Abstract
Discovering causative genetic variants in individual cases of suspected mitochondrial disease requires interrogation of both the mitochondrial (mtDNA) and nuclear genomes. Whole-exome sequencing can support simultaneous dual-genome analysis, although currently available capture kits do not target the mtDNA genome and provide insufficient capture for some nuclear-encoded mitochondrial genes. To optimize interrogation of nuclear and mtDNA genes relevant to mitochondrial biology and disease, a custom SureSelect “Mito-Plus” whole-exome library was formulated by blending RNA “baits” from three separate designs: (A) Agilent Technologies SureSelectXT 50 Mb All Exon PLUS Targeted Enrichment Kit, (B) 16-gene nuclear panel targeting sequences for known MitoCarta proteins not included in the 50 Mb All-Exon design, and (C) sequences targeting the entire mtDNA genome. The final custom formulations consisted of a 1:1 ratio of nuclear baits to which a 1 to 1,000-fold diluted ratio of mtDNA genome baits were blended. Patient sample capture libraries were paired-end sequenced on an Illumina HiSeq 2000 system using v3.0 SBS chemistry. mtDNA genome coverage varied depending on the mtDNA:nuclear blend ratio, where a 1:100 ratio provided optimal dual-genome coverage with 10X coverage for over 97.5% of all targeted nuclear regions and 1,000X coverage for 99.8% of the mtDNA genome. mtDNA mutations were reliably detected to at least an 8% heteroplasmy level, as discriminated both from sequencing errors and potential contamination from nuclear mtDNA transcripts (Numts). The “1:100 Mito-Plus Whole-Exome” Agilent capture kit offers an optimized tool for whole-exome analysis of nuclear and mtDNA genes relevant to the diagnostic evaluation of mitochondrial disease.
Keywords: Exome, Capture, Mitochondria, MitoCarta, heteroplasmy, variants, Agilent, SureSelect, HiSeq, NUMT
BACKGROUND
Whole-exome sequencing (WES) has emerged as the preferred method to identify disease genes for Mendelian disorders. Indeed, WES is proving particularly valuable for the diagnostic evaluation of individuals with phenotypically and genetically heterogeneous conditions such as suspected mitochondrial disease (McCormick et al., 2012). Mitochondrial diseases have a wide range of presenting disease manifestations, typically poor genotype-phenotype correlation of any one gene, and a wide range of phenotypically similar non-mitochondrial diseases that must be considered in the differential diagnosis for any given patient (Haas et al., 2007). Known pathogenic mutations causing mitochondrial disease have already been identified in more than 100 nuclear genes and all 37 mtDNA genes (Calvo and Mootha, 2010), although most genes have been linked to only a small number of disease cases and mutations in these known genes collectively account for less than half of cases with suspected mitochondrial disease (Calvo et al., 2012). Additional pathogenic candidates abound as an estimated 1,500 mitochondrial proteins are largely nuclear-encoded, of which the MitoCarta set of 1,034 proteins has undergone robust experimental validation and accounts for approximately 85% of all mitochondrial proteins (Pagliarini et al., 2008). The MitoCarta set includes many known disease genes, including all but 4 nuclear genes (TAZ, PUS1, RRM2B, TYM) of 77 (Calvo et al., 2012) previously linked to mitochondrial respiratory chain disease (Tucker et al., 2010) and 80 of the nuclear genes on the 101 gene sequencing panel for mitochondrial disease and related disorders that is currently available in the clinical diagnostic setting at GeneDx (Gaithersburg, MD) (Supp File 1). Targeted sequence analysis of the MitoCarta gene set together with the mtDNA genome has been estimated to be likely to identify pathogenic causes in at least 47% of all individuals with suspected primary mitochondrial disease (Calvo et al., 2012). Therefore, sequence analysis of the MitoCarta nuclear gene set, the mtDNA genome, and the entire nuclear exome can reasonably be expected to facilitate genetic diagnosis in more than half of all patients with suspected mitochondrial disease, while also presenting the simultaneous opportunity for novel disease gene discovery. Such analysis is now technically feasible by application of massively parallel sequencing methodologies that have emerged in both the research and clinical settings.
A single unified platform has not been available to reliably permit simultaneous interrogation of all known and potential causes of suspected mitochondrial disease and phenotypically overlapping disorders. Exome capture kits are not all equally designed, do not capture the same target regions, and do not all perform with the same efficiency. Indeed, the early versions of commercially available whole-exome capture kits were found to target significantly different genomic regions and to vary greatly in their overall performance (Asan et al., 2011; Kiialainen et al., 2011). In addition, no whole-exome capture kit has been optimized to provide highly reliable capture of the MitoCarta nuclear gene set nor to provide targeted capture of the mtDNA genome. While off-target capture of the mtDNA genome is inevitable in any whole-exome capture kit, this is typically highly non-reproducible with insufficient coverage to either provide reliable interrogation of the complete mtDNA genome sequence or sensitively detect heteroplasmic mtDNA mutations.
Here, we report the performance characteristics of a custom Agilent whole-exome capture that we designed to facilitate simultaneous analysis of the standard 50 Mb whole exome with optimized coverage of the complete MitoCarta nuclear gene set and the mtDNA genome. This platform provides a potential one-stop WES solution that can be applied to both research and clinical genetic diagnostic evaluations of individuals with suspected mitochondrial disease.
RESULTS and DISCUSSION
Agilent SureSelect 50 Mb standard whole-exome capture kit provides insufficient coverage for the mtDNA genome and all MitoCarta genes
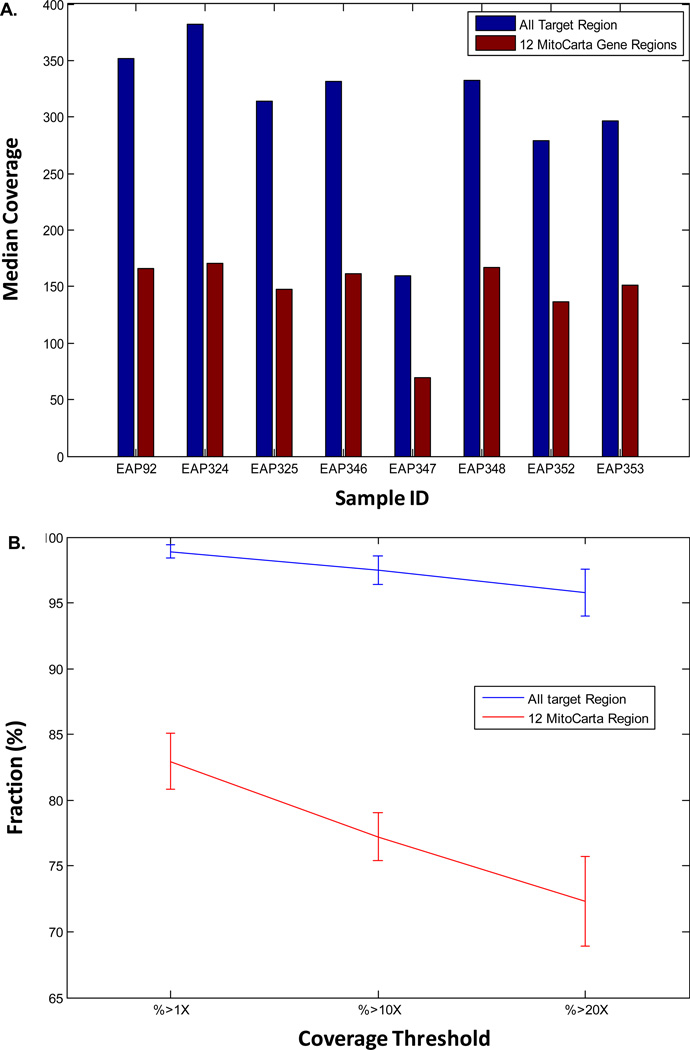
The target regions of the Agilent SureSelect 50 Mb whole-exome capture kit (“50 Mb kit”) do not include the mtDNA genome, as no probes specifically capture mtDNA. Although off-target capture from this platform does provide some mtDNA genome coverage, this is of questionable specificity and insufficient to permit reliable sequence analysis across the entirety of the mtDNA genome (data not shown). Our in silico comparison of 50 Mb kit target regions with the reference sequence gene set (NCBI RefSeq) revealed that among the 1,034 MitoCarta genes there were 12 nuclear genes (BCL2, GPX1, LYRM4, MSRB2, NDUFA11, NUDT8, PIGY, PRDX2, PRDX5, SLC25A26, TIMMI17B, ZBED5) that had less than 80% of their exonic regions covered by the target regions of the 50 Mb kit. Lack of coverage for these genes was empirically confirmed by analysis of 8 exomes captured with the standard 50 Mb kit that we sequenced in a single sample per flow cell lane on the Illumina HiSeq 2000 (Fig 1A and Supp Table 1A). The average whole-exome coverage for each sample ranged from 159.7X to 351.8X, with 98.0% to 99.1% of all target regions covered at least 1X. By comparison, the 12 MitoCarta genes in question had not only a lower average depth of coverage (range: 69.8X–170.9X) but also a markedly lower percentage of exonic regions that were covered at least 1X (range: 78.4% to 84.9%) (Fig 1B and Supp Table 1B). Experimental evidence demonstrated that lack of sequence coverage for these 12 MitoCarta nuclear genes was even more pronounced at the standard 10X and 20X depth-of-coverage cutoffs that are typically used for variant calling purposes.
Figure 1. Standard 50 Mb whole-exome capture kit has inadequate coverage of 12 nuclear genes in the 1,034 MitoCarta gene set.
(A) Median fold-coverage in 8 unrelated human blood DNA samples for all standard 50 Mb target regions and the 12 MitoCarta Gene regions that we identified to have suboptimal coverage on the standard 50 Mb whole exome design. Each sample was sequenced in a single flow cell lane on the Illumina HiSeq 2000. (B) Fraction (percent) coverage for all 50 Mb target regions and 12 MitoCarta Gene regions at varying depths of coverage from 1X to 20X. Detailed coverage data are provided in Supp Tables 1A and 1B, respectively.
SureSelect custom probe design to optimize coverage of the mtDNA genome and all MitoCarta genes
A custom SureSelect “Mito-Plus Whole-Exome” library was generated by blending RNA “baits” from 3 separate designs: (A) standard SureSelect 50 Mb Human All-Exon product that targets the nuclear exome, (B) a 16-gene panel targeting MitoCarta gene sequences that were not included in the All-Exon design, and (C) sequences targeting the entire mtDNA genome (Supp File 2). Designs B and C were created in eArray by 2X tiling across target nuclear genes or 1X tiling across the target hg19 mtDNA genome loci, respectively. Baits having significant overlap with RepeatMasker regions were excluded. For Design B, new nuclear genome baits targeted 416 additional coding regions and 186 UTRs (Supp File 3) in total for the 12 MitoCarta genes that were suboptimally targeted by the 50 Mb kit (BCL2, GPX1, LYRM4, MSRB2, NDUFA11, NUDT8, PIGY, PRDX2, PRDX5, SLC25A26, TIMMI17B, ZBED5), as well as exonic regions of 4 other MitoCarta nuclear genes present on alternative genome assemblies (C6orf136, HSD17B8, MRPS18B, TAP1). The 3 different designs were factory blended in varying molar ratios of Designs A plus B to Design C, as detailed below, for purposes of optimizing dual genome capture of mitochondrial genes.
Experimental evaluation of the optimal capture ratio of mtDNA to nuclear baits
All final custom formulations consisted of a 1:1 ratio of nuclear baits from Design A (All-Exon) to Design B (16 MitoCarta genes). Given the 1–2 log natural excess of mtDNA genomes to the nuclear genome, we sought to assess the optimal output of nuclear versus mtDNA genome sequences that retained the ability to detect low-level mtDNA variant heteroplasmy. Therefore, we experimentally evaluated a range of seven molar concentrations of all nuclear baits (Designs A plus B) to Design C (mtDNA genome) baits. Design C (mtDNA genome) baits were blended in at either an equimolar ratio or reduced concentrations of 10, 50, 100, 200, 500, or 1000-fold less than the nuclear baits. Subsequently, 9 randomly selected human blood DNA samples were selected for capture each by one of these 7 different molar ratios (labeled from A to G to indicate 1:1, 1:10, 150, 1:100, 1:200, 1:500, and 1:1000), a 1:1 ratio of Design A to Design B (with no mtDNA genome baits added), or the standard 50 Mb kit (Supp Table 2A). The 9 captured DNA samples were then sequenced in a single flow cell lane for each sample on the Illumina HiSeq 2000. Optimal coverage across the entire nuclear exome target regions was achieved for each of the 9 samples regardless of the mtDNA:nDNA molar ratio (Fig 2A and Supp Table 2A). Specifically, 99.0% to 99.4% of whole-exome nuclear target regions were covered at least 1X, with 96.0% to 98.1% of whole-exome nuclear target regions covered at least 10X (Table 1). Even at an equimolar ratio of 1:1 mtDNA:nuclear exome capture, the overall performance statistics for nuclear exome sequence coverage did not differ either in median coverage or in percentage of target regions covered at 1X, 10X, or 20X relative to either the standard 50 Mb kit alone or combined with the Design B (Mitocarta gene) nuclear probes.
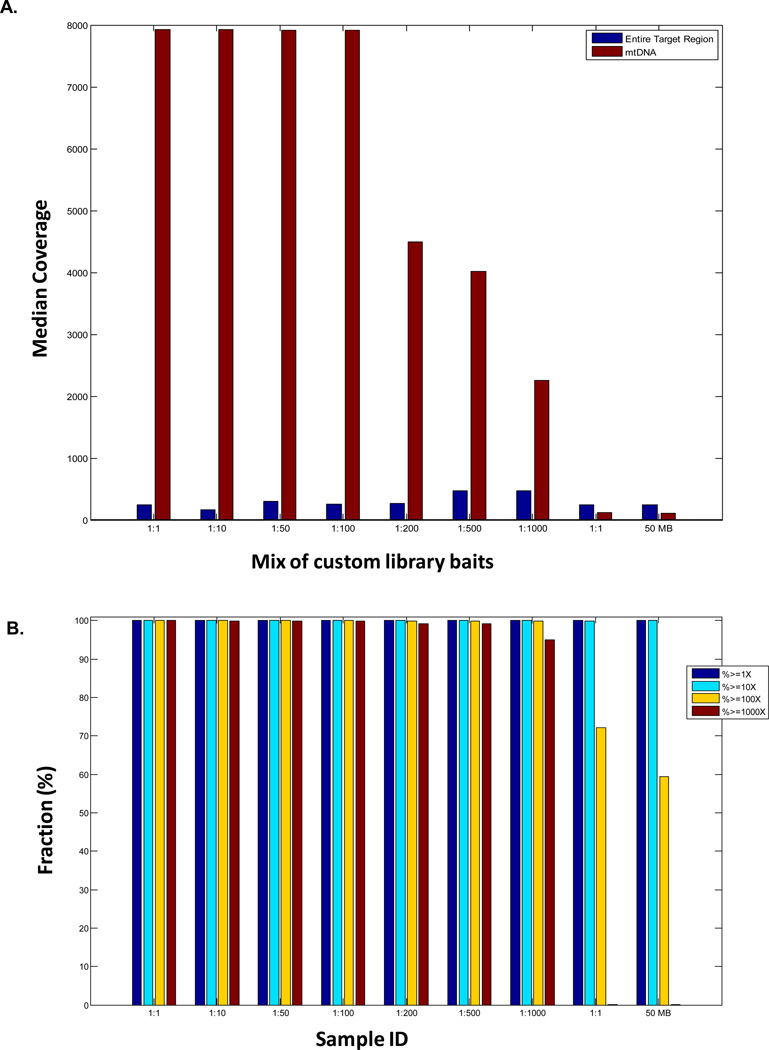
Figure 2. Coverage analysis of variable mtDNA genome to nuclear capture molar ratios.
(A) Median sequence coverage for both the nuclear exome (blue bars) and mtDNA genome (red bars) for 9 samples captured with different mixes of custom library baits. The first seven sets of bars each represent capture ratios of targeted mtDNA genome (Design C) to nuclear baits (Designs A+B), with molar ratios as labeled. The eighth set of bars indicates a 1:1 ratio of standard 50 Mb whole exome to custom MitoCarta baits for the 12 genes not adequately targeted on the initial design (Designs A+B), but no mtDNA baits were included. The ninth set of bars indicates performance of the standard 50 Mb whole exome platform (Design A). Full coverage details are provided in Supp Table 2A. (B) The fraction (percent) of the mitochondrial genome sequenced at variable depths of coverage ranging from 1X to 1000X is shown for the same 9 samples captured with the different molar ratios of custom library baits as described in Fig 2A legend. Detailed coverage data are provided in Supp Tables 2A and 2B, respectively.
Table 1. Coverage performance for both the nuclear exome and mtDNA genome with varying molar ratios of custom baits for the mtDNA genome relative to the MItoCarta optimize standard 50 Mb whole exome design.
Nuclear exome coverage was preserved regardless of mtDNA:nDNA genome capture ratio, but the mtDNA genome median and high-depth (>1,000X) coverage began to fall off at molar ratios below 1:100. Data shown included all sequence reads. Following removal of duplicate sequencing reads, the mtDNA genome coverage fall-off was even more evident beginning at 1:200 molar ratio, where 1,000X coverage was only achieved for 93.29% of the mtDNA genome.
| GENOME | COVERAGE | mtDNA:nuclear baits ratio | Nuclear baits only | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1:1 | 1:10 | 1:50 | 1:100 | 1:200 | 1:500 | 1:1000 | Mitocarta:50 Mb (Nuclear only) |
50 Mb Whole Exome |
||
| NUCLEAR | MEDIAN | 242 | 170 | 306 | 255 | 269 | 469 | 476 | 240 | 246 |
| % ≥ 1X | 99.2% | 99.0% | 99.1% | 99.4% | 99.2% | 99.2% | 99.2% | 99.4% | 99.4% | |
| % ≥ 10X | 97.1% | 96.0% | 97.2% | 97.5% | 97.3% | 98.1% | 98.1% | 97.1% | 97.5% | |
| % ≥ 20X | 95.0% | 93.1% | 95.3% | 95.5% | 95.5% | 96.9% | 96.9% | 94.9% | 95.6% | |
| mtDNA | MEDIAN | 7921 | 7921 | 7919 | 7918 | 4497 | 4013 | 2254 | 118 | 109 |
| % ≥ 1X | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| % ≥ 10X | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 99.8% | 100.0% | |
| % ≥ 100X | 100.0% | 100.0% | 100.0% | 100.0% | 99.9% | 100.0% | 99.9% | 72.2% | 59.5% | |
| % ≥ 1000X | 100.0% | 99.9% | 99.9% | 99.8% | 99.1% | 99.1% | 95.0% | 0.0% | 0.0% | |
Similarly, the standard 50 Mb kit that contained no mtDNA baits still provided some mtDNA genome coverage, which was 100% at 1X coverage and 99.99% at 10X coverage (Fig 2B and Supp Table 2B). This off-target mtDNA capture is explained by the greater natural abundance in terms of molar ratio of mtDNA to nuclear DNA. Nonetheless, such non-targeted coverage is obviously random, non-uniform, drops significantly upon analysis of 100X coverage performance, and has a minimum coverage depth of 0 to 2 reads at some mtDNA genome bases. Whereas 10X to 20X median coverage is generally acceptable for analysis of nuclear exome capture performance, a substantially higher-depth of coverage across the entire mtDNA genome is critical to permit reliable detection of low-level mtDNA variant heteroplasmy. Mixing mtDNA genome baits with nuclear baits at all 7 different ratios, from equimolar to 1 mtDNA to 1000 nDNA, all provided much improved coverage across the entire mtDNA genome. Specifically, the standard 50 Mb kit had a median 109X and mean of 133.5X mtDNA genome coverage. However, careful data analysis suggested that the optimal mtDNA:nuclear molar ratio was 1:100, where over 99.9% of the mtDNA genome was covered at least 100X, over 99.0% of the mtDNA genome was covered at least 1000X, the median coverage was 7,918X, and the minimum depth of coverage for any mtDNA base was 41X. Higher molar ratios (1:1, 1:10, 1:50) provided similar if not better mtDNA coverage as seen with 1:100, but these higher molar ratios carry the potential cost of reducing sequencing bandwidth in the nuclear target regions. Lower molar ratios (1:200, 1:500, 1:1000) demonstrated a progressive fall-off in mtDNA genome coverage, which for the 1:200 ratio was an mtDNA genome median coverage of 4,497X with only 99.1% of the mtDNA genome covered to a depth of 1000X. Therefore, we selected a 1:100 mtDNA to nuclear molar ratio for subsequent experiments.
A 1:100 molar ratio of mtDNA to nuclear baits provided optimal coverage for both the nuclear target regions and the mtDNA genome
Custom libraries with 1:100 molar ratio of mtDNA to nuclear baits were used to capture 11 exomes from human blood genomic DNA and then sequenced using one HiSeq 2000 flow cell lane per sample, with coverage statistics summarized in Fig 3a. Although capture experiments did not work as well for two samples (61p2 and 79) as they did for the other 9 samples, 1X coverage of the nuclear exome was seen for 98.4% to 99.7% of target regions for each of the 11 samples tested (Supp Table 3A). Excluding the two samples that had suboptimal performance, an average of 194X to 415X mean depth of coverage for the nuclear exome was achieved for the remaining 9 samples. Optimal mtDNA genome coverage was achieved for all 11 samples (Fig 3B), which was 99.99% to 100% of mtDNA genome bases covered at both 1X and 10X, 99.89% to 100% of mtDNA genome bases covered at 100X coverage, and 93.75% to 99.95% of mtDNA genome bases covered at 1000X for all samples. When excluding the two samples that had had suboptimal overall nuclear and mtDNA capture performance (61p2 and 79), 1000X coverage was seen at 99.6% to 99.95% of all mtDNA genome bases in each of the remaining 9 samples captured at the 1:100 mtDNA to nuclear molar ratio (Supp Table 3B).
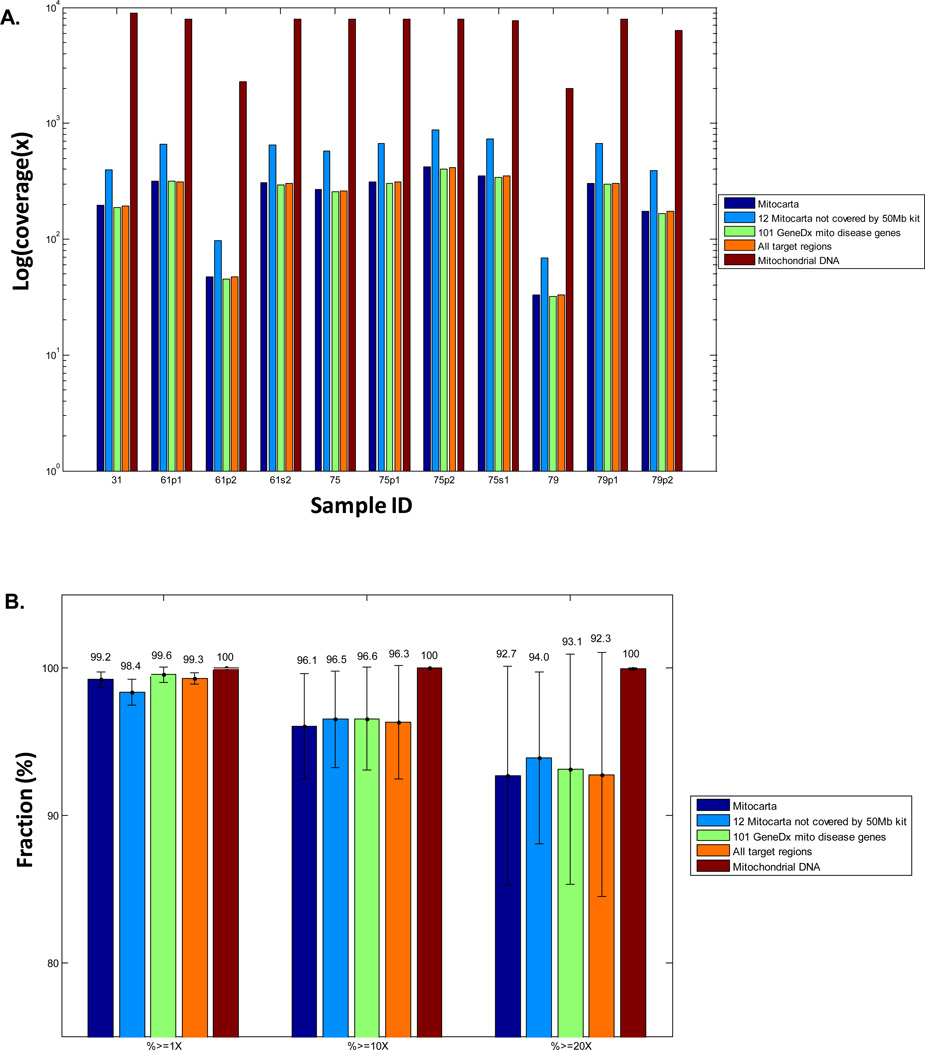
Figure 3. Coverage statistics of 1:100 Mito-Plus Whole Exome capture and Illumina HiSeq NGS analysis of 11 human blood samples.
(A) Median coverage for 5 different gene groups from left to right: all 1,034 mitocarta genes, 12 MitoCarta genes not covered adequatelsy by the standard 50 Mb whole exome kit, 101 nuclear genes on the commercially-available GeneDx Mitochondrial Disease panel (Supp File 1), all target regions, and the mtDNA genome. (B) Fraction (percent) coverage mean and standard deviation across all 11 samples for these same 5 gene groupings at 1X, 10X, and 20X depth of coverage. Detailed coverage data are provided in Supp Tables 3 and 4. Each sample was run in a single HiSeq flow cell lane for NGS analysis.
All MitoCarta nuclear genes are well-covered by the SureSelect custom 1:100 “Mito-Plus Whole-Exome” capture kit
Given the relevance of the MitoCarta nuclear gene list to candidate gene analysis in the diagnostic evaluation of suspected mitochondrial disease, we examined how well the exonic regions of 1,034 MitoCarta genes were covered on the custom 1:100 “Mito-Plus Whole Exome” capture kit. In this analysis, we looked at all exonic regions of these 1,034 MitoCarta genes, rather than just the targeted exonic regions for which we had designed new baits. At least 97.9% of exonic regions for all Mitocarta genes were covered at least 1X when including the two samples (1161p2 and 79) that had generally suboptimal coverage (Fig 3B), while more than 99.1 percent of exonic regions for all MItoCarta genes had 1X coverage in each of the 9 samples that had good overall performance (Supp Table 4A). Improved coverage was also evident for the 12 MitoCarta genes whose exons were not insufficiently covered by the 50 Mb kit design (not including the 4 genes for which we added baits for exons present on alternative assemblies), with 96.8% to 100% of all exonic regions of these genes covered at least 1X in all 11 samples (Fig 3B and Supp Table 4B). Excluding the two relatively poor-performing samples (61p2 and 79), 10X coverage was achieved for 96.8% to 98.1%, and at least 20X coverage was achieved for 95.6% to 97.5%, of the exonic regions of these 12 MItoCarta genes. Thus, these data demonstrate the improved utility of this custom capture kit for whole-exome nuclear gene sequence analysis that includes all known mitochondrial-localized proteins (MitoCarta subset) in suspected mitochondrial disease.
Since an important potential use of this custom capture platform would be in the clinical diagnostic setting to provide focused sequencing of all known mitochondrial disease genes (rather than all mitochondrial-localized proteins), we assessed the performance of our custom kit to cover 101 known mitochondrial disease genes that are currently sequenced on a clinical diagnostic basis using next generation sequencing by the “101 Mitochondrial Disease Nuclear Gene Panel” (GeneDx, Gaithersburg, MD). All 11 samples had at least 1X coverage across 98.17% to 99.93% (Supp Table 4C). Upon exclusion of the two problematic samples (61p2 and 79), 10X coverage was achieved for 97.44% to 98.76%, and at least 20X coverage for 94.35% to 98.02%, in each of the remaining 9 samples for these 101 known mitochondrial disease genes. Future work could focus on assessing patterns of specific nucleotide bases that might be systematically missed by current probes that might be captured by design of additional probes to improve capture of all bases possible in currently known, and newly recognized, mitochondrial disease genes. In addition, the same custom Design B (MitoCarta genes) and Design C (mtDNA genome) probes that we designed can be added with no alteration in expected coverage performance to the recently released v4.0 Agilent whole-exome kit, which targets the same genomic regions as the standard 50 Mb All-Exon design but is rebalanced to provide more even coverage across the 50 Mb nuclear exome (www.genomics.agilent.com).
mtDNA genome heteroplasmy detection
Sensitive detection of low-level heteroplasmic mtDNA mutations is critical to the diagnostic evaluation of suspected mitochondrial disease. While the historic “gold-standard” methodology of mtDNA genome analysis by PCR amplification and Sanger sequencing has a lower detection limit ranging between 30–50% heteroplasmy, it is widely recognized that disease may result from lower level heteroplasmy levels for some pathogenic mutations that might only be detectable with alternative molecular biology methods such as ARMS qPCR (Wang et al., 2011). Further, since heteroplasmy levels can vary between tissues in a given patient, it is desirable to achieve sensitive and reproducible detection of potential heteroplasmic mutations that are at low level in blood to avoid pursuit of invasive tissue biopsies to obtain skeletal muscle or liver in which the mutation level might be enriched. For these reasons, next generation sequencing (NGS) has emerged as the preferred molecular method for mtDNA genome analysis in the clinical diagnostic setting. However, NGS-based mtDNA genome analysis is not currently available in a single platform together with whole-exome nuclear gene analysis, but must be separately considered as a potential etiology in a given patient.
To permit low-level heteroplasmy detection, it is necessary to achieve a very high depth of coverage for the mtDNA genome. However, it is important to recognize that the lower bound of sensitivity for heteroplasmy detection is inherently dependent on several platform-specific parameters including sequencing quality and error rate. For example, with an average base quality (Q) score of 30, heteroplasmy as low as 0.1% can be detected when the base is covered to a depth of coverage over 1000X. When the average base Q score is reduced to 20, heteroplasmy levels as low as 1% can still theoretically be detected.
Sequencing platform-specific error rates directly influence the likelihood that a given mtDNA variant detected in only a small fraction of the NGS reads represents true heteroplasmy versus a sequencing-related error. The PhiX phage genome provides a robust means by which to estimate alignment errors due its genome’s simplicity and no concern for potential heteroplasmic sites. Analysis of the PhiX genome that we spiked into the Illumina HiSeq 2000 runs of Agilent Mito-Plus Whole Exome captured nuclear and mtDNA revealed a sequencing error rate of 5.79% ± 0.42%. This sequencing error is similar to the approximately 5% error rate we had previously observed when analyzing the PhiX genome that was simultaneously sequenced on the SOLiD 3.0 NGS sequencing platform of the mtDNA genome (Supp Fig 1), where the mtDNA genome was amplified by the same two long-range PCR reactions as are used for Affymetrix MitoChip v2.0 analysis (Maitra et al., 2004; Xie et al., 2011). Thus, Illumina HiSeq 2000 and SOLiD 3.0 technologies have similar rates of sequencing error rates in the 5–6% range, which represents the estimated lower bound of being able to confidently discern truly heteroplasmic mtDNA mutations from machine-generated sequencing error. Thus, low-levels of heteroplasmic mtDNA mutations can be reliably detected following different capture and sequencing technologies, but only to the limit determined by the platform-specific sequencing error rate.
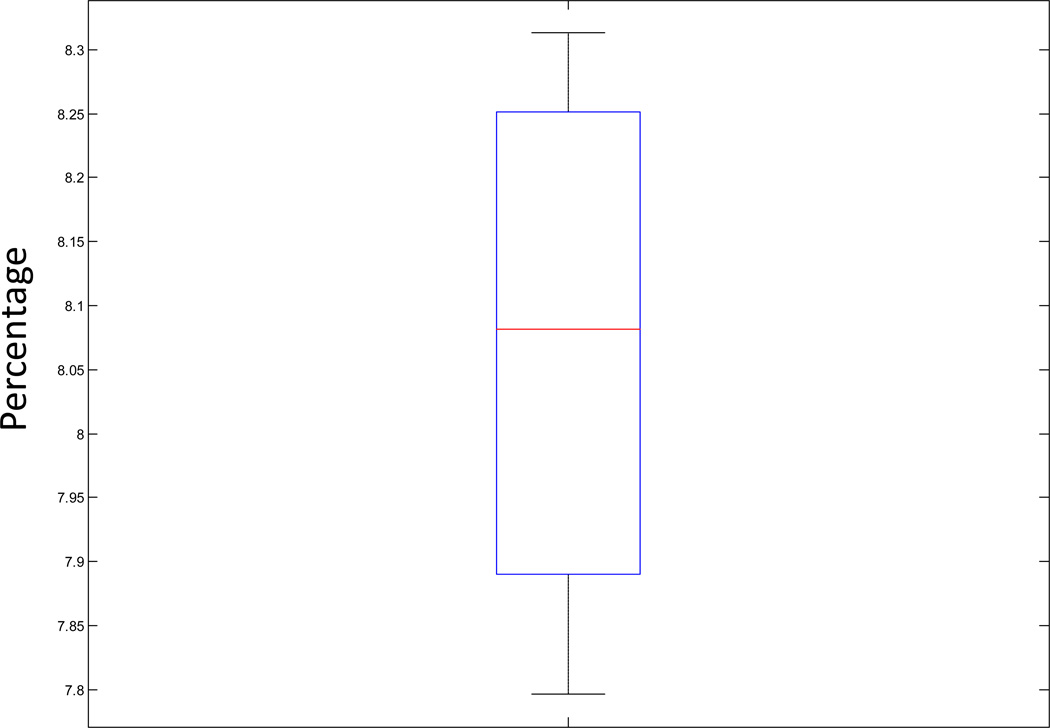
mtDNA heteroplasmy detection sensitivity by NGS is further complicated by the existence of pseudogenes in the nuclear genome that are non-functional but share strong sequence similarity with mtDNA genes (Li et al., 2012). These mtDNA pseudogenes are evolutionary remnants that result from transfer of cytoplasmic mitochondrial DNA sequences into the separate nuclear genome of a eukaryotic organism and are collectively referred to as “nuclear mitochondrial DNA transcripts” (Numt) (Mishmar et al., 2004). The analytic challenge is that an apparently heteroplasmic mtDNA mutation might instead represent off-target Numt capture that was subsequently aligned to the mtDNA genome because of the strong sequence similarities between mtDNA genes and Numts. To understand the potential influence of Numt on heteroplasmy detection sensitivity, we estimated the maximum likelihood that a seemingly heteroplasmic mutation was contaminated by a Numt. We first aligned all reads from each sample to a reference that includes all known Numts (Supp File 4), as well as the mtDNA genome. We counted the number of reads that aligned to the mtDNA genome. All reads were next aligned only to the Numts. The percentage of reads that aligned to the mtDNA genome when the Numts reference was included that can also be aligned to Numts when the mtDNA genome reference is absent provides the upper-bound estimate of the percentage of sequencing reads that align to mtDNA genome but could potentially have originated from Numt contamination. We performed this analysis for 9 randomly selected samples captured by our custom 1:100 mtDNA to nuclear whole-exome capture kit and each sequenced on one HiSeq 2000 flow cell lane, with alignment details and calculations summarized in Supp Table 5. In all 9 samples, the upper bound of Numt contribution to heteroplasmy sensitivity detection ranged from 7.80% to 8.31% (8.10 ± 0.18%) (Fig 4). Based on this observation, we can conclude with greater than 99.9% confidence that an observed heteroplasmic mutation is not from Numt contaminations if it is present in at least 8.64% of sequence reads. However, this is a very conservative estimate since we did not account for the fact that mtDNA outnumbers nuclear DNA by 1–2 log orders of magnitude (Li et al., 2012; Li and Stoneking, 2012). Thus, the true lower bound for mtDNA heteroplasmy detection sensitivity is likely much lower than 8%. Still, even 8% heteroplasmy detection sensitivity already represents a great improvement over the 30% to 50% lower bound for mtDNA heteroplasmy detection that is achieved by the “gold-standard” of Sanger sequencing. More importantly, 8% falls below the level of heteroplasmy for a pathogenic mtDNA mutation that is generally likely to cause clinical manifestations of classic mitochondrial disease. While alternative mtDNA capture approaches such as long-range PCR can provide even greater heteroplasmy sensitivity, and even large deletion detection sensitivity, by NGS analysis (Zhang et al., 2012), these data demonstrate that the Agilent custom “1:100 Mito-Plus Whole-Exome” kit offers good heteroplasmy detection sensitivity together with the distinct advantage that no separate technical or analytic methodologies for mtDNA genome sequence analysis are required at the time of sample processing for whole-exome analysis.
Figure 4. Analysis of maximal influence of Numts on reliable heteroplasmy detection sensitivity.
The boxplot conveys the ratio of reads aligned to the mitochondrial DNA genome that might originate from Numt contamination across 8 samples captured with the 1:100 mtDNA:nuclear Mito-Plus Whole-Exome custom capture kit and sequenced one sample per flow cell lane on the Illumina HISeq 2000. The length of the box represents the 25th to 75th interquartile range, the interior horizontal line represents the median, and vertical lines issuing from the box extend to the minimum and maximum values of the analysis variable. Detailed coverage data is provided in Supp Tables 5.
Technical Reproducibility
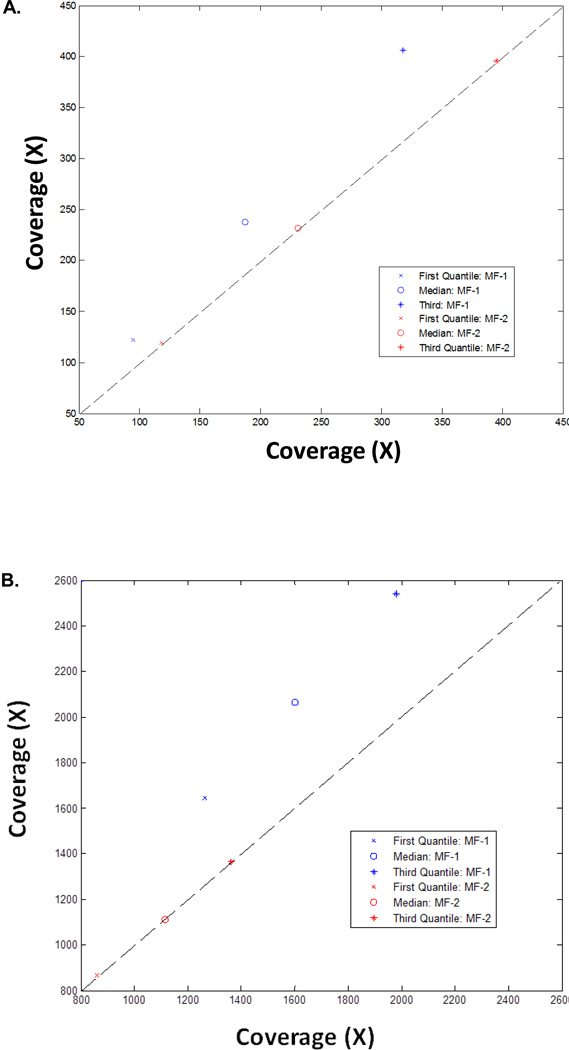
We examined the technical reproducibility of the custom “Mito-Plus whole-exome” kit to capture both nuclear exome targets and the mtDNA genome. Two capture libraries were separately prepared using the 1:500 (sample “MF1”) and 1:1000 (sample “MF2”) blend of mtDNA genome to whole-exome design using blood genomic DNA from the same mitochondrial disease patient. Each library was further split into two, differentially bar-coded, and then sequenced in separate HiSeq 2000 flow cell lanes. Therefore, this data set provides technical replicates both at the library preparation and sequencing levels. Highly reproducible coverage statistics were obtained. Overall short reads alignment characterizations/traits were strongly correlated among technical replicates for all target regions (Fig 5A) and specifically for the mtDNA genome (Fig 5B), as correlation coefficients for both analyses were approximately 1.
Figure 5. Mito-Plus Whole Exome capture reliably detects heteroplasmic mtDNA mutations.
(A) Coverage distribution statistics for replicate experiments for (A) all targeted regions, and (B) the whole mtDNA genome. MF-1 and MF-2 represent separate datasets from two flow cell lanes run on blood DNA captured with the 1:500 and 1:1000 molar ratios of mtDNA:nDNA, respectively, from the same mitochondrial disease patient having a known mtDNA-encoded ND5 gene heteroplasmic mutation (m.13513G>A). Detailed coverage data are provided in Supp Tables 6A and 6B, respectively.
In addition, this sample was used to assess the technical reproducibility of heteroplasmic mtDNA mutation detection by this platform since the sample was shown by Sanger-based sequencing to harbor a 30% heteroplasmic G to A transition mutation at position 13513 of the mt-ND5 gene. The mt-ND5 heteroplasmic mutation was present at a level of 65.0% (723 A / 1112 total reads) in the MF1-1 data set, at a level of 64.9% (803 A / 1238 total reads) in the MF1-2 data set, at a level of 63.4% (393 A / 620 total reads) in the MF2-1 data set, and at a level of 64.8% (411 A / 634 total reads) in the MF2-2 data set. Thus, heteroplasmy level determination from the mtDNA sequence data generated is highly reproducible, and likely more accurate than traditional Sanger sequencing, as is consistent with the growing recognition that NGS is becoming the new “gold-standard” for mtDNA heteroplasmy detection over Sanger sequencing (Zhang et al., 2012).
CONCLUSIONS
We have developed a custom “1:100 Mito-Plus Whole-Exome” Agilent capture kit that allows simultaneous enrichment for subsequent NGS-based sequence analysis of all currently known nuclear MitoCarta genes and the entire mtDNA genome, as is highly relevant to the diagnostic evaluation of suspected mitochondrial disease. By being embedded in a whole-exome capture kit, this mitochondrial-optimized analysis nevertheless retains the simultaneous opportunity for discovery both of phenotypically-overlapping disorders that may not directly involve the mitochondria as well as of novel disease genes. Further, our data supports that the custom “1:100 Mito-Plus Whole-Exome” design offers reliable mtDNA mutation heteroplasmy detection sensitivity together with the distinct advantage that no separate technical or analytic methodologies for mtDNA genome sequence analysis are required by the investigator at the time of sample processing for whole-exome analysis. Thus, this design holds value for providing targeted enrichment of the whole-exome for sequence-based genetic diagnosis in both research and clinical diagnostic applications where the relevance of mtDNA is well-recognized, as well as in cases where the potential contributory role of mtDNA mutations may otherwise be overlooked.
METHODS
mtDNA genome bait and blend design
Sequences targeting the entire mtDNA genome were created in eArray (Agilent) by standard 1X tiling across the target hg19 mitochondrial loci. These baits for “Design C” (mtDNA genome) were factory blended into the nuclear baits at either equimolar ratio or reduced concentration by 10, 50, 100, 200, 500, or 1000-fold less than the nuclear baits. The accession number for the Agilent mtDNA genome design bait library was ELID # 320851 (https://earray.chem.agilent.com/earray).
Nuclear mitochondrial gene set optimization
Bioinformatics analysis of the SureSelectXT 50 Mb All Exon PLUS Targeted Enrichment Kit was performed to determine the exon level coverage of 1,034 known mitochondria-localized “Human MitoCarta” genes (Pagliarini et al., 2008). Baits were designed for 16 of these nuclear genes shown to have less than 80% of their exons covered (ELID #329521) (https://earray.chem.agilent.com/earray). These baits were factory added in equimolar ratio to the SureSelectXT 50 Mb All Exon PLUS Targeted Enrichment (Agilent part number 5190-2867).
Exome Sequencing
Targeted enrichment was performed using Agilent Technologies (Santa Clara, CA) SureSelectXT 50 Mb All Exon PLUS Targeted Enrichment Kit that included custom mitochondrial genome content in varying mitochondrial:nuclear capture bait molar ratios, namely: Blend A – 1:1; Blend B – 1:10; Blend C – 1:50; Blend D – 1:100; Blend E – 1:200; Blend F – 1:500; Blend G – 1:1000. Patient sample capture libraries were prepared as described in the kit manual, and were 2 × 101 base pair paired-end sequenced on an Illumina (SanDiego, CA) HiSeq 2000 Next-Generation Sequencing system using v3.0 SBS chemistry with average flowcell lane cluster densities of ~700 - 800 K/mm2. One sample was analyzed per flowcell lane to obtain a minimum 10x read depth of ~96% for the targeted nuclear exome. The mitochondrial genome coverage varied depending on the mitochondrial:nuclear blend ratio.
Exome Data Analyses
BWA (version 0.5.9-r16) was used to align the sequence reads to the human reference genome GRCh37 downloaded from the 1000 Genomes Project website (http://www.1000genomes.org/). Samtools (version 0.1.12 or r859) was used to remove potential duplicates (with rmdup command), and make initial SNP and indel calls (with pileup command). A custom program was developed and used to further refine the SNP and indel calls. The custom program uses a false discovery rate approach to adjust raw base counts at a candidate position after Benjamini and Hochberg correction based on quality values of all bases. A coverage depth cutoff of 10X is then applied. Depth of coverage is calculated based on the alignment file using samtools.
Sequencing Error Estimation using PhiX phage genome
BWA (version 0.5.9-r16) was used to align HiSeq sequence reads to the PhiX phage genome (NC_001422.1) downloaded from NCBi. BioScope was used at default settings to align SOLiD sequence data. Samtools was applied to remove duplicates and obtain the number of high quality base reads for different strands and alternative bases at a given base position. Sequencing error rate was estimated as the sum of the number of bases different than the consensus call made by Samtools over the depth of coverage at a given base position.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the families who participated in theses studies; the Clinical Translational Research Core and the Molecular Diagnostic Laboratory at The Children’s Hospital of Philadelphia for their assistance with blood DNA extraction and storage; to Emily Place, MS, for her assistance with sample organization and processing; and to Joseph White for his assistance with data transfer and analysis. This work was funded in part by NIH award R03-DK082446 (M.J.F.) and The Foerderer Award for Excellence from the Children’s Hospital of Philadelphia (M.J.F. and X.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations or the National Institutes of Health.
LIST of ABBREVIATIONS USED
- mtDNA
mitochondrial DNA
- Numt
nuclear mitochondrial DNA transcript
- NGS
next-generation sequencing
- NCBI
National Center for Biotechnology Information
- ELID
electronic library ID
Footnotes
COMPETING INTERESTS:
M.G., O.H., and E.L. are employed by Agilent Technologies.
AUTHORS’ CONTRIBUTIONS: M.J.F., X.G., and E.L. designed the study. E.L., M.G., and O.H. designed the custom bait design for the MitoCarta and mtDNA genome probes with input from D.W., M.F., and X.G.. M.C. performed the library preparation and sequencing under the guidance of E.A.P. D.M. and D.W. tabulated Numts. E.R. performed SOLiD sequencing. X.G. and M.X. performed all NGS data bioinformatic analyses, including PhiX control analyses as well as generation of all figures and tables. M.J.F., X.G., M.X., M.C., and E.L. wrote the manuscript.
REFERENCES
- Asan Xu Y, Jiang H, Tyler-Smith C, Xue Y, Jiang T, Wang J, Wu M, Liu X, Tian G, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, Laskowski A, Garone C, Liu S, Jaffe DB, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra110. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Cohen BH. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- Kiialainen A, Karlberg O, Ahlford A, Sigurdsson S, Lindblad-Toh K, Syvanen AC. Performance of microarray and liquid based capture methods for target enrichment for massively parallel sequencing and SNP discovery. PLoS One. 2011;6:e16486. doi: 10.1371/journal.pone.0016486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schroeder R, Ko A, Stoneking M. Fidelity of capture-enrichment for mtDNA genome sequencing: influence of NUMTs. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13:R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, et al. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome research. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick E, Place E, Falk MJ. Molecular genetic testing for mitochondrial disease: From one generation to the next. Neurotherapeutics. 2012 doi: 10.1007/s13311-012-0174-1. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat. 2004;23:125–133. doi: 10.1002/humu.10304. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker EJ, Compton AG, Thorburn DR. Recent advances in the genetics of mitochondrial encephalopathies. Curr Neurol Neurosci Rep. 2010;10:277–285. doi: 10.1007/s11910-010-0112-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Venegas V, Li F, Wong LJ. Analysis of mitochondrial DNA point mutation heteroplasmy by ARMS quantitative PCR. Curr Protoc Hum Genet Chapter. 2011;19 doi: 10.1002/0471142905.hg1906s68. Unit 19 16. [DOI] [PubMed] [Google Scholar]
- Xie HM, Perin JC, Schurr TG, Dulik MC, Zhadanov SI, Baur JA, King MP, Place E, Clarke C, Grauer M, et al. Mitochondrial genome sequence analysis: a custom bioinformatics pipeline substantially improves Affymetrix MitoChip v2.0 call rate and accuracy. BMC Bioinformatics. 2011;12:402. doi: 10.1186/1471-2105-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cui H, Wong LJ. Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin Chem. 2012;58:1322–1331. doi: 10.1373/clinchem.2011.181438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.