responses are biased in favor of P25 T cells in terms of sensitivity to elicitation of effector cytokine production, even though it is this T cell population that shows limited responses to its cognate antigen under infection conditions in vivo (14). Thus, we anticipated that if we observed better effector responses from the OTII T cells in response to the recombinant PR8, this was unlikely to reflect a bias in this direction in our experimental design due to T cell-intrinsic factors.
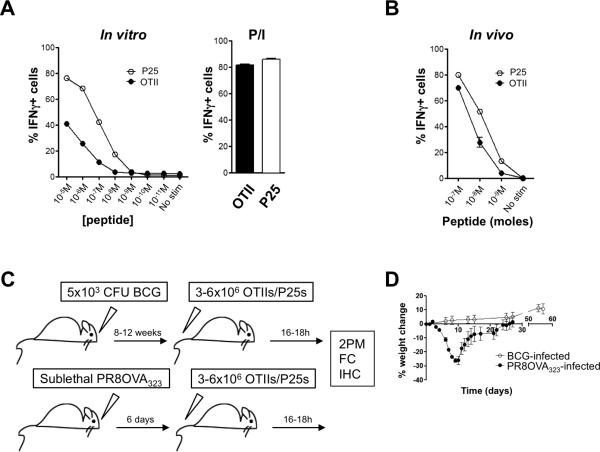
FIGURE 1.
Establishment of a robust in vivo system designed to study CD4+ T cell effector responses in lung tissue. A, Day 7 in vitro-generated effector T cells (P25 or OTIIs) were incubated with an equal number of splenocytes pulsed with the cognate antigen at the indicated concentrations for 5 hours. Percentage of IFNγ-producing T cells is represented. Bar graph to the right indicates the percentage of IFNγ-positive P25 and OTII T cells when stimulated in vitro with PMA+ionomycin. B, Naïve mice received ~1 million day 6 in vitro-generated effector T cells (P25s and OTIIs). After 18 hours, the mice received equimolar amounts of both OVA323 and Ag85b. Two hours later, the mice were sacrificed and the percentage IFNγ-producing transferred cells in the spleen was determined. Data are representative of at least 2 independent experiments, done in triplicates. Symbols and bars indicate mean with indicated standard error of the mean. C, Schematic representation of the experimental setup. 2PM=two-photon microscopy; FC=Flow cytometry; IHC=immunohistochemistry. D, Percent weight loss over time. Symbols indicate mean of at least 5 animals, and error bars the standard error of the mean.
Having established the antigen response hierarchy of OTII and P25 T cells, we proceeded to develop model systems that would allow us to answer the questions delineated above (Fig. 1C, D). For pulmonary BCG infections, we chose a dose of 5×103 CFUs, which is a hundred times lower than the dose we previously used for intravenous infections, given that that 1% of intravenously administered mycobacteria arrive in the lung (32-34). Pulmonary infection with mycobacteria led to formation of mature granulomas defined as macrophage agglomerates in the lung by 8-12 weeks and at this time point most of the BCG organisms were confined to these structures (Fig. S2A). It has also been reported that the number of effector CD4+ T cells in the lung peaks at 8-12 weeks after aerosol infection using a 5 times lower dose of BCG than we employed here (33). Additionally, we found that transferring effector CD4+ T cells at earlier time points when granulomas hadn't formed yet resulted in poor effector cell recruitment to the lung (data not shown). For these reasons, we chose to study the 8-12 week period post infection. This infection ran an indolent clinical course, with no weight loss and with minimal pulmonary pathology upon gross examination (Fig. S2B). In a time frame ranging from 8 to 12 weeks post-infection, mice received in vitro-generated effector CD4+ T cells derived from P25 (BCG-specific) and OTII (antigen-unspecific) TCR transgenic animals. For influenza infections we chose a sublethal dose, which led to significant weight loss and morbidity (Fig. 1C, D) and pulmonary pathology (Fig. S2B). Day 6 to 7 after infection represents the peak of the CD4+ and CD8+ T cell adaptive response to influenza (35) and therefore day 7 was chosen as the experimental end point. One day prior to analysis, mice received OTII T cells as well as P25 T cells as the non-antigen specific control population.
Effector T cells display distinct effector behavior in lungs infected with different agents
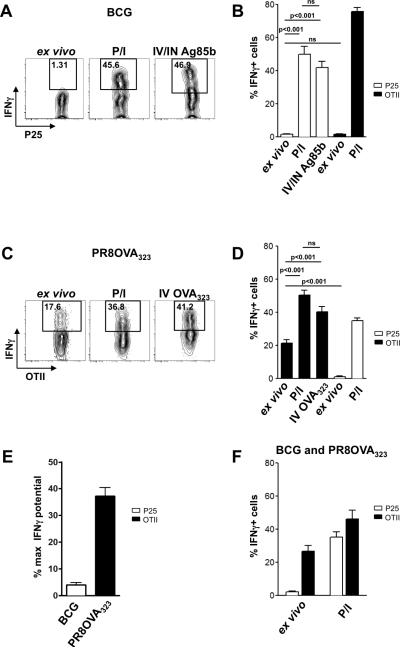
To examine CD4+ T cell effector responses to mycobacterial antigens in the lung, mice were infected with BCG intranasally and 8-12 weeks later, received in vitro-generated Th1-polarized effector P25 and OTII T cells. Sixteen to 18 hours after transfer, the mice were sacrificed, the lungs processed, and transferred cells analyzed by flow cytometry without ex vivo stimulation. Under these conditions, very few P25 cells (1-2%) were producing inflammatory cytokines, such as IFNγ (Fig. 2A, B) or TNFα (Fig. S3A), as assessed by intracellular staining. However, greater than 50% of these cells had the potential to produce cytokine when they were stimulated in vitro with PMA+ionomycin or in vivo with peptide (Fig. 2A, B, Fig. S3A), indicating cytokine production in the granuloma by effector T cells is limited by activation conditions in the infected tissue and not by T cell-intrinsic constraints on cytokine production capacity. In contrast to our previous analysis in liver (14), obtaining a peptide-dependent increase in cytokine production under these conditions required delivery of the antigen by both the i.v. and i.n. routes. Control OVA-specific T cells in these mice also behaved similarly, in that they produced virtually no cytokine when examined directly upon extraction from the tissue, but were able to do so when maximally stimulated (Fig. 2B, black bars). In BCG-infected lungs, we did not detect any statistically-significant differences in cytokine responses between antigen-specific and non-specific effectors recovered from the tissue (Fig. 2B). Although we conducted these experiments when the number of CD4+ T cells in the granulomas is reported to reach a peak (33), another study suggested that cytokine production reaches a maximum earlier after infection (36), and it was also possible that providing more antigen in the form of a higher inoculum of bacteria might change the outcome. We therefore infected mice systemically with a hundred times higher BCG dose (5 × 105 CFU) and studied earlier time points after intranasal infection. Even two weeks post systemic BCG infection with this higher dose, very few (~5%) P25 cells in the lungs produced IFNγ (Fig. S3B). Additionally, we infected mice intranasally with a one hundred times greater dose (5×105 CFUs) and examined the CD4+ T cell effector cytokine response over time and compared this response to that generated in mice infected with the lower dose of 5×103 CFUs. Even with this higher dose, as compared to what we saw in animals infected with the low dose, we detected no increase in cytokine response from the antigen-specific T cells when examined directly ex vivo at the indicated time points (Fig. S3C). These studies at early time points and with higher infectious doses indicate that the low fraction of cytokine-producing antigen-specific cells seen with BCG is a general characteristic of the response to this organism and not an artifact of the particular conditions used for the bulk of our experiments.
FIGURE 2.
Effector CD4+ T cells display distinct effector behavior in lungs infected with different agents. A and C, Representative FACS plots showing the percentage of all transferred cells producing IFNγ, as measured by intracellular cytokine stain, when taken from BCG- (A) or Influenza-infected mice (C). P/I=PMA+ionomycin. IN=intranasally. IV=intravenously. Numbers indicate percentages. Gating strategy for lung single cell suspensions involved gating on live cells with a generous forward and side scatter gate, then gating on CD45+ cells, followed by gating on the transferred T cell population of interest. Finally, CD4 by IFNγ was plotted. Negative gate set based on isotype control for each experimental condition. B and D, Quantification of the percentage of IFNγ+ P25 T cells or OTII T cells, when taken directly ex vivo from the lungs, after 4 hours of in vitro PMA+ionomycin stimulation, or 2 hours after in vivo administration of cognate antigen. Responses of the control effector population not recognizing antigen are shown in the same graph depicted by different color bars. Data are represented as mean and standard error of the mean. Statistics performed using the unpaired Student's t test. E, Data represented as percentage of maximal IFNγ production potential, obtained by comparing percentage IFNγ+ ex vivo to percentage IFNγ+ after PMA+ionomycin stimulation. ns=non-significant. Data are representative of at least 4 independent experiments with 3-5 mice per group. Data represented as mean with standard error of the mean. F, Mice infected with BCG 11 weeks prior were also infected with PR8OVA323. 7 days post PR8OVA323 infection (and thus 12 weeks post BCG infection), effector T cells were transferred into the animals. Eighteen hours after transfer, transferred T cells were isolated and the percentage of IFNγ-producing cells (gated on P25s or OTIIs) was determined. Data are representative of 2 independent experiments with 3 mice per group. Data represented as mean with standard error of the mean.
In contrast, cells isolated from influenza-infected mice behaved quite differently. A significant proportion of the isolated antigen-specific OTII T cells were actively producing cytokines (20-30%), as measured by IFNγ (Fig. 2C, D) or TNFα intracellular staining (Fig. S3D). This fraction, however, was still lower than the maximal potential for the population, as stimulation with either PMA+ionomycin in vitro or cognate peptide administration in vivo led to a greater frequency (40-70%) of cytokine-containing T cells (Fig. 2C, D, Fig. S3D). On the other hand, control P25 T cells not specific for any influenza virus-encoded antigen, barely produced cytokine when taken directly ex vivo, but did respond to pharmacologic reagents (Fig. 2D, white bars). The quantitative differences in ex vivo effector cytokine production by antigen-specific T cells seen in the BCG-infected vs. the influenza-infected mice were even more apparent when we looked at effector cytokine production represented as a fraction of the maximal potential (Fig. 2E). Examined in this way, it was evident that a much smaller proportion of T cells in BCG-infected mice specific for a mycobacterial antigen are stimulated to produce cytokines in situ as compared with the T cells in the influenza-infected mice specific for an antigen encoded by this virus.
To test whether the inflammatory environment could be the basis for the difference in cytokine production in response to BCG and influenza infection, the ex vivo cytokine production by OTII and P25 T cells was compared in animals co-infected with BCG and PR8OVA323. Both OTII and P25 T cells isolated from co-infected animals maintained the cytokine production profile typical of their counterparts isolated from either BCG- or PR8OVA323-infected animals (Fig. 2F), arguing against the inflammatory environment as the basis for the low effector cytokine production in response to BCG infection. In fact, the maximal potential reached by the control (antigen-unspecific) T cell populations in the two infection models after transfer suggest that the milieu was more anti-inflammatory in influenza than BCG-infected lungs. The OTII T cells resident in the BCG-infected lungs maintained the ability to produce cytokine to an extent similar to what the cells were able to achieve prior to transfer (70-80% in both cases) (Fig. S3E and Fig. 2B, black bars). The P25 T cells in the influenza-infected mice, on the other hand, when maximally stimulated, did not reach the same level of cytokine production as they could prior to transfer into the infected animals (40-50% versus 70-80% respectively) (Fig. S3F and Fig. 2D, white bars). These findings are consistent with earlier reports of anti-inflammatory activities in the lungs of influenza-infected animals (37-39), making even more striking the disparity between high rates of specific responses in influenza-infected tissue as compared to BCG-infected lungs. This disparity was not simply the result of employing TCR-transgenic effector cells in these studies, because similar differences were seen among endogenously responding T cells (Fig. S3G).
Differences in CD4+ T cell effector function are mirrored by differences in dynamic behavior
Cytokine-producing cell frequency and arrested cell frequency were closely matched in liver granulomas (14), consistent with a model in which only those T cells engaging APCs with enough TCR ligand to cause signaling arrest migration and initiate an effector cytokine response. An alternative model has been proposed for tissue effector responses in which T cells sum subthreshold signals acquired by sequential interaction with many presenting cells until activation is achieved, leading to cytokine production without migration arrest (40, 41). Such a scenario would predict a marked difference between the fraction of cytokine-producing cells and the fraction of arrested antigen-specific T cells in each of these infection models. We therefore examined effector CD4+ T cell dynamic behavior in the lung.
For imaging, we developed a lung explant model (Fig. S1). An intravital microscopy approach (where the lung is imaged in a live animal either by surgical exposure via a thoracotomy or through an implanted window), although theoretically preferable, has substantial limitations with regards to our question. Both BCG- and influenza-infected lung tissues are quite heterogeneous with respect to the distribution of cells and infected areas. Because of this heterogeneity, the ability to scan large areas of lung tissue is a critical requirement for avoiding sampling errors, and this capacity is provided with explants and not the presently available intravital techniques.
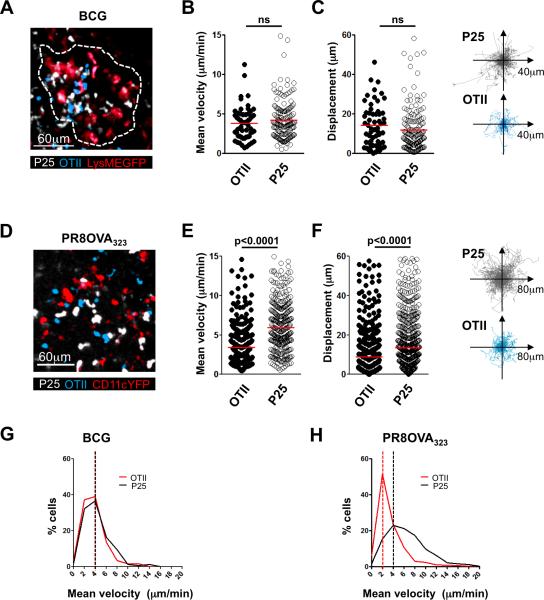
Similar to our previously published data examining liver granulomas in systemically-infected mice (14), both antigen-specific and non-specific effector T cells were found to accumulate in the lung granulomas (Fig. 3A). The mean velocities of the P25 and the OTII effector populations in the lung tissue were not significantly different (Fig. 3B, Movie S1), and in line with this, total T cell displacement over the duration of imaging was also not significantly different between the two lymphocyte populations in the same tissue sample (Fig. 3C, Movie S1).
FIGURE 3.
Differences in CD4+ T cell effector function are mirrored by differences in dynamic behavior. A-C, Lung BCG granuloma image acquired by two-photon microscopy and analysis thereof. D-F, Image of inflammatory patch in an influenza-infected lung acquired by two-photon microscopy and analysis thereof. A and D, Maximum projection rendering of imaging volume. Colors of the words correspond to colors in the image, here and throughout. In panel A, the dotted line delineates the margins of the granuloma in the image. B and E, Mean velocity of OTIIs and P25s; red line indicates mean value. C and F, Total displacement over the duration of the imaging session (red line indicates mean value) and representation of the tracks of OTIIs and P25s over the duration of the movie, all translated to the same starting point. G and H, Frequency distribution of P25 and OTII velocities in BCG (G) and influenza-infected tissue (H); bin width of 2μm. Dashed lines indicate mode of the distributions. ns=non-significant. All data are representative of at least 3 similar experiments, with at least 3 mice per group. Statistics performed using the unpaired Student's t test.
To determine if these findings applied to other infectious settings, we conducted similar experiments using influenza A as the pulmonary pathogen, an infectious situation in which we anticipated that antigen abundance would not be limiting given the cytokine response data reported above. Six days after infection with PR8OVA323, mice received OTII T cells as the antigen-specific population as well as P25 T cells as the control population. Sixteen to 18 hours later, lungs of influenza-infected mice were imaged using the explant technique described above. Both CD4+ T cell populations accumulated in the PR8-infected lungs 18-20 hours post transfer (Fig. 3D). In contrast to what was seen in mycobacteria-infected pulmonary tissue, 2-photon imaging of explanted lungs showed a significant difference in migration dynamics between the OTII T cells specific for a virally-encoded antigen and the control P25 T cells. OTII T cells moved at a significantly slower mean velocity when compared to the P25 T cells in the same lung analyzed at the same time (Fig. 3E, Movie S2). Total displacement was also significantly less when comparing the antigen-specific T cell population to the control T cells (Fig. 3F, Movie S2). We also analyzed the difference in dynamic behavior between antigen-specific and non-specific effector T cells in the BCG- and influenza-infected lungs by determining the frequency of cells moving at different velocities and representing these data as a frequency distribution (Fig. 3G, H). Both the antigen-specific population and the control population displayed a similar velocity distribution in the BCG-infected lung (Fig. 3G); in contrast, the OVA-specific OTII T cells showed a distribution skewed towards slower velocities in infected lungs expressing their cognate antigen OVA as compared to the antigen-unspecific P25 T cells in this same setting (Fig. 3H). This alternative way of displaying the data shows more clearly that most OVA-specific effector T cells in the influenza/OVA-infected lungs are moving at velocities considered to represent arrest in migration (at about 2μm/min). Overall, these findings, which were in sharp contrast to the BCG infection model, suggested that the influenza-infected lung provided more opportunity for TCR engagement and signaling by the antigen-specific T cells. Furthermore, the significantly greater fraction of antigen-specific T cells displaying migration arrest in conjunction with a greater proportion of these T cells producing inflammatory cytokines in influenza-infected lungs compared to BCG-infected lungs were suggestive of an inverse relationship between cell migration and productive antigen sensing.
Local antigen abundance dictates T cell effector function as well as dynamic behavior
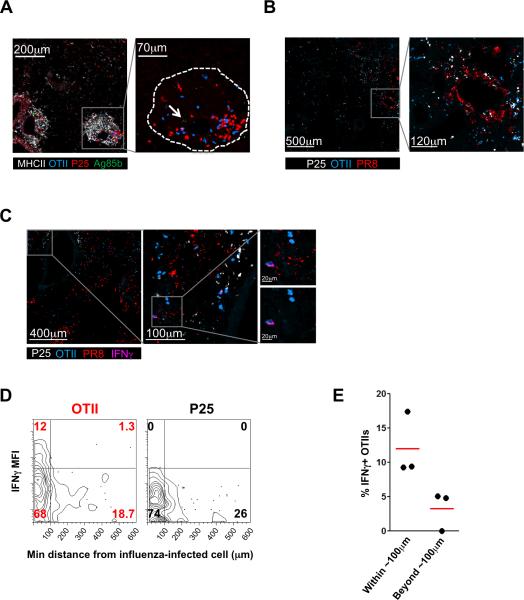
Additional experiments were conducted to further examine the hypothesis that differences in antigen abundance play a major role in determining the distinct fraction of effector T cells responding in the two infection models. Because providing more peptide in vivo enabled larger fractions of the OTII and especially the P25 T cells to produce cytokines (Fig. 2B, D), it did not seem that the number of APCs or MHCII expression by APCs in the tissue was a limiting factor. To look more directly at the role of antigen abundance, we stained paraformaldehyde-fixed lung frozen sections for the presence of Ag85b and PR8 (due to the lack of an antibody that reliably identifies OVA323 peptide, we employed a polyclonal serum against PR8, using polyclonal anti-RSV serum as a specificity control (Fig. S4)). Ag85b was restricted to the center of granulomas and present in small amounts, as previously reported (14) (Fig. 4A). On the other hand, influenza protein staining was much stronger and more widespread in the lung (Fig. 4B). While this staining represents the protein source of antigenic peptides and not the processed, MHC-bound fraction, these data do suggest that there is greater availability of the influenza-encoded antigen for generation of pMHC ligands, in accord with the above findings.
FIGURE 4.
Antigen abundance dictates T cell effector and dynamic behavior. A and B, Immunofluorescent staining of BCG-infected mouse lung with anti-Ag85b antibody (arrow indicates Ag85b stain) (A) or of influenza-infected mouse lung with anti-PR8 antibody (B). P25 and OTII T cells were labeled with an intracellular dye prior to transfer. In panel A, the dotted line delineates the margins of the granuloma in the image. C, IFNγ staining in an influenza-infected mouse lung. Insets shown are magnifications of the areas in the grey boxes for panels A-C. D, Two-dimensional contour plot depicting data from fluorescent immunohistochemical analysis of influenza-infected mouse lungs, after importing position and mean fluorescent intensity data obtained in Imaris into FlowJo, as described in the Methods section. Data are representative of two independent experiments. E, Percentage of IFNγ-producing OTIIs within the population of cells that reside either beyond or within 100μm of influenza-infected cells. Each dot represents one experiment and the red line represents the mean.
To determine whether in this virally-infected tissue with widespread inflammation and antigen presence cytokine production was focal and targeted, we examined if a finer-grained relationship existed between antigen (virus) location within the lung tissue and effector T cell cytokine production by determining the location of IFNγ-producing cells in relation to virally-infected cells (Fig. 4C). Through histo-cytometric image analysis (30), we found a non-random distribution of the IFNγ-producing antigen-specific CD4+ T cells, nearly all of which were located within about 100μm of the influenza-infected cells (Fig. 4D, E). Thus, while both antigen-specific and non-specific effector CD4+ T cells are located in the general area of infection, nearly all IFNγ-producing antigen-specific cells are within 100μm of the infected cells, and most imaged antigen-specific cells in these virally-infected areas show an arrest of migration. We conclude that both influenza-specific and non-specific effector CD4+ T cells are efficiently recruited to infected areas where the level of antigen presentation is sufficient to induce influenza-specific T cell arrest and IFNγ production. Although not all the T cells extracted from the entire influenza-infected lung show IFNγ production without restimulation, about ~70% of the antigen-specific cells are in the regions of high antigen density (Fig. 4D), and of the cells that are in these regions, approximately 50% are arrested (Fig. 3H). This 35% of the total OTII T cells corresponds closely to the fraction of T cells making IFNγ directly ex vivo. Hence, in the case of influenza infection, there is a strong relationship between antigen-induced stopping and cytokine response, as we previously observed in the liver granuloma model (14).
Discussion
Here we present a careful analysis of effector CD4+ T cell dynamic behavior and cytokine production in mycobacterium vs. influenza-infected lungs in an effort to determine whether our previous report of severe antigen restriction leading to very limited CD4+ T cells responses in mycobacterial liver granulomas was generalizable to other organs (the lung) and if the same limitation in T cell responses was seen with an acute severe infection.
We found that as in BCG or Mtb-infected liver, Ag85b-specific CD4+ T cells showed very limited activation in BCG-infected lungs, with only a few percent of the cells showing TCR-mediated migration arrest or containing IFNγ or TNFα at the time of extraction, although most of the T cells could make cytokine if stimulated ex vivo or in vivo using antigenic peptide. In striking contrast, OTII CD4+ T cells specific for a peptide encoded by recombinant influenza virus exhibited substantial migration arrest in infected lungs and a corresponding high frequency of cytokine-producing cells. Using quantitative image analysis by histo-cytometry, we could demonstrate that a large fraction of effector T cells in influenza-infected lungs reside within 100μm of a rich source of viral protein antigen and that the majority of IFNγ-producing T cells are within this radius, with a very close correspondence between migration arrest and cytokine production. Given our evidence that the P25 T cells specific for Ag85b are more sensitive to antigen display than OTII T cells, these findings clearly indicate that antigen paucity restricts the effector T cell response in mycobacterial infection, but that such a limitation is specific to only a subset of pathogens.
We assessed T cell dynamic behavior in lung tissue using an explant approach to lung imaging. This technique does not require excessive ex vivo manipulation of the organ (such as inflation with agarose gel, or sectioning of the tissue), allows for imaging of immune cell behavior in the lung for up to two hours, and permits survey of a much larger volume of tissue than intravital approaches using windows. With this technique, we not only generated data showing the same correlation between migration arrest and in vivo cytokine production in influenza-infected lung tissue as seen in mycobacterial liver granulomas (14), but also made additional observations related to the effects of infection state on T cell motility in general. For example, the P25 T cells in the influenza-infected tissue lacking their cognate antigen moved at a greater mean velocity than did OTII T cells in BCG-infected lungs lacking the relevant antigen. This reduced velocity of control T cells in BCG-infected lungs might partially be the result of the dense myeloid environment that T cells must migrate through in granulomas (14). These data emphasize the importance of ensuring the presence of a control population in all imaging experiments, as the comparison can then be made with a cell population exposed to the same physical and chemical environmental cues as the cellular population of interest. They also indicate that the factors influencing T cell motility are quite divergent in the two infectious states and suggest it will be of importance to characterize this distinct milieu and the impact that slower T cell effector motility has on the clearance of pathogen (42). Computational studies have indicated that the rate of pathogen detection, a parameter that is highly dependent in turn on effector cell migration velocity, plays a key role in determining the balance between pathogen eradication vs. continued expansion (43, 44).
Our model involves the transfer of in vitro-activated effector T cells. Although we could explore P25 function and behavior starting with small numbers of transferred naïve cells in the liver infection model in which mature granulomas appear within 2-3 weeks, genetic incompatibilities between the presently available P25 TCR transgenic animals and the C57BL/6 recipients preclude such studies over the longer (8-12 week) time frame required for granuloma development in the lung infection model. However, we have performed experiments in influenza-infected mice where the effector T cells have been generated in vivo, by transferring naïve T cells prior to infection at numbers close to what would be expected to be the precursor frequency of several hundred cells per mouse (45). These experiments recapitulated the data obtained with the in vitro-generated effector T cells (data not shown), namely, that a substantial fraction of all effector T cells actively engaged in effector function. Additionally, to address the question of whether the use of transferred effector cells had an impact on the fraction of cytokine-producing cells recovered, we also analyzed the endogenous CD4+ T cell pool for the production of cytokine. We again found that cytokine production was significantly higher in the influenza-infected mice than in the case of the BCG-infected animals.
Taken together, these findings are in agreement with our hypothesis that antigen abundance is in fact responsible for regulating T cell dynamic behavior in peripheral tissue, with greater antigen presence associated with a significant reduction in motility and this in turn associated with greater cytokine production. Although some investigators have proposed that summation of shorter contacts can also lead to T cell activation under certain conditions (40), our data are in accord with observations made by others (5, 6, 8, 14) that stopping and prolonged TCR stimulation most closely correlates with cytokine production. Additionally, we show that although antigen-unspecific effector CD4+ T cells are effectively recruited to the effector site, the inflammatory environment does not lead to substantial antigen-unspecific effector responses and that effector responses are strictly dependent on the presence of antigen at the effector site. This conclusion is supported by the lack of cytokine production by the antigen-unspecific effector control populations under infectious conditions, the cytokine production profile displayed by the antigen-specific cells under co-infection conditions, and the cytokine responses seen when using in vivo peptide to maximize effector responses by one of the two effector populations. We speculate that the lack of antigen-unspecific effector function may be an evolutionarily acquired defense mechanism designed to minimize the deleterious effects of excessive cytokine production in vital organs, such as the lung. Finally, using a new method for multicolor quantitative immunohistochemistry (30), we demonstrate that cytokine-producing effector CD4+ T cells are in close proximity to infected cells, further supporting the idea that cytokine production is focal, specific, and antigen-driven.
The ex vivo flow cytometric analysis of whole tissue homogenates we utilize here provides information about cellular behavior at a population level, but does not provide information on single cell behavior in the tissue of interest that can be directly linked to the dynamic behavior of that cell. Unfortunately, existing gene expression reporter systems that encode long-lived cytoplasmic fluorescent proteins do not provide an accurate picture of cytokine expression because they persist for much longer than cytokine is made and secreted by activated T cells (46). However, by combining an ex vivo approach with in vivo imaging we have been able to gain insight into the origin of the heterogeneity of effector T cell migratory and functional behavior at the effector site, providing evidence that antigen density plays a major role in the extent of the response by tissue-homing effector T cells and that this parameter differs markedly in infections with diverse pathogens. More widespread and robust antigen presentation in the setting of an acute pulmonary influenza infection leads to engagement and activation of a larger fractional percentage of the effector T cell pool compared to a chronic non-resolving mycobacterial infection. Nevertheless, even during acute pulmonary influenza infection, effector function may not reach its maximal potential. This insight suggests that one way to improve local effector responses is to increase antigen display in the infected site. This approach may be constrained, however, by difficulty in targeting such antigen to focal sites of pathogen replication rather than broadly within a tissue, organ, or even the whole body, with the risk of inducing immunopathology due to over-exuberant responses. Additional studies will be needed to determine if it is possible to balance the host protective effects of such antigen administration with the possibility of disruption of tissue homeostasis.
Supplementary Material
Acknowledgements
We thank P. Thomas and P. Doherty for kindly providing PR8 strains, as well as C. Feng and A. Sher for providing wild type and RFP-expressing BCG strain Pasteur. The authors would also like to thank Marlene Brandes for helpful discussions and for sharing technical expertise. This work was supported by the Intramural Research Program, Clinical Center and National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- 1.Bousso P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nature reviews. Immunology. 2008;8:675–684. doi: 10.1038/nri2379. [DOI] [PubMed] [Google Scholar]
- 2.Bousso P, Robey E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nature immunology. 2003;4:579–585. doi: 10.1038/ni928. [DOI] [PubMed] [Google Scholar]
- 3.Germain RN, Robey EA, Cahalan MD. A decade of imaging cellular motility and interaction dynamics in the immune system. Science. 2012;336:1676–1681. doi: 10.1126/science.1221063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrickson SE, von Andrian UH. Single-cell dynamics of T-cell priming. Current opinion in immunology. 2007;19:249–258. doi: 10.1016/j.coi.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomaus I, Kawakami N, Odoardi F, Schlager C, Miljkovic D, Ellwart JW, Klinkert WE, Flugel-Koch C, Issekutz TB, Wekerle H, Flugel A. Effector T cell interactions with meningeal vascular structures in nascent autoimmune CNS lesions. Nature. 2009;462:94–98. doi: 10.1038/nature08478. [DOI] [PubMed] [Google Scholar]
- 6.Beattie L, Peltan A, Maroof A, Kirby A, Brown N, Coles M, Smith DF, Kaye PM. Dynamic imaging of experimental Leishmania donovani-induced hepatic granulomas detects Kupffer cell-restricted antigen presentation to antigen-specific CD8 T cells. PLoS pathogens. 2010;6:e1000805. doi: 10.1371/journal.ppat.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fife BT, Pauken KE, Eagar TN, Obu T, Wu J, Tang Q, Azuma M, Krummel MF, Bluestone JA. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nature immunology. 2009;10:1185–1192. doi: 10.1038/ni.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filipe-Santos O, Pescher P, Breart B, Lippuner C, Aebischer T, Glaichenhaus N, Spath GF, Bousso P. A dynamic map of antigen recognition by CD4 T cells at the site of Leishmania major infection. Cell host & microbe. 2009;6:23–33. doi: 10.1016/j.chom.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Kawakami N, Nagerl UV, Odoardi F, Bonhoeffer T, Wekerle H, Flugel A. Live imaging of effector cell trafficking and autoantigen recognition within the unfolding autoimmune encephalomyelitis lesion. J Exp Med. 2005;201:1805–1814. doi: 10.1084/jem.20050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JV, Jiang N, Tadokoro CE, Liu L, Ransohoff RM, Lafaille JJ, Dustin ML. Two-photon laser scanning microscopy imaging of intact spinal cord and cerebral cortex reveals requirement for CXCR6 and neuroinflammation in immune cell infiltration of cortical injury sites. Journal of immunological methods. 2010;352:89–100. doi: 10.1016/j.jim.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matheu MP, Beeton C, Garcia A, Chi V, Rangaraju S, Safrina O, Monaghan K, Uemura MI, Li D, Pal S, de la Maza LM, Monuki E, Flugel A, Pennington MW, Parker I, Chandy KG, Cahalan MD. Imaging of effector memory T cells during a delayed-type hypersensitivity reaction and suppression by Kv1.3 channel block. Immunity. 2008;29:602–614. doi: 10.1016/j.immuni.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaeffer M, Han SJ, Chtanova T, van Dooren GG, Herzmark P, Chen Y, Roysam B, Striepen B, Robey EA. Dynamic imaging of T cell-parasite interactions in the brains of mice chronically infected with Toxoplasma gondii. Journal of immunology. 2009;182:6379–6393. doi: 10.4049/jimmunol.0804307. [DOI] [PubMed] [Google Scholar]
- 13.Wilson EH, Harris TH, Mrass P, John B, Tait ED, Wu GF, Pepper M, Wherry EJ, Dzierzinski F, Roos D, Haydon PG, Laufer TM, Weninger W, Hunter CA. Behavior of parasite-specific effector CD8+ T cells in the brain and visualization of a kinesis-associated system of reticular fibers. Immunity. 2009;30:300–311. doi: 10.1016/j.immuni.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egen JG, Rothfuchs AG, Feng CG, Horwitz MA, Sher A, Germain RN. Intravital imaging reveals limited antigen presentation and T cell effector function in mycobacterial granulomas. Immunity. 2011;34:807–819. doi: 10.1016/j.immuni.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bold TD, Banaei N, Wolf AJ, Ernst JD. Suboptimal activation of antigen-specific CD4+ effector cells enables persistence of M. tuberculosis in vivo. PLoS pathogens. 2011;7:e1002063. doi: 10.1371/journal.ppat.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladel CH, Daugelat S, Kaufmann SH. Immune response to Mycobacterium bovis bacille Calmette Guerin infection in major histocompatibility complex class I- and II-deficient knockout mice: contribution of CD4 and CD8 T cells to acquired resistance. European journal of immunology. 1995;25:377–384. doi: 10.1002/eji.1830250211. [DOI] [PubMed] [Google Scholar]
- 17.Kamijo R, Le J, Shapiro D, Havell EA, Huang S, Aguet M, Bosland M, Vilcek J. Mice that lack the interferon-gamma receptor have profoundly altered responses to infection with Bacillus Calmette-Guerin and subsequent challenge with lipopolysaccharide. J Exp Med. 1993;178:1435–1440. doi: 10.1084/jem.178.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flynn JL, Chan J. Immunology of tuberculosis. Annual review of immunology. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 19.Caruso AM, Serbina N, Klein E, Triebold K, Bloom BR, Flynn JL. Mice deficient in CD4 T cells have only transiently diminished levels of IFN-gamma, yet succumb to tuberculosis. Journal of immunology. 1999;162:5407–5416. [PubMed] [Google Scholar]
- 20.Mogues T, Goodrich ME, Ryan L, LaCourse R, North RJ. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2001;193:271–280. doi: 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hmama Z, Gabathuler R, Jefferies WA, de Jong G, Reiner NE. Attenuation of HLADR expression by mononuclear phagocytes infected with Mycobacterium tuberculosis is related to intracellular sequestration of immature class II heterodimers. Journal of immunology. 1998;161:4882–4893. [PubMed] [Google Scholar]
- 22.Wojciechowski W, DeSanctis J, Skamene E, Radzioch D. Attenuation of MHC class II expression in macrophages infected with Mycobacterium bovis bacillus Calmette-Guerin involves class II transactivator and depends on the Nramp1 gene. Journal of immunology. 1999;163:2688–2696. [PubMed] [Google Scholar]
- 23.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunology and cell biology. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 24.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 25.Wolf AJ, Desvignes L, Linas B, Banaiee N, Tamura T, Takatsu K, Ernst JD. Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med. 2008;205:105–115. doi: 10.1084/jem.20071367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cellular immunology. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 27.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726. [PubMed] [Google Scholar]
- 28.Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, Nussenzweig MC. Visualizing dendritic cell networks in vivo. Nature immunology. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 29.Thomas PG, Brown SA, Morris MY, Yue W, So J, Reynolds C, Webby RJ, Doherty PC. Physiological numbers of CD4+ T cells generate weak recall responses following influenza virus challenge. Journal of immunology. 2010;184:1721–1727. doi: 10.4049/jimmunol.0901427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-Cytometry: A Method for Highly Multiplex Quantitative Tissue Imaging Analysis Applied to Dendritic Cell Subset Microanatomy in Lymph Nodes. Immunity. 2012 doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T Cell-Positive Selection Uses Self-Ligand Binding Strength to Optimize Repertoire Recognition of Foreign Antigens. Immunity. 2012 doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper AM, Callahan JE, Keen M, Belisle JT, Orme IM. Expression of memory immunity in the lung following re-exposure to Mycobacterium tuberculosis. Tubercle and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 1997;78:67–73. doi: 10.1016/s0962-8479(97)90017-4. [DOI] [PubMed] [Google Scholar]
- 33.Palendira U, Bean AG, Feng CG, Britton WJ. Lymphocyte recruitment and protective efficacy against pulmonary mycobacterial infection are independent of the route of prior Mycobacterium bovis BCG immunization. Infection and immunity. 2002;70:1410–1416. doi: 10.1128/IAI.70.3.1410-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung YJ, LaCourse R, Ryan L, North RJ. ‘Immunization’ against airborne tuberculosis by an earlier primary response to a concurrent intravenous infection. Immunology. 2008;124:514–521. doi: 10.1111/j.1365-2567.2007.02803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swain SL, Dutton RW, Woodland DL. T cell responses to influenza virus infection: effector and memory cells. Viral immunology. 2004;17:197–209. doi: 10.1089/0882824041310577. [DOI] [PubMed] [Google Scholar]
- 36.Saxena RK, Weissman D, Saxena QB, Simpson J, Lewis DM. Kinetics of changes in lymphocyte sub-populations in mouse lungs after intrapulmonary infection with M. bovis (Bacillus Calmette-Guerin) and identity of cells responsible for IFNgamma responses. Clinical and experimental immunology. 2002;128:405–410. doi: 10.1046/j.1365-2249.2002.01839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinstry KK, Strutt TM, Swain SL. Regulation of CD4+ T-cell contraction during pathogen challenge. Immunological reviews. 2010;236:110–124. doi: 10.1111/j.1600-065X.2010.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nature reviews. Immunology. 2012;12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nature medicine. 2009;15:277–284. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Celli S, Garcia Z, Bousso P. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J Exp Med. 2005;202:1271–1278. doi: 10.1084/jem.20051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Jin B, Henrickson SE, Perelson AS, von Andrian UH, Chakraborty AK. How antigen quantity and quality determine T-cell decisions in lymphoid tissue. Molecular and cellular biology. 2008;28:4040–4051. doi: 10.1128/MCB.00136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egen JG, Rothfuchs AG, Feng CG, Winter N, Sher A, Germain RN. Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity. 2008;28:271–284. doi: 10.1016/j.immuni.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Karlin A, Loike JD, Silverstein SC. Determination of the critical concentration of neutrophils required to block bacterial growth in tissues. The Journal of experimental medicine. 2004;200:613–622. doi: 10.1084/jem.20040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budhu S, Loike JD, Pandolfi A, Han S, Catalano G, Constantinescu A, Clynes R, Silverstein SC. CD8+ T cell concentration determines their efficiency in killing cognate antigen-expressing syngeneic mammalian cells in vitro and in mouse tissues. The Journal of experimental medicine. 2010;207:223–235. doi: 10.1084/jem.20091279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol. 2012;188:4135–4140. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003;198:1069–1076. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.