Abstract
Human infections with arthropod-borne Rickettsia species remain a major global health issue, causing significant morbidity and mortality. Epidemic typhus due to Rickettsia prowazekii has an established reputation as the ‘scourge of armies’, and as a major determinant of significant ‘historical turning points’. No suitable vaccines for human use are currently available to prevent rickettsial diseases. The unique lifestyle features of rickettsiae include obligate intracellular parasitism, intracytoplasmic niche within the host cell, predilection for infection of microvascular endothelium in mammalian hosts, association with arthropods and the tendency for genomic reduction. The fundamental research in the field of Rickettsiology has witnessed significant recent progress in the areas of pathogen adhesion/invasion and host immune responses, as well as the genomics, proteomics, metabolomics, phylogenetics, motility and molecular manipulation of important rickettsial pathogens. The focus of this review article is to capture a snapshot of the latest developments pertaining to the mechanisms of rickettsial pathogenesis and immunity.
Keywords: endothelium, genomics, innate immunity, metabolomics, pathogenesis, plasmids, proteomics, Rickettsia, spotted fever, typhus
Rickettsial diseases: old & new
The genus Rickettsia comprises etiologic agents of Rocky Mountain spotted fever (RMSF) and Mediterranean spotted fever (MSF), caused by Rickettsia rickettsii and Rickettsia conorii, respectively, and typhus syndromes, namely epidemic and endemic typhus due to infection with Rickettsia prowazekii and Rickettsia typhi, respectively. Historically, rickettsial diseases have had a longstanding reputation as severe human infectious diseases leading to death, disability and the ‘scourge of armies’ during World Wars I and II. In general, rickettsiae are small, aerobic, coccobacillary a-proteobacteria with a number of unusual features, exemplified by their strict intracellular lifestyle within host cell cytoplasm, fastidious growth requirements, association with arthropods with the ability to be maintained in nature via transovarial and trans-stadial transmission, and adaptability to maintain lifecycles involving vertebrate and invertebrate hosts. Although pathogenic Rickettsia species have traditionally been divided into spotted fever and typhus groups, a detailed contemporary phylogenetic analysis proposed by Gillespie et al. has now classified more than 20 validated species within the genus Rickettsia into four groups [1], including the ancestral group (Rickettsia bellii and Rickettsia canadensis; associated with ticks); the typhus group (R. prowazekii and R. typhi; associated with fleas and lice); the spotted fever group (SFG; Rickettsia aeschlimanii, Rickettsia africae, R. conorii, Rickettsia heilongjiangensis, Rickettsia helvetica, Rickettsia honei, Rickettsia japonica, Rickettsia massiliae, Rickettsia montanensis, Rickettsia parkeri, Rickettsia peacockii, Rickettsia rhipicephali, R. rickettsii, Rickettsia sibirica and Rickettsia slovaca; which are associated with ticks) and finally a transitional group (Rickettsia australis, Rickettsia akari and Rickettsia felis; associated with ticks, mites and fleas).
Among rickettsial diseases known to human-kind, RMSF is the most severe SFG rickettsiosis, with significant mortality rates of 20–30% in the absence of timely intervention with the appropriate antibiotic. As a potentially fatal tick-borne infection, RMSF is an illness notifiable to the CDC in the USA, for which the reported annual incidence has risen from less than two to approximately seven cases per million people between 2000 and 2007. Fortunately, the case fatality rates have continued to decline in the postantibiotic era. In central and southern America, RMSF is endemic in various coastal, urban, suburban and deep forest regions of Argentina, Brazil, Costa Rica, Colombia, Panama and Mexico. MSF due to R. conorii is endemic in the Mediterranean basin, and is generally considered to be a milder disease than RMSF; however, adult mortality rates as high as 21% have been reported recently, and variations in R. conorii strain virulence are highlighted by death rates as high as 29% with Israeli spotted fever compared with 13% with the Malish strain [2]. Another important aspect of R. conorii transmission to humans is the presence of a characteristic ‘eschar’ (also known as tache-noire) at the site of tick bite [3,4], a phenomenon also known to occur in patients suffering from other spotted fever rickettsioses, and supported by the recent rare finding of an abdominal eschar in a fatal case of RMSF in Costa Rica [5]. Furthermore, the ability of several other spotted fever species such as R. aeschlimanii, R. massiliae, R. helvetica and R. slovaca, which were considered to be nonpathogenic, to cause human disease is also being recognized. Although the first confirmed human case of infection with R. parkeri was reported in 2004, the pathogen was first isolated in 1937 from Gulf Coast ticks Amblyomma maculatum in Texas [6]. Furthermore, the possibility that rickettsiae can be transmitted by previously unsuspected arthropod vectors in areas with very low prevalence of human rickettsioses also suggests the pathogens’ exploitation of mechanisms to adapt to new ecological niches and maintain their virulence [7].
Owing to its historic impact during times of war, reports of outbreaks as late as the 1990s, and potential for rapid spread through body lice among human populations under poor socioeconomic circumstances, contributing to disruption of hygiene, epidemic typhus caused by R. prowazekii also remains an important human threat. Available estimates attribute more than 3 million human deaths in Russia alone, during the wake of World War I and the Russian revolution, to louse-transmitted R. prowazekii infection. Although historic epidemics occurred in relation to World Wars I and II, epidemic typhus outbreaks in refugee camps of Burundi, and evidence for re-emergence in Algeria, Peru and Russia, have been reported in the not-so-distant past [8]. With a mortality rate of up to 60% in untreated cases, epidemic typhus is a severe infectious disease of humans, often difficult to differentiate from other infectious etiologies owing to early ‘flu-like’ symptoms. Also, R. prowazekii is the only rickettsial species able to persist subclinically in convalescent patients and later manifest as a relatively milder form of disease called Brill–Zinsser disease, or recrudes-cent typhus. Taken together, human rickettsioses continue to pose a significant strain on human health across the globe [8,9], and no suitable vaccines are currently available to prevent these infections. There also exists precedence, as well as historical evidence, for the deliberate misuse of highly pathogenic species R. prowazekii as a biological war strategy [10]. Yet another important concern is the potential for natural emergence and/or intentional development of strains that display resistance to antibiotics used for therapy [11]. Accordingly, R. prowazekii remains on the CDC list of select agents and National Institute of Allergy and Infectious Diseases list of category B priority pathogens, whereas other pathogenic rickettsiae are classified as category C, for fundamental and translational research aimed against the bioterror threats.
In their natural lifecycle, rickettsiae are known to associate with arthropod vectors. Accordingly, human rickettsial infections involve exposure to the infected vectors. With the exception of epidemic typhus due to R. prowazekii, which can be spread among close communities of people through body lice feeding on an infected person and then passing the infection to others, humans primarily serve as accidental, dead-end hosts. Transovarial and trans-stadial passage of SFG rickettsiae in tick vectors ensures their survival without the need for a multihost reservoir system. Tick-borne rickettsiae are transmitted to humans by tick salivary secretions when a tick attaches to the host and takes a blood meal. The characteristic features of SFG rickettsioses typically include the formation of an ‘eschar’ at the site of tick attachment and feeding, and later development of a petechial/maculopapular rash, initially on the outer parts of the body, such as extremities, followed by spread to the trunk. By contrast, transmission of epidemic typhus to humans involves contamination of bite sites by the feces of infected lice. SFG rickettsiae cause varying degrees of harm to their tick vectors; however, lice harboring R. prowazekii are unable to survive the infection, and usually die within a couple of weeks. In humans, a number of rickettsial species within the spotted fever and typhus groups cause diseases with apparently similar clinical manifestations, but varying severity.
It is now well established that pathogenic rickettsiae preferentially infect vascular endothelial cells lining the small- and medium-sized vessels during infection of their mammalian hosts. Mononuclear phagocytes represent a minor target in disseminated infection, but are the major cell types infected with rickettsiae in the eschar where arthropods inoculate rickettsiae into the dermis of the skin. Identified at the end of the nineteenth century and considered to be a seemingly simple and passive monolayer of cells guarding blood vessels, the vascular endothelium represents a highly evolved and dynamic system that not only acts as a barrier between the extravascular and intravascular tissues, but also actively regulates critically important physiological functions, including maintenance and regulation of vascular permeability, wound repair, tethering, rolling, extravasation of leukocytes and the coagulation pathway. Therefore, unsurprisingly, the common denominators for the predominant pathological sequelae and major complications of rickettsial diseases include endothelial cell activation and dysfunction, which lead to compromised vascular integrity and loss of barrier function, manifesting as noncardiogenic pulmonary edema, acute respiratory distress, complications of the CNS and failure of multiple organ systems, yet abnormalities, such as disseminated intravascular coagulation, are very rarely seen in severe, complicated cases of infection.
Although the major host target during human rickettsial infections is the vascular endothelium, the majority of initial studies aimed at establishing intracellular growth requirements and/or patterns were conducted with either L929 or Vero cells, both of which are fibroblastic and nonphagocytic in nature [12]. Vascular endothelial cells, whose primary function is to maintain vascular homeostasis, have now emerged as key immunoreactive cells involved in host defense and inflammation, in part owing to their ability to synthesize and secrete many growth factors, cytokines, chemokines, adhesion molecules and vasoactive substances capable of exerting significant autocrine and/or paracrine effects on the microvascular, as well as other target cell, functions [13]. It is encouraging to note that despite their reputation as ‘difficult to work with’ pathogens because of their obligate intra-cellular nature, lack of well-defined systems for molecular manipulation and genetic intractability, and the need for specialized infrastructure, including biosafety level 3 containment to work with live organisms, the last few years have witnessed impressive progress in nearly all aspects of Rickettsiology, including sequencing of a series of rickettsial genomes leading to more robust phylogenetics, comparative genomics, proteomics, genetic manipulation, discovery of rickettsial plasmids and other important aspects of rickettsial biology. The knowledge of basic molecular and cellular concepts of rickettsial pathogenesis and immunity has also advanced significantly. With appreciation for a series of recent reviews elegantly summarizing different topics pertaining to human rickettsioses [14–16], and an entire volume dedicated to Rickettsiales as intracellular pathogens [17], this article attempts to primarily focus on the latest advances and progress in our understanding of mechanisms underlying host–pathogen interactions and rickettsial pathogenesis/immunity.
Adhesion to & entry into host cells
In order to survive, multiply and successfully establish the infection, Rickettsia needs to adhere to and invade target host cells. Early investigations of adhesion-invasion mechanisms revealed that drug-induced alterations of host cell cyto-skeleton or inactivation of rickettsiae have significant adverse effects on their entry into host cells. Owing to the fact that viability of the invading bacteria and metabolic activity of the host cell were both determined as the requisites for intracellular uptake of rickettsiae, the process was accordingly termed ‘induced phagocytosis’ [18,19]. SFG rickettsiae possess two well-characterized, surface-exposed proteins, known as OmpA and OmpB, whereas OmpA is not found on typhus group organisms. Out of these, OmpA was initially identified as a protein critical for R. rickettsii adhesion to host cells [20]. Other putative rickettsial adhesins, encoded by gene Adr1 (RC1281) in R. conorii and Adr2 (RP828) in R. prowazekii were subsequently identified by a proteomics-based analysis and proposed to be involved in rickettsial invasion [21].
Recent bioinformatics analyses have revealed at least 17 surface cell antigens (Sca), encoding proteins similar to autotransporter proteins, involved in rickettsial adhesion to host cell receptors. The four identified are Sca0 (OmpA), Sca1, Sca2 and Sca5 (OmpB), which have been shown to play important roles in rickettsial adhesion and/ or invasion [15]. Sca1 is an adhesin for R. conorii to attach to mammalian epithelial and endothelial cells, but has no role in invasion [22]. Sca2 is expressed in R. conorii and is conserved among many SFG species, and participates in mediating interactions with target mammalian cells [23]. Sca2 in R. parkeri plays a role as a formin-like mediator of actin-based motility, indicating that some of the bacterial Sca proteins may interact with host proteins to promote survival of rickettsiae [24]. Sca4 of rickettsiae colocalizes with vinculin in cells at the site of focal adhesions, and binds and activates vinculin through two vinculin-binding sites that are conserved in all rick-ettsial species [25]. Interestingly, Ku70, a subunit of nuclear DNA-dependent protein kinase localized in the cytoplasm as well the plasma membrane, serves as a receptor for rickettsial OmpB and, thus, plays an important role in R. conorii internalization [26,27]. Evidence in support of this finding further suggests that ubiquitination of host cell Ku70 occurs during rickettsial infection by recruitment of a ubiquitin ligase c-cbl to the sites of entry [28]. Host cell actin polymerization also plays an important role in rickettsial internalization, as indicated by potential contributions of upstream signaling mechanisms involving Cdc42 (a GTPase), phosphoinositide 3-kinase, c-Src and possibly other protein tyrosine kinases, resulting in the activation of Arp2/3 complex [29,30]. A recent report further documents the participation of clathrin and caveolin-2-dependent endocytosis in the internalization process of rickettsiae [28]. OmpA-mediated rickettsial adherence to endothelium is shown to be dependent upon its interaction with α2β1 integrins on the host cell surface. This study also demonstrates that rickettsial adhesion is dependent on a discontinuous RGD motif of OmpA [31]. Taken together, it appears that multiple interactions between the rickettsial ligands on its outer membrane and host cell surface entities serving as their specific receptor counterparts, and consequent activation of downstream signaling mechanisms, are critical for the process of entry and acquisition of an intracellular host niche.
The internalization of rickettsiae into host cells is a quick and efficient process that occurs within a few minutes after initial contact. Rickettsiae likely utilize membranolytic proteins, hemolysin C and phospholipase D, encoded by tlyC and pldA genes, respectively, to disrupt the phagosomal membranes and gain access to host cytosol [32–34]. A phospholipase A2 (PLA2)-like activity is also predicted to be involved in rick-ettsial entry into the host cells [35]. Published evidence renders support to this hypothesis by showing that the gene RT0522 (pat2) in R. typhi encodes PLA2 activity. RT0522 protein is expressed during R. typhi infection of Vero76 cells, possesses a bona fide PLA2 activity and is secreted into the host cell cytoplasm [36]. More recently, RT0590 (pat1) encoded by R. typhi has also been shown to have PLA2 activity, which is required for adherence and entry into host cells during infection. Unlike RT0522 (pat2), which appears to be pseudogenized in 76% of Rickettsia genomes, R. typhi pat1 is ubiquitously present in all rickettsial genomes and, despite having undetectable transcript levels, it is expressed, secreted into host cytosol, and functionally activated by unidentified host activator(s) for its PLA2 activity [37].
Pathogenic rickettsiae infect and replicate in vitro in a number of different cell types, including fibroblasts, HeLa cells and other cancer cell types [26,38,39]. However, a unique property displayed by rickettsiae in vivo is their affinity to preferentially infect vascular endothelial cells lining the small- and medium-sized blood vessels in humans and in established animal models of infection [40–44]. As a consequence, rickettsiae invade, infect and disseminate through the endothelium damaging vascular networks.
Actin-based motility: intracellular movements & intercellular dissemination
Once intracellular, SFG rickettsiae form a polar actin tail, which not only facilitates their movements within and from cell-to-cell, but also plays a key role in the establishment, dissemination and pathogenesis of resulting diseases. On the other hand, typhus organisms either do not display actin-based motility, as is the case with R. prowazekii, or have erratic motility patterns exhibited by R. typhi [45,46]. Thus, the primary cell-to-cell spread mechanism utilized by R. prowazekii involves the necrotic lysis of a heavily-infected cell, as a consequence of intracellular replication by binary fission, leading to significantly higher levels of accumulation in comparison to spotted fever rickettsiae and resultant host cell lysis [44]. For R. typhi, cell-to-cell spread is significantly reduced due to shorter actin tail lengths; however, the overall rate of movement is very similar when compared with R. rickettsii [45]. A Wiskott–Aldrich syndrome protein (WASP) termed RickA is sufficient to direct actin-based motility via activation of actin nucleation and Arp2/3 complex recruitment. As expected, RickA (RC0909) is present in R. conorii, but not found in R. prowazekii [47]. A notable exception among SFG species lacking actin-based motility is R. peacockii, a species nonpathogenic to humans. At the molecular level, the gene rickA in R. peacockii is disrupted by a transposable active insertion sequence element named ISRpe1, which contains the open reading frame (ORF) sequences for recombinase/transposase, and the capability to transcriptionally express the transposase [48]. The actin-based motility in R. typhi, although erratic in comparison to linear movements of spotted fever rickettsiae, in the absence of RickA suggests the possibility that redundant actin-polymerization mechanisms exist in pathogenic rickettsiae. This hypothesis is further substantiated by the presence and expression of RickA in Rickettsia raoultii, a recently discovered spotted fever species isolated from Dermacentor ticks. R. raoultii encodes a 565 amino acid protein highly homologous to RickA in other rickettsiae, but still displays a defect in actin-based motility during in vitro infection of L929 cells [49]. R. bellii, belonging to the ancestral group of rickettsiae, encodes an ortholog of RickA, frequently colonizes the host nucleus and has been shown to form actin tails for movement into the host cell nucleus. Upon entering into the nucleus devoid of an Arp2/3 complex, which is required for actin tail formation, the bacterium apparently gets trapped, resulting in the abolishment of actin-based motility. Presumably, R. bellii utilizes RickA to enter into the host nucleus where it multiplies locally until the nuclear membrane is disrupted, releasing the bacteria [50]. The sequence divergence between R. bellii and R. conorii RickA might explain the failure for the lack of detection of RickA expression in crude bacterial extracts of R. bellii using a monoclonal antibody against R. conorii RickA [49]. Actin tails are also observed in R. australis [51] and R. felis [52], both of which belong to the transitional group of Rickettsia. Furthermore, RickA expression in R. felis has been demonstrated by Western blotting of crude extracts [49]. Based on these findings illustrating the presence of actin-based motility in Rickettsia belonging to different groups (SFG, transitional group and ancestral group), with the exception of typhus rickettsiae, it is tempting to speculate that actin-based motility represents an important lifestyle feature for the genus Rickettsia, which during the course of evolution appears to have been lost in the typhus species while being retained in the other subgroups [53].
RickA is a nucleation-promoting factor capable of activating the Arp2/3 complex and inducing actin polymerization in vitro [46,47]; however, rickettsial comet tails display an unbranched organization and absence of Arp2/3 sub units. In addition, evidence for motility in cells expressing verprolin, cofilin and acidic domain of neuronal-WASP suggest that, unlike Listeria and Shigella, rickettsial actin-based motility also involves an Arp2/3-independent mechanism [54–56]. In support of this notion, recent studies have further implicated an autotransporter protein Sca2 in Arp2/3-independent motility of R. rickettsii and R. parkeri [24,57]. A number of SFG species, including R. africae, R. akari, R. australis, R. conorii and R. rickettsii encode full-length Sca2 containing an N-terminal Sec-dependent secretion signal, four putative WASP homology (WH2) domains, proline-rich regions (similar to formins) and autotransporter domains on the C-terminus. By contrast, R. prowazekii only expresses a truncated version of Sca2 lacking the first three WH2 domains, proline-rich segments and secretory signal sequence. Predictably, R. typhi also encodes shortened Sca2 (1483 amino acids compared with 1821 amino acids in SFG rickettsiae) with overlapping autotransporter and WH2 domains, but divergent formin homology 1 domain and secretion signal [57]. R. prowazekii effector, RalF, also contains a proline-rich region, and can potentially serve as a formin modulator to play a role in actin polymerization [58]. These novel findings, suggest that motile Rickettsia species in the SFG utilize eukaryotic formin-like properties of Sca2 as a primary mechanism for moving within and spreading from cell to cell. This may also explain the lack of motility in R. canadensis, which lacks actin-based motility and possesses an intact RickA, but has a number of deletions in the N-terminal domain of Sca2 [24,49,51].
Secretion systems of intracellular rickettsiae
Intracellular bacteria are known to possess sophisticated secretion machineries that allow them to deliver effector proteins into the host cell and modulate function(s) of various protein(s) with an ultimate goal of subverting and/or exploiting eukaryotic host functions to gain a survival advantage. In this context, bacteria use membrane-associated transporter systems known as type IV secretion systems (T4SSs) to accomplish delivery of effectors into target host cells. These secretion systems are known to play a role in the transfer of DNA to other bacteria and host cells, in both acquisition of genetic material from, as well as release into, the extracellular environment, and in the injection of toxin(s), virulence factors or other effector mediators into the host cytoplasm. The prototypical T4SS of the pathogen Agrobacterium tumefaciens delivers oncogenic nucleoproteins into plant cells and is composed of 11 VirB proteins, namely VirB1–VirB11, in addition to a coupling protein VirD4 with nucleoside triphosphatase activity [59]. The VirB proteins in this system are divided into channel components (VirB6–VirB10), energetic components (VirB4 and VirB11 as nucleoside triphosphatases) and the pilus-associated components (represented by VirB2, VirB3 and VirB5). From the treasure trove of data arising from the sequencing and annotation of entire genomes, the presence of most, if not all, of the T4SS proteins has now been documented in pathogenic rickettsiae. For example, a total of 15 ORFs closely related to prototypical T4SS of A. tumefaciens were identified in the R. typhi genome, which represent orthologs of VirD4, VirB3, VirB10, VirB11, two copies each of VirB4, VirB8 and VirB9, and five different genes with homology to VirB6 [60]. Recently, a comparative bioinformatics approach aimed at identifying the components of the T4SS further identified the presence of much shorter sequences of VirB1, VirB2 and VirB7 in 13 rickettsial genomes [61]. Notably, the VirD4 gene is highly conserved, particularly within the five motifs present in the C-terminal domain, and is implicated in the binding to effector molecules of T4SS. VirB5, which is required for pilus assembly, is lacking, and it is possible that this gene may have been purged from the rickettsial genomes due to their obligatory intracellular lifestyle with host cytosol as the primary host niche and the tendency for reductive evolution [61]. Except for a report documenting their presence in R. felis [52], pili are not observed in other rickettsial species [1,61]. Furthermore, pili associated with R. felis cannot be encoded by rick-ettsial T4SS genes, because the gene encoding the minor pilus subunit (a homolog of VirB5) is absent in rickettsial genomes, an observation further supported by the identification of only the major pilus subunit homolog (VirB2), which has been shown to be surface presented in Anaplasma marginale [1,61,62]. R. typhi genome encodes two VirB9 genes (VirB9a and VirB9b) sharing similarity in their N-terminal domain and containing predicted signal peptides. The C-terminal domain of VirB9b (annotated as TrbG), which is required for interactions with VirB7 and VirB7-like lipoproteins, is highly truncated, contains only the residues belonging to the first β-strand and cannot possibly assist in the assembly of T4 pore [61,63]. However, gene fusion assays reveal that the signal peptide of VirB9b and three VirB6 homologs (VirB6c, VirB6d and VirB6e) are functional and capable of mediating secretion in Escherichia coli as a surrogate host [64]. Except for the absence of VirB6e in R. massiliae and a split VirB6d gene in R. bellii, all rickettsial genomes encode five copies of VirB6 gene (VirB6a through VirB6e), which are highly conserved across all genomes. It is important to consider, however, that VirB6 genes constitute the most variable components of the T4SS due to a very high degree of divergence between the five copies of VirB6 genes, exemplified by approximately 80% divergence in VirB6/TrbL domains. It is, therefore, possible that this divergence in VirB6 genes might result in the formation of multiple channels for DNA import/export, leading to increased lateral gene transfer between organisms [61,63,65,66]. Regarding other components of the T4SS, the VirB2 forms part of the T4 channel. Interestingly, the VirB2 homologs of Anaplasma phagocytophilum are differentially expressed in tick and human cells, yet there is no report on the expression of T4SS genes for Rickettsia species in host cells [61–63]. Despite being located directly upstream of the VirB8b gene, VirB7 could not initially be annotated in rickettsial genomes, owing to its small size and divergence from other bacterial VirB7 and VirB7-like proteins. However, computational and manual alignment of RP288 and its orthologs from other bacteria identified a Cys lipoprotein-processing site and a potential P[ILV]NK motif to be conserved in all proteins, suggesting that VirB7 might be functional in rickettsial genomes [61,63]. In spite of recent progress in the identification and detailed computational analysis of T4SS genes in Rickettsia species [63], the temporal and spatial regulation of VirB/D transcriptome in mammalian and arthropod hosts remains unresolved. Also, the translocated effector molecules and the mechanisms by which these effectors subvert eukaryotic cellular processes during infection remain almost completely obscure. Based on considerable homology with an effector secreted by L. pneumophila via its Dot/Icm type IVB secretion system, one of the rickettsial effectors described so far is RalF encoded by R. prowazekii [67]. The LpRalF from Legionella pneumophila is a translocated effector containing an N-terminal Sec7 domain, as well a capping domain over the active site, both of which exhibit 46% similarity with the similar regions in RpRalF of R. prowazekii. Intriguingly, the Sec7 domains of both LpRalF and RpRalF display similar catalytic activity to activate Arf1; however, the two proteins have different effector functions in the host cells. LpRalF is an ADP-ribosylation guanine nucleotide exchange factor for the Arf family of small GTPases involved in the regulation of transport functions in eukaryotic cells, whereas RpRalF is involved in the modulation of actin dynamics at the plasma membrane [58]. Overall, these recent findings provide important new insights into the manipulation and exploitation of host cell functions to facilitate the invasion and establishment of disease.
Evidence emanating from the genome sequence data further suggests that, in addition to VirB/D T4SS, rickettsiae also utilize other types of secretion apparatus including the Sec translocation system, components of the two-arginine trans-location and type V autotransporter pathways, and type I secretion system. The three major structural features of the type 1 secretion system are an outer membrane protein belonging to the TolC family, a periplasmic membrane fusion protein and an ATP-binding transporter associated with the inner protein. A recent study demonstrates that a RARP-1 conserved among all rickettsial genomes, is cotranscribed with adjacent genes RT0217 encoding a hypothetical protein and RT0216 (TolC) and secreted by R. typhi in a TolC-dependent manner, suggesting a potentially important role for this mechanism in rickettsial virulence. Furthermore, when expressed in E. coli, both the N-terminal signal peptide and C-terminal ankyrin (Ank) repeats are essential for RARP-1 secretion [68].
Innate & adaptive host immune responses
As discussed above, a unique and salient feature of rickettsiae is the tropism for microvascular endothelium bordering the blood vessels, leading to disseminated inflammation, loss of barrier function and altered vascular permeability – collectively referred to as rick-ettsial vasculitis. A critical knowledge gap in this regard is that interactions of pathogenic rickettsiae with their preferred primary ‘host niche’ during human disease are poorly understood. Therefore, acquisition of new knowledge of early beneficial host responses aimed at facilitating the clearance of infection prior to its taking hold (‘nip in the bud’ strategy) and dissemination through the vasculature is highly significant. It is important to identify, mechanistically understand and then strategically stimulate novel innate immune responses to curtail the establishment within the endothelium and subsequent systemic dissemination of rickettsiae from primary target host cells.
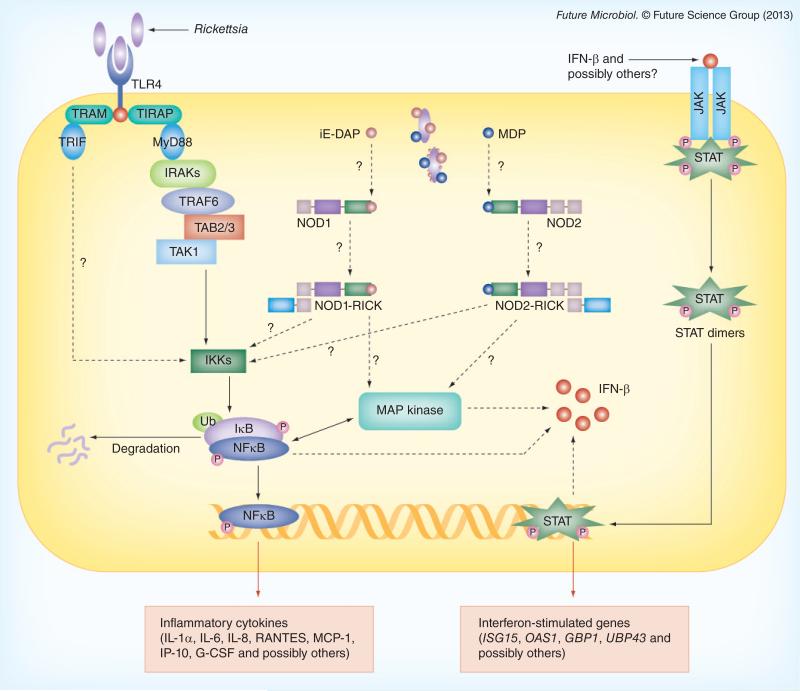
Infection of vascular endothelial cells with R. rickettsii and R. conorii results in the activation of NF-κB and p38 mitogen-activated protein kinase/activating transcription factor-2, as well as expression and secretion of a plethora of cytokines and chemokines. Rickettsiae also exploit manipulation of host cell death mechanisms, as illustrated by apoptosis of infected cells consequential of the inhibition of infection-induced activation of NF-κB. Follow-up studies to identify upstream signaling mechanisms further reveal an essential role for NF-κB activation in the maintenance of mitochondrial integrity, maintenance of the balance between pro- and anti-apoptotic proteins, and prevention of activation of caspase cascade during R. rickettsii infection of cultured human endothelial cells [69]. In addition, ample published evidence also suggests that endothelial cells infected with R. rickettsii or R. conorii display induced expression of prothrombotic, proadhesive and proinflammatory genes, the hallmark features of ‘endothelial activation’ (summarized in Figure 1 and [3,42–44,69,70]). Originally discovered and identified for their roles in antiviral defense, interferons are key signaling cytokines now established as primers, activators and inhibitors of innate and adaptive immune responses. Type 1 interferons in humans and mice include a number of IFN-α subtypes in addition to a single species of IFN-β. IFN-γ, on the other hand, stimulates antiviral function, and activates dendritic cells and macrophages to strengthen innate immune responses in the presence of IL-12 and IL-18 [71]. Not surprisingly, viruses exploit a plethora of strategies to interfere with the host type I IFN response. Intriguing recent discoveries on the relevance of IFN-β in the pathogenesis of certain bacterial infections, however, justify the classification of type I IFNs as ‘more than just antiviral’. During bacterial infections, IFN signaling defends the host by integrating early innate immune responses with later events governed by adaptive immunity. Initial studies employing R. prowazekii interactions with fibroblast-like L929 cells demonstrated the production of IFNs-α and -β, but not IFN-γ in vitro. Interestingly, secreted IFN-α as well as IFN-β display inhibitory activity against replication of vesicular stomatitis virus and R. prowazekii [72,73]. The growth of R. prowazekii is also adversely affected by IFN-γ in a variety of host cells [74–76]. In addition, IFN-γ has a potentiating effect on the inhibition of R. conorii growth by TNF-α in HEp-2 cells [76]. IFN-γ plays a protective role in host defense during infection of susceptible mouse strains with Rickettsia species [41,77–79]. Available evidence further indicates that early immune responses during R. conorii infection are also mediated by NK cells and appear to be regulated by IFN-γ [80].
Figure 1. Signaling mechanisms underlying Rickettsia-induced transcriptional activation and potential pathogen-associated molecular patterns as the determinants of host cell response and fate.
Potential interactions, in other words predictions that remain to be tested at this stage, are indicated by dotted lines. Since these have not yet been investigated, they are indicated by question marks. Solid lines indicate interactions/connections that are known and proven experimentally.
Clinically, patients with serologically confirmed diagnosis of MSF (also known as boutonneuse fever) have significantly higher plasma levels of IFN-g during the acute phase of the disease, when compared directly with the convalescent phase [81]. Similar results are also seen during the acute phase of severe Japanese spotted fever caused by R. japonica [82,83]; however, the levels of serum IFN-γ do not exhibit any changes during the course of African tick-bite fever caused by R. africae [84]. Patients with mild/moderate MSF show increased intralesional expression of IFN-γ, IL-10 and inducible nitric oxide synthase, which play a critical role in antirickettsial immune responses at the site of infection. Furthermore, high levels of IFN-γ mRNA positively correlate with the production of indoleamine-2,3-dioxygenase, a mechanism implicated in inhibiting the growth of rickettsiae, and in accordance with the absence of rickettsiae in the blood [85]. Taken together, the available evidence suggests a protective role for IFN-γ in rickettsial diseases in humans, likely via activation of intracellular bactericidal mechanisms.
STAT proteins possess the unique dual ability for transduction of extracellular signals as well as direct regulation of transcription. It is now well documented that activation of different STAT proteins is essential for the antiviral activities of IFNs [86]. STAT1-deficient mice exhibit no overt developmental abnormalities, but display a complete lack of responsiveness to either IFN-α or IFN-γ and are highly sensitive to infection by microbial pathogens and viruses [87]. In humans, a natural heterozygous germline STAT1 mutation is associated with enhanced susceptibility to mycobacterial, but not viral disease [88]. Available evidence further supports the notion that inherited STAT1 deficiency in humans results in impaired response to IFN-α/β and lethal viral disease [89]. It has been recognized that human CMV inhibits MHC II expression by disruption of the JAK–STAT pathway to avoid immuno surveillance and establish persistence [90]. Similarly, replication of HCV in Huh7 cells and infected liver interferes with STAT3 expression and STAT1/STAT2 activation to facilitate disease progression [91]. In comparison, the knowledge pertaining to the involvement of JAK–STAT signaling in the pathogenesis of bacterial infections has remained relatively obscure. Upregulation of the host's innate antimicrobial response to Chlamydia trachomatis, an obligately intra cellular pathogen, is dependent on the JAK–STAT1 pathway [92]. Coordinated upregulation of the JAK–STAT pathway due to peptidoglycan derived from Lactobacillus species has also been documented [93]. On the other hand, Ehrlichia chaffeensis, another obligately intracellular bacterium of monocytes and macrophages, inhibits IFN-γ-mediated tyrosine phosphorylation of JAK and STAT through protein kinase A activity [94], and suppresses the transcription of JAK1 and STAT1 during the first hour of infection of THP-1 cells [95]. Therefore, E. chaffeensis apparently interferes with IFN-mediated activation of macrophages by downregulating the JAK–STAT pathway. Similarly, entero hemorrhagic E. coli O157:H7 disrupts STAT-1-mediated IFN-γ signal transduction in epithelial cells [96].
Signal transduction pathways governing the host's immune response to microbial pathogens are invariably complex and multithreaded. Attempts to unravel such complexities in host– pathogen interplay have led to the discovery of IFN-β as another key component in host response to infection. Activated by type 1 IFNs, the JAK–STAT signaling cascade is indispensable for mediating innate immune responses, yet there is a complete lack of information regarding its role in rickettsial diseases. In this context, converging new evidence from complementary experimental approaches demonstrates that Rickettsia activation triggers selective and differential activation of STAT proteins in vascular endothelium [97], and further supports the premise that both IFN-β and STAT1 interfere with rickettsial replication for the benefit of the host (Figure 1). Intriguingly, STAT1 activation can be primarily attributed to the expression and secretion of IFN-β by infected endothelium and IFN-β inhibits rickettsial replication in host cells in an STAT-1-dependent manner [98]. Since the transcription enhancer of IFN-β promoter binds to NF-κB, ATF-2 and IFN regulatory factors (IRFs) 3 and 7 [99], the Rickettsia-induced IFN-β response may well be the outcome of one specific mechanism or possibly a combination of these mechanisms. In this context, our initial findings reveal no evidence for Ser396 phosphorylation as an indicator of IRF3 activation [98], but suggest increased expression of IRF7 and IRF9, implicating the potentially unique involvement of either one or both of these IRFs during R. conorii infection. Further analysis of downstream targets regulated by the JAK–STAT pathway, including IFN inducible genes, STAT target genes, negative regulators of JAK–STAT pathway and SH3/SH2 adaptor proteins using pathway-specific PCR arrays reveals that IFN-regulated genes, such as ISG15, OAS1 and GBP1, are stimulated by R. conorii infection [100]. Among these, GBP1 and OAS1 are stimulated by both type I and type II IFNs, whereas ISG15 is induced predominantly by type I IFNs. In particular, our data not only suggest robust expression of ISG15 mRNA and protein in R. conorii-infected endothelial cells, but also yield preliminary evidence of its participation in the post-translational modification of other cellular proteins via ISGylation [101]. Since ISG15 has primarily been implicated in antiviral immune responses [102], determination of its role(s) in intracellular bacterial infections and in the vascular system are both highly significant and innovative avenues worthy of further detailed investigation.
Interfering with the proliferation of rickettsiae in nonphagocytic host cells, an environment conducive to their growth and dissemination and protection from host defense mechanisms, poses an important challenge. In a mouse model of spotted fever rickettsiosis, it has been shown that IFN-γ and TNF-α originating from macrophages, NK cells and T lymphocytes, stimulate nitric oxide production in endothelial cells, which is responsible for antirickettsial activity [79]. Rickettsial killing in macrophages also entails limitation of tryptophan availability through increased degradation by indoleamine-2,3-dioxygenase activity, whereas in peripheral blood monocytes and endothelial cells activated by cytokines, hydrogen peroxide is also involved in the killing of intracellular rickettsiae [103]. Fc-dependent antibody-mediated protection against R. conorii infection of endothelium and macrophages occurs through trapping rickettsiae within the phagosome and resultant phagolysosomal processing mediated by nitric oxide, reactive oxygen species and L-tryptophan starvation [104].
Animal models of R. australis, R. conorii and R. typhi infection have been exploited for systematic investigations of pathological and biochemical aspects of human rickettsial diseases because they closely mimic disseminated infection of the vasculature and other salient features of rickettsioses in humans. A murine and a nonhuman primate model of epidemic typhus describe the infection with virulent Breinl strain of R. prowazekii [105,106]. Such experimental systems provide useful guidance to further our understanding of the host–pathogen relationship during disease, and to compensate for the lack of well-characterized mutants of any rickettsial species, with the exception of a recently reported phospholipase D mutant of R. prowazekii [107]. Thus, in vivo comparative studies using specific strains with varying degrees of virulence within a particular species (e.g., virulent Breinl strain vs attenuated Madrid E strain of R. prowazekii) may yield important new insights into rickettsial pathogenesis and virulence as well as host innate immune mechanisms. Indeed, established animal models of spotted fever rickettsioses, which closely mimic the salient manifestations of human disease, have been employed to identify mechanistic aspects of innate and adaptive immunity, cytokine-mediated endothelial activation and control of intracellular infection. While CD4 and CD8 T cells are involved in protective immunity, recruitment of lymphocytes and macrophages have been implicated in rickettsial clearance from the foci of infection [108,109]. In comparison with their wild-type counterparts, mice lacking class 1 MHC are more than 50,000-fold more susceptible to a lethal infection with R. australis. Similarly, the murine hosts lacking genes for IFN-γ and perforin are also highly susceptible (greater than 100-fold), suggesting a crucial role for cytotoxic T lymphocytes in host defense mechanisms; IFN-γ and perforin-mediated activities contribute only partly to the antirickettsial effect of cytotoxic T lymphocytes [108]. Expression of CXCL9 (Mig) and CXCL10 (IP-10), known CXCR3 ligands, and chemokines targeting CD8+ T cells and CD4+ T-helper cells, is increased in tissues of experimentally infected mice and in biopsy specimens of patients with RMSF [109]. Yet, antibody-mediated neutralization of CXCL9 or CXCL10 in R. conorii-infected hosts or experimental infection of CXCR3-deficient mice with R. australis had no effect on the disease outcome or clearance of rickettsiae from the target host tissues [110]. Furthermore, mouse and human T cells have been implicated in executing immunological cross-protection between R. conorii and R. typhi, lending support to the concept of cross-reactive T cell epitopes among spotted fever versus typhus group rickettsiae [111]. Intriguingly, intracellular rickettsiae in bone marrow-derived dendritic cells are found to be localized in the cytosol and phagosomes, which may enhance the priming function of these cells via activation of CD4+ and CD8+ T cells, respectively, via antigen presentation through MHC class II and class I pathways. Differential interactions with dendritic cells during in vivo infection with rickettsiae may also have an important role in determining the host susceptibility, based on the suppression of adaptive immune response in susceptible C3H mice and stimulation of a protective response in C57BL6 mice. Furthermore, it has been suggested that R. conorii infection induces maturation of dendritic cells in vitro by robust upregulation of the expression of cell surface molecules, such as CD40, CD80, CD86 and MHC-II, as well as production of proinflammatory cytokines. Adoptive transfer of such stimulated dendritic cells into susceptible naive mice yields protection against challenge with the lethal infectious dose by inhibiting rickettsial proliferation and dissemination into target host tissues via augmentation of the immune response through increased activity of CD4+, CD8+ and NK cells [112,113].
Toll-like receptors (TLRs) are pattern recognition receptors that play a vital role in the induction of antimicrobial genes in some instances, and control of adaptive immune responses in others. Indeed, it is now well established that TLRs interact with different combinations of adaptor proteins to activate MAPK and NF-κB pathways in a cell type-specific and context-dependent manner to drive particular immune responses [114,115]. The contributions of different TLRs in bacterial infections studied so far, however, depend on the site of infection and the characteristics of invading pathogens. In general, TLR4 recognizes lipopolysaccharide, whereas TLR2 interacts with many different microbial products, including peptidoglycan and cell-wall lipoproteins [115]. A. phagocytophilum, an obligately intracellular bacterium closely related to rickettsiae, activates NF-κB in macrophages via TLR2 [116]; however, in vivo infection is controlled by T- and/or B-cell immunity without the requirement for TLRs and adaptor protein MyD88 [117]. Recent clinical evidence implicates a potential role for TLR4 receptor polymorphisms in determining disease severity during human infections with R. conorii [118], and studies in an animal model have further implicated involvement of TLR4-specific immune responses in rickettsial clearance from the infected host [113,119,120].
Another important microbial detection system that plays a critical role in innate immune responses is the nucleotide oligomerization domain (NOD)-like receptors. NOD1 and NOD2 are predominantly located in the cytoplasm and also act to govern signaling mechanisms regulating NF-κB and MAP kinase pathways. NOD1 and NOD2 recognize peptidoglycan fragments containing meso-diaminopimelic acid and muramyl dipeptide, respectively [121]. Although rickettsiae occupy an ‘intracytoplasmic niche’ for their growth and replication within the host cell, making their recognition and potential interactions with cytosolic NOD proteins highly likely, the potential importance of NOD signaling during rickettsial infection of endothelial cells has not yet been explored. Since rickettsiae contain genes for lipopolysaccharide and peptidoglycan biosynthesis and possess both in their membrane structures [44,122], and vascular endothelial cells constitutively express TLR4 and NOD1, we hypothesize that TLRs and NODs play an important role in determining the extent of host cell transcriptional activation. Accordingly, targeted comprehensive comparative analyses of the roles of TLR2, TLR4 and NOD proteins in the interplay between host endothelium and pathogenic rickettsiae of varying virulence should be conducted in future studies.
New rickettsial genomes & comparative genomics
Comparative genomics of multiple closely related species/strains provides a high-impact tool for the evaluation of gene fluxes in an evolutionary context, leading to the estimation of variations within the genomes and better understanding of the phylogenetic relatedness and taxonomy of the organism. The applicability of comparative genomics to Rickettsia species as human pathogens acquired significance with the seminal publication of the complete genome sequence for R. prowazekii [123], which was shown to share a significant (nearly 18%) sequence homology with mitochondrial proteins mainly involved in ATP synthesis and transport, tricarboxylic acid cycle, ATP/ADP translocation systems and respiratory chain complexes [123]. Since then, a total of 51 rickettsial genomes belonging to over 20 recognized species have been sequenced, the majority of which have been extensively reviewed [124], and are listed in Table 1. Of note, while the genome of R. prowazekii, one of the highly pathogenic Rickettsia species, still remains the smallest (1.11 Mb) among all rickettsial species to date, with 835 ORFs; the Rickettsia endosymbiont of Ixodes scapularis (REIS) is the largest (>2.1 Mb), with 2309 predicted ORFs [123,125]. Accordingly, genome reduction in Rickettsia is considered to be a major driving force governing the evolution of pathogenesis/virulence mechanisms and acquisition of intracellular niche. In addition, gene loss has been implicated as a predominant player in shaping the rickettsial genomes [126,127]. Additional molecular mechanisms such as frameshift mutations, fragmentation of ORFs, mobile genetic elements and insertion/deletions are also known to create pseudogenes in several bacterial genomes [128]; for example, R. conorii is known to harbor hundreds of pseudogenes and gene fragments [129]. Although obligate intracellular bacteria are known to have high coding density, for example Carsonella ruddii, a secondary endosymbiont of aphids, encodes 97% of its genome [130], Rickettsia species maintain a high amount of noncoding DNA, which predominantly contains pseudo-genes and partial gene fragments. Based on the whole genome analysis, strain-variable ORFs present in R. conorii are expected to code for a median protein size of 173 amino acids, yet the median size of core genes (present in the majority of rickettsial genomes) is 284 amino acids. Thus, the short protein size of strain-variable proteins is thought to be influenced by the presence of pseudogenes and recently acquired proteins [129]. The existence and roles for bacterial noncoding RNAs in gene regulation is also well documented [131,132], and regulatory RNAs have been implicated in several processes including pathogenicity [133,134]. Recently, R. prowazekii has been shown to carry stem loop structures preceding homopolymeric poly(T) stretches, predominantly occurring in bacterial termination sites. Furthermore, the discovery of anti-sense RNAs, mostly generated by lack of proper termination between neighboring genes, implicates the existence of noncoding RNA in its genome [135]. Although small RNAs have yet to be fully characterized in Rickettsia, their occur-rence in other intracellular pathogens such as Listeria monocytogenes [136] and C. trachomatis [137] has been reported.
Table 1.
Genome statistics and plasmid content of Rickettsia species subdivided into different subgroups†.
| Rickettsia species | Size (Mb) | RAST CDS | GC (%) | Plasmids |
|---|---|---|---|---|
| Spotted fever group | ||||
| R. africae | 1.29 | 1545 | 32.4 | 1 |
| R. conorii | 1.27 | 1578 | 32.4 | - |
| R. heilongjiangensis | 1.28 | 1562 | ND | - |
| R. helvetica | 1.42 | 1739 | 32.2 | 1 |
| R. honei | 1.27 | 1614 | 32.4 | - |
| R. japonica | 1.28 | 1575 | 32.7 | 1 |
| R. massiliae (2)‡ | 1.28-1.38 | 1601-1721 | 32.5 | 1 |
| R. montanensis | 1.28 | 1513 | 32.6 | - |
| R. parkeri | 1.30 | 1604 | 32.4 | - |
| R. peacockii | 1.31 | 1558 | 32.6 | 1 |
| R. philipi | 1.29 | 1570 | 32.5 | - |
| R. rhipicephali | 1.31 | 1612 | 32.3 | 1 |
| R. rickettsii (8)‡ | 1.26-1.29 | 1547-1595 | 32.4-32.6 | - |
| R. sibirica | 1.25 | 1554 | 32.5 | - |
| R. slovaca (2)‡ | 1.28 | 1598-1611 | 32.5 | - |
| R. endosymbiont of Ixodus scapularis | 2.1 | 2404 | 32.8 | 4 |
| Candidatus R. amblyommii | 1.48 | 1856 | 32.4 | 3 |
| Typhus group | ||||
| R. prowazekii (8)‡ | 1.11 | 892-924 | 29 | - |
| R. typhi (3)‡ | 1.11 | 875-892 | 28.9 | - |
| Transitional group | ||||
| R. australis | 1.32 | 1565 | 32.3 | 1 |
| R. akari | 1.23 | 1437 | 32.3 | - |
| R. felis | 1.59 | 1810 | 32.5 | 1 |
| Ancestral group | ||||
| R. bellii (2)‡ | 1.52-1.53 | 1612-1657 | 31.6 | - |
| R. canadensis (2)‡ | 1.15-1.16 | 1130-1230 | 31-31.1 | - |
Based on the phylogenetic classification proposed by Gillespie et al. [1]. The genome size, RAST, CDS and GC content of all available species and strains, but not subspecies listed in PATRIC database [183], are included. The number of plasmids only accounts for those reported in the published literature. Note that the genome statistics from the following strains belonging to different species are used for compiling the table: R. africae ESF-5; R. akari strain Hartford; R. australis strain Cutlack; R. bellii strains OSU 85-389 and RML369-C; R. canadensis strains CA410 and McKiel; R. conorii strain Malish 7; R. endosymbiont of Ixodes scapularis; R. felis URRWXCal2; R. heilongjiangensis 054; R. helvetica C9P9; R. honei RB; R. japonica YH; R. massiliae strains MTU5 and AZT80; R. montanensis strain OSU 85-930; R. parkeri strain Portsmouth; R. peacockii strain Rustic; R. philipii strain 364D; R. prowazekii strains BuV67-CWPP, Chernikova, Dachau, GvV257, Katsinyian, Madrid E, Rp22 and RpGv24; R. rhipicephali strain 3-7-female6-CWPP; R. rickettsii strains Sheila Smith, Arizona, Brazil, Colombia, Hauke, Hino, Hlp#2 and Iowa; R. sibirica 246; R. slovaca strains 13-B and D-CWPP; R. typhi strains B9991CWPP, TH1527 and Wilmington; and Candidatus R. amblyommii strain GAT-30V.
Represents the number of strains for which the complete genome sequence is available and genome statistics are used for compiling the data.
CDS: Coding sequences; GC: Guanine-cytosine content; ND: Not determined; PATRIC: Pathosystems Resource Integration Center; RAST: Rapid annotation using subsystems technology.
Horizontal gene transfer (HGT) is a key mechanism by which bacteria acquire genes from other related bacteria, leading to innovation and evolution of their genomes resulting in the adaptability to acquire and establish new niches [138]. Until recently, this mechanism has been projected to play a limited role in the evolution of Rickettsia due to their intracellular lifestyle. However, the relatively recent availability of several new rickettsial genomes, such as those belonging to R. massiliae, R. peacockii and REIS, in conjunction with comparative genomic analysis, have supported the notion that HGT plays a more prominent role in rickettsial genomes than previously thought. Accordingly, several lines of evidence establishing the role for HGT in rick-ettsial evolution have now been published and extensively reviewed [1,50,125,126]. The genome of REIS (the largest to be sequenced to date) carries more than 650 transposons, mobile genetic elements and Rickettsiales amplified genetic elements, suggesting that these elements might have seeded rickettsial genomes with genes from other bacteria [125]. Based on single-gene phylogenetic approaches, 79 genes of R. felis were also shown to be a result of gene recombination events resulting from HGT [139]. In this regard, the occurrence of Ank, leucine-rich repeats and tetratricopeptide repeats (TPR) containing proteins in most rickettsial genomes, including those of R. felis, R. bellii and REIS (albeit in numbers lower than those present in other intracellular pathogens such as Legionella and Amoebophilus), is also intriguing as these repeats, known to be commonly present in eukaryotic hosts, might have been acquired by an ancestral rickettsial genome during the process of host–pathogen coevolution. Not surprisingly, proteins containing Ank and TPR repeats have been implicated in the pathogenesis of several obligate intracellular pathogens [140,141]. Recent evidence demonstrates that R. typhi secretes an Ank rich protein (RARP1) during infection of the mammalian host cells, and RARP1 is an effector molecule secreted into the host cellular milieu in a TolC-dependent manner [68].
Toxin–antitoxin (TA) modules have been identified in several free-living bacteria and implicated in programmed cell death [142]. Again, in light of the obligate intracellular nature of Rickettsia, the existence of TA modules in its genome was thought to be uncommon. However, comparative genomic approaches have identified five TA modules (relBE, phd/doc, vapBC/vag, mazEF and parDE) to be present in all rickettsial genomes, excluding those belonging to the typhus group [66]. VapC, a rickettsial toxin, was found to be secreted into the host cytoplasm during infection, and to act on the host cell through RNase activity [143]. To date, ORFs for 56 toxins and 86 antitoxins have been predicted in several rickettsial genomes, and R. africae contains ten TA modules [144]. Thus, gene loss leading to reductive genome sizes and TA modules presumably represent two major evolutionary tactics defining the degree of virulence in rickettsial pathogens. It is, however, necessary to delineate the intricacies of rickettsial genomes in further detail to identify and characterize such virulence factors and define their roles in the pathogenesis and immune pathways.
Genome-wide transcriptome analysis of mRNA isolated from R. conorii eschars revealed that approximately 15% of the genes, mostly belonging to the DNA repair and osmotic stress pathways, are upregulated in comparison to rickettsiae grown in Vero cells and a majority of these genes are unique to the SFG group [145]. On the other hand, global transcriptional profiling of R. rickettsii suggested that except for sudden and dramatic changes in temperature (37 vs 4°C), which triggers differential regulation of 56 genes, the rickettsial gene expression profiles display only minimal variations during mild temperature changes, iron limitation and growth in different host-cell backgrounds [146]. Exposure of R. prowazekii to heat shock (37 vs 42°C) also upregulates (>twofold) 23 genes, of which 57% are characterized as heat shock inducible genes [147]. Since Rickettsia are known to encode a lower number of transcriptional regulators when compared with free-living microbes, it is possible that the genes acquired during early stages of rickettsial evolution conferred functions facilitating the invasion and adaptation to different hosts, and upon finding a suitable host environment, most of the nonessential transcriptional response regulators were deliberately lost [129].
Traditionally, rickettsial classification and taxonomic delineations were predominantly based on the geographic distribution of human diseases, vectors and other biological reservoirs, serology, energy production and biosynthesis, and strain stability and maintenance [3,58]. Recently, advanced molecular approaches, such as single gene phylogeny based on 16S rRNA, gltA, ompA, ompB, intergenic spacers (16S-23S, dksA-xerC, nuppA-purC and rpmE-tRNAfMet), geno-typing approaches involving single nucleotide polymorphisms and variable nucleotide tandem repeats have been applied to the identification of rickettsial species and strains [148–152]. Although intracellular bacteria tend to coevolve with their host(s), a recent study comparing the 16S rRNA and 18S rRNA sequences from Rickettsia species and their respective hosts, found a high degree of incongruence between both of the phylogenies, suggesting a diverse host range for rickettsial species [126,153]. Furthermore, concatenation of 731 core genes from ten rickettsial genomes yields a robust phylogeny, which is congruent with the most recent classification of Rickettsia species [66]; thus, availability of complete genomes has provided better resolution for understanding rickettsial phylogeny and evolution.
Plasmids & molecular manipulation
Similar to most other intracellular bacteria, Rickettsia species were considered to be evolving mainly by gene loss rather than gene gain from other related bacteria. From an indepth analysis of complete genome sequences of several Rickettsia species, which clearly display signatures of mobile genetic elements, acquired foreign DNA and plasmids, it is now appreciated that several mechanisms, including gene loss, HGT, transposon elements and plasmids, play an important role in shaping the rickettsial genomes. Plasmids are likely more common in rickettsial genomes, and out of more than 20 Rickettsia species sequenced to date, ten (R. massiliae, R. africae, R. peacockii, R. rhipecephali, Rickettsia amblyommii, R. felis, R. japonica, R. helvetica, R. australis and REIS) are known to harbor between one and four plasmids (Table 1). As expected, these plasmids harbor genes encoding for DNA replication and partitioning, putative environmental and host adaptive proteins, such as heat shock proteins, and putative virulence factors, such as patatin [1,154,155]. R. bellii and R. akari also contain putative conjugative plasmids([Eremeeva, Madan, Dasch, Unpublished Data] presented at the 20th meeting of American Society for Rickettsiology), whereas R. felis plasmid pRFδ is presumed to be an artifact, in light of the rationale that two divergent plasmids with identical origins of replication are highly unlikely to be retained and maintained in any bacteria [61]. Among the sequenced plasmids encoded by different rickettsiae, the plasmid pRAF present in R. africae is the smallest (~12.4 kb) and encodes for ten proteins, whereas the pREIS2 plasmid in REIS is the largest (~66.8 kb) with the potential to code for 83 predicted proteins. The plasmids of REIS also contain several copies of Rickettsiales amplified genetic element components. Notably, genes encoding TraAITi and TraDTi (involved in the processing of plasmid DNA before conjugation) are also present, indicating the occurrence of HGT during the course of REIS evolution [52,124,125,156]. Both pREIS2 and pREIS4 contain multigene regions predominantly composed of pseudogenes, which are also incorporated into the REIS chromosome at several locations, indicating an integral role for plasmids in shaping the rickettsial genomes [125]. R. australis, the causative agent of Queensland tick typhus, also harbors a plasmid encoding Ank, TPR domain containing proteins, TraAITi and TraDTi genes, transposases, site-specific recombinases and several other hypothetical proteins [157]. Plasmids of different rickettsial species, such as R. monacensis (pRM plasmid) and R. amblyommii (pRAM plasmid) have been successfully used for the construction of shuttle vectors required for genetic manipulation of Rickettsia.
Genetic manipulation of organisms offers a promising approach for functional characterization of proteins with unknown function. Standard approaches for alteration of bacterial genomes include homologous recombination as well as site-directed and transposon-based mutagenesis. The different approaches used for rickettsial genome modification have been extensively discussed, allowing us to refer the reader to the recent book chapter by Munderloh et al. [158]. Owing to difficulties associated with the applicability of standard approaches of molecular genetics, a number of investigations focused on the functional aspects of rickettsial proteins utilizing the strategy of heterologous expression in E. coli and Salmonella enterica serovar Typhymurium. By using E. coli metK deletion mutant as a host, R. prowazekii metK was expressed and shown to functionally complement the E. coli MetK protein, thereby confirming the functionality of R. prowazekii metK in the synthesis of S-adenosylmethionine [159]. However, the metK ORF is disrupted by inactivating mutations in some rickettsial strains and the function of metK in these genomes is compensated by EamA, an S-adenosylmethionine transporter [160]. The pld and tlyC genes of R. prowazekii are implicated in phagosomal escape during infection based on their expression in S. enterica serovar Typhimurium, revealing a potentially important role for the R. prowazekii pld gene in the establishment of infection [34]. By using a rifampin resistance marker, the pld gene was knocked out in R. prowazekii by site-directed mutagenesis approach, allowing for the selection of viable R. prowazekii mutants lacking phospholipase D activity. Intriguingly, these mutants were still able to exit the phagosome in RAW 264.7 cells, possibly due to the presence of redundant phospholipase activities; for example, R. typhi genes RT0590 (pat1) and RT0522 (pat2) are both shown to encode for PLA2 activity [37,107,161]. To date, this is the only published report showing the successful manipulation and description of a rickettsial mutant using site-directed mutagenesis approach. However, recent studies using transposon (Tn5 and Himar1) mutagenesis have successfully generated mutant strains in Rickettsia species leading to the characterization of rickettsial proteins predicted to be involved in plaque formation and pathogenesis [162,163]. An important consideration here is that although vectors used for site-directed and transposon-based mutagenesis are considered to be valuable tools for genetic manipulation, these vectors do not replicate within the Rickettsia and are only transiently active. In addition to shuttle vectors using the parA and dnaA-like genes involved in DNA replication and partitioning, intervening sequences from R. amblyommii plasmids pRAM18 and pRAM32 have been generated and successfully tested for replication within the Rickettsia genomes, paving the way for analysis of rickettsial genes, promoters and regulatory elements in their native hosts [164]. Plasmid vector pRAM18dRGA represents a shuttle vector derived from R. amblyommii plasmids pRAM18 and pRAM32, encodes a green fluorescent protein and rifampin resistance marker, and also carries a red fluorescent protein cloned into the multiple cloning site (MCS). R. montanensis stably transformed with this plasmid was found to express both green and red fluorescent proteins. This plasmid could also be transformed into R. prowazekii (known to lack plasmids), indicating that Rickettsia species may have the capacity to harbor and stably replicate the extra-chromosal elements [165]. However, attempts to transform R. massiliae, which harbors a native plasmid with parA gene exhibiting homology to that of the shuttle vector, with pRAM18dRGA failed; however, the shuttle vector containing parA–dnaA region from pRAM23 (a R. amblyommii plasmid carrying parA nonhomologous to that of R. massiliae native plasmid) could be successfully transformed, indicating that the degree of homology between parA genes is critical for maintaining plasmid complements in rickettsiae [164]. Furthermore, an improved variant of the previously described pMW1650 plasmid vector containing Himar1 transposon [166], now known as Rickettsia insertion and expression plasmid, contains two MSCs; MCS1 and 2, required for cloning of two individual proteins under different promoters. A recent study reports on the cloning of green fluorescent protein under the control of ompA promoter in MCS1 and FLAG epitope-tagged rickA under its native promoter in MCS2 of the Rickettsia insertion and expression plasmid. The resultant construct was subsequently transformed into R. parkeri, leading to the detection of RickA and GFPuv proteins by Western blotting and fluorescence imaging, respectively [167]. Thus far, the complementation of target genes in mutant strains generated by transposon mutagenesis or homologous recombination has not been reported in Rickettsia species. The availability of self-replicating plasmids will presumably enable the complementation of mutant genes capable of being expressed under their native promoters, leading to further defining the role of rickettsial proteins in their native hosts under both in vitro and in vivo conditions.
Proteomics: recent studies on protein methylation in R. prowazekii
As genetic manipulation of Rickettsia is hampered by the lack of robust genetic tools to assign protein function, proteomics approaches using 2D-PAGE, liquid chromatography-mass spec-trometry (MS)/MS and nano-liquid chromatography-MS/MS are gaining significance to identify rickettsial proteins expressed during infection and pathogenesis. By a global proteomic approach, 91 proteins that are mostly virulence-related surface proteins, such as OmpA, OmpB, RickA and β-peptide of R. parkeri, were identified to be differentially expressed during human infection and five antigens (OmpA, OmpB, IF2, FtsZ and cysteinyl-tRNA synthetase) were found to be present in the serum of patients infected with R. parkeri [168]. A recent study comparing the proteome of R. prowazekii grown in different cell lines found differential expression of its proteins, and the most striking differences were observed in universal stress protein UspA-like nucleotide binding protein and deoxyuridine 5′-triphosphate nucleotidohydrolase, which are highly expressed when grown in murine fibroblast L929 cell line. In addition, enoyl-(acyl carrier protein) reductase, a protein involved in fatty acid biosynthesis, was highly expressed when grown in I. scapularis ISE6 cell line, indicating that Rickettsia have the inherent ability to differentially regulate its proteome depending on the host [169].
Post-translational modification of bacterial proteins is also gaining significance as modified proteins play a key role in signal transduction and regulatory processes, protein stability, molecular interactions and localization, and several modifications, such as phosphorylation, N- and O-glycosylation, methylation and acetylation of bacterial proteins, have been reported [170]. Rickettsial genomes encode several two-component systems, such as PleC–PleD, EnvZ–OmpR and NtrY–NtrX, indicating protein phosphorylation to be a common phenomenon of rickettsial proteins. Two R. prowazekii lysine methytransferases (PKMT1 and PKMT2) have been shown to be involved in the methylation of lysine residues in the passenger domain of the OmpB, a cell surface protein that is present in all rickettsial species and plays a critical role in attachment and invasion of the host during infection. While PKMT1 (RP789) from R. prowazekii Madrid E strain (avirulent) can catalyze mono-, di- and trimethylation of OmpB, PKMT2 (RP027-028) from R. prowazekii strain Rp22 (virulent) can catalyze trimethylation. However, these results should not be extrapolated to compare the degree of virulence between the strains because R. prowazekii Rp22 also carries a homolog of PKMT1, which can potentially complement the function of PKMT2 [171]. Lysine methylation is hypothesized to enhance the partial charges of ε-amino groups of OmpB, which is otherwise highly acidic, affecting the electrostatic interactions and hydrogen bonding during protein– protein interactions, and also potentially altering the interaction with host surface proteins during invasion and establishment of infection [171]. The role of lysine methylation on invasion, innate immune activation and cell signaling needs to be determined to gain further insight to correlate the role of rickettsial protein modifications with pathogenesis.
Since Rickettsia have highly reduced genomes, the gene composition retained in their genomes is predicted to encode key proteins involved in several functions, such as biosynthetic processes, regulation of stress responses, invasion and infection, and innate immune activation. To gain a better understanding of the function and regulation of these proteins in rickettsial genomes, structural studies hold critical importance; so far, the structure of 12 proteins, including R. rick-ettsii cold shock protein, R. prowazekii fumarate hydratase (an enzyme involved in catalyzing the third step in TCA cycle) and R. prowazekii FabG (a 3-ketoacyl-[acyl-carrier-protein] reductase involved in fatty acid biosynthesis pathway, and considered to be a potential target for drug development) have been resolved and are available in Protein Data Bank [172–174,201].
Metabolomics
Rickettsia and eukaryotic mitochondrion exhibit striking similarities between their gene repositories and share genes involved in energy transport and metabolism [123]. Being an obligate intracellular pathogen growing solely in nutrient-rich host cell cytoplasm, rickettsiae are traditionally presumed to have leaky cell membranes allowing for passive exchange of nutrients [175]. With the availability of complete genome sequences and application of comparative genomic approaches, it is now unequivocally established that Rickettsia species encode several unique transport machineries involved in the exchange and transport of end products of host cell metabolic pathways [176,177]. Five tlc genes (tlc1, tlc2, tlc3, tlc4 and tlc5) belonging to the family of transmembrane transporters have been annotated and found to be conserved in all rickettsial genomes sequenced to date. Among these, R. prowazekii Tlc1, an ATP/ADP translo-case, has so far been best characterized and implicated in the energy exchange via maintenance of ATP/ADP equilibrium between bacterial cell and host cytosol, thus providing ready energy to the bacterium [176]. Rickettsiae also harbor a ribonucleotide reductase, an enzyme required for the conversion of nucleoside triphosphates into deoxynucleoside triphosphates, but lack the metabolic machinery to synthesize nucleoside triphosphates. The transmembrane transport proteins Tlc4 and Tlc5 are found to transport CTP/UTP/GDP and GTP/GDP, respectively, from host-cell cytosol into the bacterium and aid in nucleic acid biosynthesis. Another group of transmembrane transporters include the GlpF and GlpT proteins involved in the translocation of glycerol and snglycerol-3-phosphate required for phospholipid biosynthesis. Despite lacking genes involved in glycolysis and gluconeogenesis, R. prowazekii encodes GpsA, a functional enzyme required for synthesis of glycerol-3-phosphate using dihydroxyacetone phosphate as the substrate. However, the mechanisms by which dihydroxy-acetone phosphate is transported into the bacteria from host cell cytoplasm are still unclear, and it is likely that Rickettsia might harbor a complex transport network for exploiting and transporting both intermediate and end products of several biochemical pathways from the host cytosol [177]. Interestingly, R. conorii is shown to differentially regulate the expression of SpoT paralogs, which are involved in the signaling and regulation of other proteins during nutrient starvation [178]. As the availability of nutrients is not a limiting factor during early stages of infection, Rickettsia potentially upregulates the transport machinery and upon reaching a threshold (increase in bacterial burden) the bacteria might downregulate its genes by expressing signaling molecules, enabling its survival even in nutrient-depleted niches. It is possible that during the course of evolution, SpoT paralogs may have diverged functionally to detect a wide array of signaling molecules during poor nutrient conditions.
Conclusion & future perspective
Despite more than 100 years of study, the mechanisms by which Rickettsia escapes the phagosome and establishes a successful intracellular infection still remain to be fully understood. The obligate intracellular nature of rickettsiae residing and replicating within the host cell cytosol mandates that host-derived factors play a key role in their multiplication and virulence. The use of recently established genetic approaches to manipulate the host cells, for example RNAi, will prove to be helpful in dissecting the host–pathogen cross-talk and provide a better understanding of the role of host proteins in the establishment of infection. Considerable progress has recently been made concerning the role of TLR4 and associated signaling intermediates in the regulation of immune response genes; however, the potential roles of other TLRs, including those localized to the cytoplasm and the inflammasome complex, still remain elusive. A recent study has shown that Brucella abortus, a facultative intra cellular bacterium, is recognized by several ASC-dependent inflammasomes and a functional Brucella T4SS as crucial for inflammasome activation [179]. Given the fact that Rickettsia gain entry into nonphagocytic host cells by induced phagocytosis followed by a quick escape from within the phagosome, it would be reasonable to assume that at least a few intracellular bacteria are initially processed through the autophagic cascade and/or undergo destructive lysis releasing their DNA into host cytosol. In addition, rickettsiae also possess genes required to encode for type IV and type I secretion systems. It is thus possible that pathogen-associated molecular patterns, such as nucleic acids, cell wall components and effector molecules delivered through the secretion machineries, for example RalF in R. prowazekii and RARP1 in R. typhi, may be subject to sensing by the designated host cytosolic receptors and surveillance mechanisms. In this regard, further details on the potential roles of inflammasome signaling cascades will enhance our understanding of the mechanisms involved in innate immune activation and host–pathogen inter-relationships.
Until recently, functional genomics approaches involving genetic manipulation technique(s) to understand the roles of rickettsial proteins in adhesion, invasion and virulence have been fraught with a number of difficulties imposed by the occupancy of an intracellular niche and lack of suitable vector systems. However, recent development of plasmid shuttle vectors and the application of transposon-based mutagenesis and homologous recombination approaches for generating gene deletion mutants in Rickettsia hold significant potential for providing valuable information on the functional importance of known genes and to potentially assign functions to a series of hypothetical proteins present in rickettsial genomes. To date, 51 complete genome sequences belonging to more than 20 different recognized Rickettsia species are available in various database domains, and several comparative genomic studies have been conducted to understand the gene architecture and driving forces involved in shaping these genomes [66,125,126]. It is now well established that HGT plays a crucial role in introducing new genetic material, often containing ORFan genes (genes with no detectable homologs), into bacterial genomes, and several lines of evidence suggest that rickettsial genomes contain signatures of HGT [65,138,180,181]. Rickettsiae also contain several species- and lineage-specific predicted proteins for which the biological function is still unknown. Studies focusing on the potential roles of these unique proteins, some of which might play a role in pathogenesis, are also warranted for better understanding of the pathways and mechanisms involved in the onset, establishment, progression and manifestation of rickettsial diseases. Proteins present in obligate intra-cellular bacteria are known to have a tendency to interact with a higher number of other interacting partners involved in multiple pathways in comparison to their homologs in free-living bacteria, implying that proteins in obligatory pathogens might have multiple functions [182]. Another important feature is that gene loss is considered to be a primary attribute involved in the evolution of rickettsial genomes. Intriguingly, R. prowazekii has approximately 835 predicted ORFs, yet is considered to be one of the most virulent species among all Rickettsia. Proteomic approaches, such as protein arrays, affinity purification and biomolecular fluorescence imaging should provide critical insights into the expression profile, function(s) and inter-actome of rickettsial proteins, an outcome that is essential to fully understand the sophisticated means by which these pathogens exploit host cell processes to ensure their survival and cause diseases of varying severity. Finally, we anticipate that these studies and other future breakthrough developments will expand our knowledge into important host defense mechanisms as well as bacterial repertoires to subvert and circumvent host responses that define the biology of Rickettsia survival and parasitism, which should direct the design of effective strategies to combat these infections.
Executive summary.
Rickettsial diseases: old & new
■ Human rickettsioses are now recognized to occur across the globe and encompass infections attributed to a combination of previously known, newly emerging and re-emerging pathogenic Rickettsia species.
■ Advanced phylogenetics has classified rickettsial agents into ancestral, spotted fever, transitional and typhus subgroups, with additional groups likely to emerge in the near future. The major rickettsial diseases include Rocky Mountain spotted fever (Rickettsia rickettsii), Mediterranean spotted fever (Rickettsia conorii), epidemic typhus (R. prowazekii) and endemic typhus (Rickettsia typhi).
Adhesion to & entry into host cells
■ Rickettsial OmpB interactions with host cell Ku70 and OmpA binding to cell surface integrins α2β1 facilitate bacterial entry, suggesting the exploitation of redundant mechanisms to ensure host cell adhesion and invasion.
■ The mechanisms governing quick escape from the phagosome into host cytosol remain to be better understood, but likely involve membranolytic activities of hemolysin, phospholipase D and phospholipase A2.
Actin-based motility
■ Once intracellular, spotted fever rickettsiae (R. rickettsii, R. conorii and Rickettsia parkeri) form polar actin tails for intracellular motility and intercellular spread. R. prowazekii lacks this property, whereas R. typhi displays erratic actin-based motility.
■ RickA and Sca2 have been implicated in rickettsial movements within and between host cells, although recent evidence indicates that RickA may be dispensable.
Secretion systems
■ Rickettsiae possess a number of genes coding for Vir proteins as components of a type IV secretion system, although Vir proteins involved in the formation of the pilus assembly appear to be missing. A TolC-dependent secretion of RARP-1, an ankyrin repeat effector molecule, has recently been documented in R. typhi.
■ To the best of our knowledge, only two rickettsial effectors, namely RalF in R. prowazekii and RARP-1 in R. typhi, have been recognized so far. Other putative effectors, such as RickA, which lacks a secretory signal sequence, and VapC, a toxin, are also predicted to be secreted via as yet unknown mechanism(s).
Innate & adpative host immune responses
■ In humans and established murine models of infection, endothelial cells bordering the small- and medium-sized vessels are the preferred major targets of pathogenic rickettsiae.
■ Human endothelial cells infected in vitro with both spotted fever and typhus rickettsiae undergo ‘endothelial activation’, as demonstrated by a switch from basal, nonthrombogenic phenotype to increased expression of procoagulant and proadhesive molecules.
■ Rickettsiae activate host cell NF-κB and exploit its antiapoptotic effects to prevent/delay host cell death in order to maintain their ‘intracellular niche’.
■ Recent evidence implicates IFN-β-mediated activation of JAK–STAT signaling mechanism and downstream interferon-stimulated genes in ‘antiviral-like’ immune responses of endothelial cells to R. rickettsii and R. conorii.
■ Rickettsiae stimulate the maturation and activation of dendritic cells, the adoptive transfer of which is protective against a lethal challenge in a mouse model of R. conorii infection.
■ NK cells are involved in early antirickettsial immune repsonse, and T cells mediate reciprocal crossprotective immunity between spotted fever and typhus group rickettsiae.
New rickettsial genomes & comparative genomics
■ Analysis of a number of rickettsial genomes representing different species and/or strains suggests two common themes as major determinants of their genetic makeup: horizontal gene transfer and genomic reduction.
■ Presence of a considerable number of ankyrin, leucine-rich, and tetratricopeptide repeats, especially in Rickettsia felis, R. bellii and Rickettsia endosymbiont of Ixodes scapularis genomes, indicates that proteins containing such repeats may also be secreted and serve as ‘nucleomodulins’ to alter the patterns of cellular gene expression to directly influence host responses.
Plasmids & molecular manipulation
■ Multiple species within the family Rickettsiaceae have now been shown to harbor plasmids, which may play a role in implementing important host adaptation mechanisms, such as temperature variations, and carry putative virulence factors.
■ Development of a replicating plasmid represents an important milestone, as this plasmid may serve as a useful tool to overexpress or complement proteins without altering the genomic context of the bacterium.
■ The shuttle vector system(s) to introduce transposons into the bacterium should prove to be instrumental in identifying and establishing specialized protein functions.
Proteomics & metabolomics
■ As expected, rickettsiae display the ability for differential expression of proteins/enzymes in mammalian versus tick host cell environments.
■ Rickettsial proteins also undergo post-translational modification(s), as suggested by methylation of lysine residues in the passenger domain of OmpB by PKMT1 and PKMT2. However, the biological relevance of these changes in the regulation of protein function(s) and host–pathogen interactions remains largely unexplored.
Acknowledgments
The authors gratefully acknowledge all financial support from the National Institute of Allergy and Infectious Diseases and the National Heart, Lung, and Blood Institute of the NIH through ongoing and completed research project grants.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations of financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References/Website
Papers of special note have been highlighted as:
• of interest
• of considerable interest
- 1.Gillespie JJ, Beier MS, Rahman MS, et al. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS ONE. 2007;2(3):e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Sousa R, Franca A, Doria Nobrega S, et al. Host- and microbe-related risk factors for and pathophysiology of fatal Rickettsia conorii infection in Portuguese patients. J. Infect. Dis. 2008;198(4):576–585. doi: 10.1086/590211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 1997;10(4):694–719. doi: 10.1128/cmr.10.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker DH, Occhino C, Tringali GR, Di Rosa S, Mansueto S. Pathogenesis of rickettsial eschars: the tache noire of boutonneuse fever. Hum. Pathol. 1988;19(12):1449–1454. doi: 10.1016/s0046-8177(88)80238-7. [DOI] [PubMed] [Google Scholar]
- 5.Arguello AP, Hun L, Rivera P, Taylor L. A fatal urban case of Rocky Mountain spotted fever presenting an eschar in San Jose, Costa Rica. Am. J. Trop. Med. Hyg. 2012;87(2):345–348. doi: 10.4269/ajtmh.2012.12-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paddock CD, Sumner JW, Comer JA, et al. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 2004;38(6):805–811. doi: 10.1086/381894. [DOI] [PubMed] [Google Scholar]
- 7.Dumler JS, Walker DH. Rocky Mountain spotted fever–changing ecology and persisting virulence. N. Engl. J. Med. 2005;353(6):551–553. doi: 10.1056/NEJMp058138. [DOI] [PubMed] [Google Scholar]
- 8.Badiaga S, Brouqui P. Human louse-transmitted infectious diseases. Clin. Microbiol. Infect. 2012;18(4):332–337. doi: 10.1111/j.1469-0691.2012.03778.x. [DOI] [PubMed] [Google Scholar]
- 9.Richards AL. Worldwide detection and identification of new and old rickettsiae and rickettsial diseases. FEMS Immunol. Med. Microbiol. 2012;64(1):107–110. doi: 10.1111/j.1574-695X.2011.00875.x. [DOI] [PubMed] [Google Scholar]
- 10.Azad AF, Radulovic S. Pathogenic rickettsiae as bioterrorism agents. Ann. NY Acad. Sci. 2003;990:734–738. doi: 10.1111/j.1749-6632.2003.tb07452.x. [DOI] [PubMed] [Google Scholar]
- 11.Walker DH. Principles of the malicious use of infectious agents to create terror: reasons for concern for organisms of the genus Rickettsia. Ann. NY Acad. Sci. 2003;990:739–742. doi: 10.1111/j.1749-6632.2003.tb07453.x. [DOI] [PubMed] [Google Scholar]
- 12.Winkler HH. Rickettsia species (as organisms). Ann. Rev. Microbiol. 1990;44:131–153. doi: 10.1146/annurev.mi.44.100190.001023. [DOI] [PubMed] [Google Scholar]
- 13.Young MR. Endothelial cells in the eyes of an immunologist. Cancer Immunol. Immunother. 2012;61(10):1609–1616. doi: 10.1007/s00262-012-1335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14■.Mansueto P, Vitale G, Cascio A, et al. New insight into immunity and immunopathology of rickettsial diseases. Clin. Dev. Immunol. 2012;2012:967852. doi: 10.1155/2012/967852. [Summary of our current understanding of the mechanisms of pathogenesis and immunity during rickettsial infections.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan YG, Riley SP, Martinez JJ. Adherence to and invasion of host cells by spotted fever group Rickettsia species. Front. Microbiol. 2010;1:139. doi: 10.3389/fmicb.2010.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botelho-Nevers E, Socolovschi C, Raoult D, Parola P. Treatment of Rickettsia spp. infections: a review. Expert Rev. Anti Infect. Ther. 2012;10(12):1425–1437. doi: 10.1586/eri.12.139. [DOI] [PubMed] [Google Scholar]
- 17.Palmer GH, Azad AF, editors. Intracellular pathogens II: Rickettsiales. ASM Press; Washington DC, USA: 2012. pp. 1–443. [Google Scholar]
- 18.Walker TS, Winkler HH. Penetration of cultured mouse fibroblasts (L cells) by Rickettsia prowazeki. Infect. Immun. 1978;22(1):200–208. doi: 10.1128/iai.22.1.200-208.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker TS. Rickettsial interactions with human endothelial cells in vitro: adherence and entry. Infect. Immun. 1984;44(2):205–210. doi: 10.1128/iai.44.2.205-210.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 1998;24(5):289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 21.Renesto P, Samson L, Ogata H, et al. Identification of two putative rickettsial adhesins by proteomic analysis. Res. Microbiol. 2006;157(7):605–612. doi: 10.1016/j.resmic.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YG, Martinez JJ. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 2010;78(5):1895–1904. doi: 10.1128/IAI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect. Immun. 2009;77(12):5272–5280. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 2010;12(11):1057–1063. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park H, Lee JH, Gouin E, Cossart P, Izard T. The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J. Biol. Chem. 2011;286(40):35096–35103. doi: 10.1074/jbc.M111.263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez JJ, Seveau S, Veiga E, Matsuyama S, Cossart P. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123(6):1013–1023. doi: 10.1016/j.cell.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 27.Martinez JJ. Invasion of the mammalian host: early events at the cellular and molecular levels. In: Palmer GH, Azad AF, editors. Intracellular Pathogens II: Rickettsiales. ASM Press; Washington, DC, USA: 2012. pp. 142–153. [Google Scholar]
- 28.Chan YG, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 2009;11(4):629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J. Cell. Sci. 2004;117(Pt 21):5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- 30.Rydkina E, Turpin LC, Sahni SK. Activation of p38 mitogen-activated protein kinase module facilitates in vitro host cell invasion by Rickettsia rickettsii. J. Med. Microbiol. 2008;57(Pt 9):1172–1175. doi: 10.1099/jmm.0.47806-0. [DOI] [PubMed] [Google Scholar]
- 31■.Hillman RD, Jr, Baktash YM, Martinez JJ. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with alpha2beta1 integrin. Cell. Microbiol. 2012;15(5):727–741. doi: 10.1111/cmi.12068. [Documents the role of host cell surface integrins in rickettsial adhesion and invasion. The C-terminal region of OmpA, a surface cell antigen (Sca0) present only in spotted fever group is shown to interact with α2β1 integrin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radulovic S, Troyer JM, Beier MS, Lau AO, Azad AF. Identification and molecular analysis of the gene encoding Rickettsia typhi hemolysin. Infect. Immun. 1999;67(11):6104–6108. doi: 10.1128/iai.67.11.6104-6108.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renesto P, Dehoux P, Gouin E, Touqui L, Cossart P, Raoult D. Identification and characterization of a phospholipase D-superfamily gene in rickettsiae. J. Infect. Dis. 2003;188(9):1276–1283. doi: 10.1086/379080. [DOI] [PubMed] [Google Scholar]
- 34.Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 2005;73(10):6668–6673. doi: 10.1128/IAI.73.10.6668-6673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman DJ, Santucci LA, Meyers N, Sekeyova Z. Penetration of host cells by Rickettsia rickettsii appears to be mediated by a phospholipase of rickettsial origin. Infect. Immun. 1992;60(7):2733–2740. doi: 10.1128/iai.60.7.2733-2740.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman MS, Ammerman NC, Sears KT, Ceraul SM, Azad AF. Functional characterization of a phospholipase A2 homolog from Rickettsia typhi. J. Bacteriol. 2010;192(13):3294–3303. doi: 10.1128/JB.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahman MS, Gillespie JJ, Kaur SJ, et al. Rickettsia typhi possesses phospholipase A2 enzymes that are involved in infection of host cells. PLoS Pathog. 2013;9(6):e1003399. doi: 10.1371/journal.ppat.1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clifton DR, Rydkina E, Huyck H, et al. Expression and secretion of chemotactic cytokines IL-8 and MCP-1 by human endothelial cells after Rickettsia rickettsii infection: regulation by nuclear transcription factor NF-kappaB. Int. J. Med. Microbiol. 2005;295(4):267–278. doi: 10.1016/j.ijmm.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Walker DH. Targeting Rickettsia. N. Engl. J. Med. 2006;354(13):1418–1420. doi: 10.1056/NEJMcibr055756. [DOI] [PubMed] [Google Scholar]
- 40.Walker DH, Popov VL, Wen J, Feng HM. Rickettsia conorii infection of C3H/HeN mice. A model of endothelial-target rickettsiosis. Lab. Invest. 1994;70(3):358–368. [PubMed] [Google Scholar]
- 41.Walker DH, Popov VL, Feng HM. Establishment of a novel endothelial target mouse model of a typhus group rickettsiosis: evidence for critical roles for gamma interferon and CD8 T lymphocytes. Lab. Invest. 2000;80(9):1361–1372. doi: 10.1038/labinvest.3780144. [DOI] [PubMed] [Google Scholar]
- 42.Olano JP. Rickettsial infections. Ann. NY Acad. Sci. 2005;1063:187–196. doi: 10.1196/annals.1355.031. [DOI] [PubMed] [Google Scholar]
- 43.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat. Rev. Microbiol. 2008;6(5):375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- 44.Hackstadt T. The biology of rickettsiae. Infect. Agents Dis. 1996;5(3):127–143. [PubMed] [Google Scholar]
- 45.Heinzen RA. Rickettsial actin-based motility: behavior and involvement of cytoskeletal regulators. Ann. NY Acad. Sci. 2003;990:535–547. doi: 10.1111/j.1749-6632.2003.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 46.Jeng RL, Goley ED, D'Alessio JA, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell. Microbiol. 2004;6(8):761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 47.Gouin E, Egile C, Dehoux P, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427(6973):457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- 48.Simser JA, Rahman MS, Dreher-Lesnick SM, Azad AF. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol. Microbiol. 2005;58(1):71–79. doi: 10.1111/j.1365-2958.2005.04806.x. [DOI] [PubMed] [Google Scholar]
- 49.Balraj P, El Karkouri K, Vestris G, Espinosa L, Raoult D, Renesto P. RickA expression is not sufficient to promote actin-based motility of Rickettsia raoultii. PLoS ONE. 2008;3(7):e2582. doi: 10.1371/journal.pone.0002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogata H, La Scola B, Audic S, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2(5):e76. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinzen RA, Hayes SF, Peacock MG, Hackstadt T. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect. Immun. 1993;61(5):1926–1935. doi: 10.1128/iai.61.5.1926-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogata H, Renesto P, Audic S, et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3(8):e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch MD, Reed SCO, Haglund CM. Establishing intracellular infection: escape from the phagosome and intracellular colonization (Rickettsiaceae). In: Palmer GH, Azad AF, editors. Intracellular Pathogens II: Rickettsiales. ASM Press; Washington, DC, USA: 2012. pp. 154–174. [Google Scholar]
- 54.Gouin E, Gantelet H, Egile C, et al. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell. Sci. 1999;112(Pt 11):1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- 55.Harlander RS, Way M, Ren Q, Howe D, Grieshaber SS, Heinzen RA. Effects of ectopically expressed neuronal Wiskott–Aldrich syndrome protein domains on Rickettsia rickettsii actin-based motility. Infect. Immun. 2003;71(3):1551–1556. doi: 10.1128/IAI.71.3.1551-1556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe. 2010;7(5):388–398. doi: 10.1016/j.chom.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 2010;78(5):2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58■■.Alix E, Chesnel L, Bowzard BJ, et al. The capping domain in RalF regulates effector functions. PLoS Pathog. 2012;8(11):e1003012. doi: 10.1371/journal.ppat.1003012. [Suggests independent evolution of bacterial effector (RalF) in Rickettsia prowazekii, and further establishes that RalF modules actin dynamics via its interactions with the host plasma membrane.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voth DE, Broederdorf LJ, Graham JG. Bacterial type IV secretion systems: versatile virulence machines. Future Microbiol. 2012;7(2):241–257. doi: 10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLeod MP, Qin X, Karpathy SE, et al. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 2004;186(17):5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, et al. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE. 2009;4(3):e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sutten EL, Norimine J, Beare PA, et al. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect. Immun. 2010;78(3):1314–1325. doi: 10.1128/IAI.01207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gillespie JJ, Brayton KA, Williams KP, et al. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 2010;78(5):1809–1823. doi: 10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ammerman NC, Rahman MS, Azad AF. Characterization of SEC-translocon-dependent extracytoplasmic proteins of Rickettsia typhi. J. Bacteriol. 2008;190(18):6234–6242. doi: 10.1128/JB.00794-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blanc G, Ogata H, Robert C, Audic S, Claverie JM, Raoult D. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 2007;17(11):1657–1664. doi: 10.1101/gr.6742107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillespie JJ, Williams K, Shukla M, et al. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE. 2008;3(4):e2018. doi: 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295(5555):679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 68■■.Kaur SJ, Rahman MS, Ammerman NC, et al. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J. Bacteriol. 2012;194(18):4920–4932. doi: 10.1128/JB.00793-12. [First evidence for the role of a rickettsial SEC–TolC pathway in the delivery of RARP-1, an ankyrin repeat containing protein, into the host cytosol. This study also shows a critical role for the C-terminal ankyrin repeat region in RARP-1 secretion by Rickettsia typhi.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sahni SK, Rydkina E. Host–cell interactions with pathogenic Rickettsia species. Future Microbiol. 2009;4(3):323–339. doi: 10.2217/FMB.09.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahni SK, Rydkina E, Simpson-Haidaris PJ. Innate immune response and inflammation: roles in pathogenesis and protection (Rickettsiaceae). In: Palmer GH, Azad AF, editors. Intracellular Pathogens II: Rickettsiales. ASM Press; Washington, DC, USA: 2012. pp. 243–269. [Google Scholar]
- 71.Schroder K, Hertzog PJ, Ravasi T, Hume DA. nterferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75(2):163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 72.Turco J, Winkler HH. Interferon-alpha/beta and Rickettsia prowazekii: induction and sensitivity. Ann. NY Acad. Sci. 1990;590:168–186. doi: 10.1111/j.1749-6632.1990.tb42219.x. [DOI] [PubMed] [Google Scholar]
- 73.Turco J, Winkler HH. Selection of alpha/beta interferon- and gamma interferon-resistant rickettsiae by passage of Rickettsia prowazekii in L929 cells. Infect. Immun. 1990;58(10):3279–3285. doi: 10.1128/iai.58.10.3279-3285.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turco J, Winkler HH. Effect of mouse lymphokines and cloned mouse interferon-gamma on the interaction of Rickettsia prowazekii with mouse macrophage-like RAW264.7 cells. Infect. Immun. 1984;45(2):303–308. doi: 10.1128/iai.45.2.303-308.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turco J, Winkler HH. Gamma-interferon-induced inhibition of the growth of Rickettsia prowazekii in fibroblasts cannot be explained by the degradation of tryptophan or other amino acids. Infect. Immun. 1986;53(1):38–46. doi: 10.1128/iai.53.1.38-46.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manor E, Sarov I. Inhibition of Rickettsia conorii growth by recombinant tumor necrosis factor alpha: enhancement of inhibition by gamma interferon. Infect. Immun. 1990;58(6):1886–1890. doi: 10.1128/iai.58.6.1886-1890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, Jerrells TR, Spitalny GL, Walker DH. Gamma interferon as a crucial host defense against Rickettsia conorii in vivo. Infect. Immun. 1987;55(5):1252–1255. doi: 10.1128/iai.55.5.1252-1255.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walker DH, Olano JP, Feng HM. Critical role of cytotoxic T lymphocytes in immune clearance of rickettsial infection. Infect. Immun. 2001;69(3):1841–1846. doi: 10.1128/IAI.69.3.1841-1846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng HM, Popov VL, Walker DH. Depletion of gamma interferon and tumor necrosis factor alpha in mice with Rickettsia conorii-infected endothelium: impairment of rickettsicidal nitric oxide production resulting in fatal, overwhelming rickettsial disease. Infect. Immun. 1994;62(5):1952–1960. doi: 10.1128/iai.62.5.1952-1960.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Billings AN, Feng HM, Olano JP, Walker DH. Rickettsial infection in murine models activates an early anti-rickettsial effect mediated by NK cells and associated with production of gamma interferon. Am. J. Trop. Med. Hyg. 2001;65(1):52–56. doi: 10.4269/ajtmh.2001.65.52. [DOI] [PubMed] [Google Scholar]
- 81.Vitale G, Mansueto S, Gambino G, et al. The acute phase response in Sicilian patients with boutonneuse fever admitted to hospitals in Palermo, 1992–1997. J. Infect. 2001;42(1):33–39. doi: 10.1053/jinf.2000.0758. [DOI] [PubMed] [Google Scholar]
- 82.Mahara F. Japanese spotted fever: report of 31 cases and review of the literature. Emerg. Infect. Dis. 1997;3(2):105–111. doi: 10.3201/eid0302.970203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwasaki H, Mahara F, Takada N, Fujita H, Ueda T. Fulminant Japanese spotted fever associated with hypercytokinemia. J. Clin. Microbiol. 2001;39(6):2341–2343. doi: 10.1128/JCM.39.6.2341-2343.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jensenius M, Ueland T, Fournier PE, et al. Systemic inflammatory responses in African tick-bite fever. J. Infect. Dis. 2003;187(8):1332–1336. doi: 10.1086/368415. [DOI] [PubMed] [Google Scholar]
- 85.de Sousa R, Ismail N, Nobrega SD, et al. Intralesional expression of mRNA of interferon-gamma, tumor necrosis factor-alpha, interleukin-10, nitric oxide synthase, indoleamine-2,3-dioxygenase, and RANTES is a major immune effector in Mediterranean spotted fever rickettsiosis. J. Infect. Dis. 2007;196(5):770–781. doi: 10.1086/519739. [DOI] [PubMed] [Google Scholar]
- 86.Heim MH. Intracellular signalling and antiviral effects of interferons. Dig. Liver Dis. 2000;32(3):257–263. doi: 10.1016/s1590-8658(00)80831-2. [DOI] [PubMed] [Google Scholar]
- 87.Meraz MA, White JM, Sheehan KC, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK–STAT signaling pathway. Cell. 1996;84(3):431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 88.Dupuis S, Dargemont C, Fieschi C, et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293(5528):300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 89.Dupuis S, Jouanguy E, Al-Hajjar S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003;33(3):388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 90.Miller DM, Cebulla CM, Sedmak DD. Human cytomegalovirus inhibition of major histocompatibility complex transcription and interferon signal transduction. Curr. Top. Microbiol. Immunol. 2002;269:153–170. doi: 10.1007/978-3-642-59421-2_10. [DOI] [PubMed] [Google Scholar]
- 91.Larrea E, Aldabe R, Molano E, et al. Altered expression and activation of signal transducers and activators of transcription (STATs) in hepatitis C virus infection: in vivo and in vitro studies. Gut. 2006;55(8):1188–1196. doi: 10.1136/gut.2005.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lad SP, Fukuda EY, Li J, De La Maza LM, Li E. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J. Immunol. 2005;174(11):7186–7193. doi: 10.4049/jimmunol.174.11.7186. [DOI] [PubMed] [Google Scholar]
- 93.Sun J, Shi YH, Le GW, Ma XY. Distinct immune response induced by peptidoglycan derived from Lactobacillus sp. World J. Gastroenterol. 2005;11(40):6330–6337. doi: 10.3748/wjg.v11.i40.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee EH, Rikihisa Y. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 1998;66(6):2514–2520. doi: 10.1128/iai.66.6.2514-2520.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang JZ, Sinha M, Luxon BA, Yu XJ. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 2004;72(1):498–507. doi: 10.1128/IAI.72.1.498-507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ceponis PJ, McKay DM, Ching JC, Pereira P, Sherman PM. Enterohemorrhagic Escherichia coli O157:H7 disrupts Stat1-mediated gamma interferon signal transduction in epithelial cells. Infect. Immun. 2003;71(3):1396–1404. doi: 10.1128/IAI.71.3.1396-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sahni SK, Kiriakidi S, Colonne MP, Sahni A, Silverman DJ. Selective activation of signal transducer and activator of transcription (STAT) proteins STAT1 and STAT3 in human endothelial cells infected with Rickettsia rickettsii. Clin. Microbiol. Infect. 2009;15(Suppl. 2):303–304. doi: 10.1111/j.1469-0691.2008.02248.x. [DOI] [PubMed] [Google Scholar]
- 98■■.Colonne PM, Eremeeva ME, Sahni SK. Beta interferon-mediated activation of signal transducer and activator of transcription protein 1 interferes with Rickettsia conorii replication in human endothelial cells. Infect. Immun. 2011;79(9):3733–3743. doi: 10.1128/IAI.05008-11. [Implicates IFN-β, a type I interferon, in autocrine/paracrine stimulation of host endothelium to trigger JAK–STAT signaling and downstream ‘antiviral’-like innate immune responses, such as the induction of ISG15 and other genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129(6):1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Colonne PM, Sahni A, Sahni SK. Suppressor of cytokine signaling protein SOCS1 and UBP43 regulate the expression of type I interferon-stimulated genes in human microvascular endothelial cells infected with Rickettsia conorii. J. Med. Microbiol. 2013;62(Pt 7):968–979. doi: 10.1099/jmm.0.054502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Colonne PM, Sahni A, Sahni SK. Rickettsia conorii infection stimulates the expression of ISG15 and ISG15 protease UBP43 in human microvascular endothelial cells. Biochem. Biophys. Res. Commun. 2011;416(1–2):153–158. doi: 10.1016/j.bbrc.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao C, Collins MN, Hsiang TY, Krug RM. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 2013;21(4):181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feng HM, Walker DH. Mechanisms of intracellular killing of Rickettsia conorii in infected human endothelial cells, hepatocytes, and macrophages. Infect. Immun. 2000;68(12):6729–6736. doi: 10.1128/iai.68.12.6729-6736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feng HM, Whitworth T, Popov V, Walker DH. Effect of antibody on the Rickettsia–host cell interaction. Infect. Immun. 2004;72(6):3524–3530. doi: 10.1128/IAI.72.6.3524-3530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gonder JC, Kenyon RH, Pedersen CE., Jr Epidemic typhus infection in cynomolgus monkeys (Macaca fascicularis). Infect. Immun. 1980;30(1):219–223. doi: 10.1128/iai.30.1.219-223.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bechah Y, Capo C, Grau GE, Raoult D, Mege JL. A murine model of infection with Rickettsia prowazekii: implications for pathogenesis of epidemic typhus. Microbes Infect. 2007;9(7):898–906. doi: 10.1016/j.micinf.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 107.Driskell LO, Yu XJ, Zhang L, et al. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect. Immun. 2009;77(8):3244–3248. doi: 10.1128/IAI.00395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Valbuena G, Feng HM, Walker DH. Mechanisms of immunity against rickettsiae. New perspectives and opportunities offered by unusual intracellular parasites. Microbes Infect. 2002;4(6):625–633. doi: 10.1016/s1286-4579(02)01581-2. [DOI] [PubMed] [Google Scholar]
- 109.Valbuena G, Bradford W, Walker DH. Expression analysis of the T-cell-targeting chemokines CXCL9 and CXCL10 in mice and humans with endothelial infections caused by rickettsiae of the spotted fever group. Am. J. Pathol. 2003;163(4):1357–1369. doi: 10.1016/S0002-9440(10)63494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Valbuena G, Walker DH. Effect of blocking the CXCL9/10-CXCR3 chemokine system in the outcome of endothelial-target rickettsial infections. Am. J. Trop. Med. Hyg. 2004;71(4):393–399. [PubMed] [Google Scholar]
- 111.Valbuena G, Jordan JM, Walker DH. T cells mediate cross-protective immunity between spotted fever group rickettsiae and typhus group rickettsiae. J. Infect. Dis. 2004;190(7):1221–1227. doi: 10.1086/423819. [DOI] [PubMed] [Google Scholar]
- 112.Fang R, Ismail N, Soong L, et al. Differential interaction of dendritic cells with Rickettsia conorii: impact on host susceptibility to murine spotted fever rickettsiosis. Infect. Immun. 2007;75(6):3112–3123. doi: 10.1128/IAI.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jordan JM, Woods ME, Feng HM, Soong L, Walker DH. Rickettsiae-stimulated dendritic cells mediate protection against lethal rickettsial challenge in an animal model of spotted fever rickettsiosis. J. Infect. Dis. 2007;196(4):629–638. doi: 10.1086/519686. [DOI] [PubMed] [Google Scholar]
- 114.Sumbayev VV, Yasinska IM. Role of MAP kinase-dependent apoptotic pathway in innate immune responses and viral infection. Scand. J. Immunol. 2006;63(6):391–400. doi: 10.1111/j.1365-3083.2006.001764.x. [DOI] [PubMed] [Google Scholar]
- 115.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13(5):816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 116.Choi KS, Scorpio DG, Dumler JS. Anaplasma phagocytophilum ligation to toll-like receptor (TLR) 2, but not to TLR4, activates macrophages for nuclear factor-kappa B nuclear translocation. J. Infect. Dis. 2004;189(10):1921–1925. doi: 10.1086/386284. [DOI] [PubMed] [Google Scholar]
- 117.Von Loewenich FD, Scorpio DG, Reischl U, Dumler JS, Bogdan C. Frontline: control of Anaplasma phagocytophilum, an obligate intracellular pathogen, in the absence of inducible nitric oxide synthase, phagocyte NADPH oxidase, tumor necrosis factor, Toll-like receptor (TLR)2 and TLR4, or the TLR adaptor molecule MyD88. Eur. J. Immunol. 2004;34(7):1789–1797. doi: 10.1002/eji.200425029. [DOI] [PubMed] [Google Scholar]
- 118.Balistreri CR, Candore G, Lio D, et al. Role of TLR4 receptor polymorphisms in Boutonneuse fever. Int. J. Immunopathol. Pharmacol. 2005;18(4):655–660. doi: 10.1177/039463200501800406. [DOI] [PubMed] [Google Scholar]
- 119.Jordan JM, Woods ME, Olano J, Walker DH. The absence of Toll-like receptor 4 signaling in C3H/HeJ mice predisposes them to overwhelming rickettsial infection and decreased protective Th1 responses. Infect. Immun. 2008;76(8):3717–3724. doi: 10.1128/IAI.00311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jordan JM, Woods ME, Soong L, Walker DH. Rickettsiae stimulate dendritic cells through Toll-like receptor 4, leading to enhanced NK cell activation in vivo. J. Infect. Dis. 2009;199(2):236–242. doi: 10.1086/595833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. Nod-like proteins in inflammation and disease. J. Pathol. 2008;214(2):136–148. doi: 10.1002/path.2271. [DOI] [PubMed] [Google Scholar]
- 122.Pang H, Winkler HH. Analysis of the peptidoglycan of Rickettsia prowazekii. J. Bacteriol. 1994;176(3):923–926. doi: 10.1128/jb.176.3.923-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Andersson SG, Zomorodipour A, Andersson JO, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396(6707):133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 124.Gillespie JJ, Nordberg EK, Azad AF, Sobral BWS. Phylogeny and comparative genomics: the shifting landscape in the genomics era. In: Palmer GH, Azad AF, editors. Intracellular Pathogens II: Rickettsiales. ASM Press; Washington, DC, USA: 2012. pp. 84–141. [Google Scholar]
- 125.Gillespie JJ, Joardar V, Williams KP, et al. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 2012;194(2):376–394. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Merhej V, Raoult D. Rickettsial evolution in the light of comparative genomics. Biol. Rev. Camb. Philos. Soc. 2011;86(2):379–405. doi: 10.1111/j.1469-185X.2010.00151.x. [DOI] [PubMed] [Google Scholar]
- 127.Georgiades K, Raoult D. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin–antitoxin modules. PLoS ONE. 2011;6(3):e17962. doi: 10.1371/journal.pone.0017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311(5768):1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 129.Fuxelius HH, Darby AC, Cho NH, Andersson SG. Visualization of pseudogenes in intracellular bacteria reveals the different tracks to gene destruction. Genome Biol. 2008;9(2):R42. doi: 10.1186/gb-2008-9-2-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakabachi A, Yamashita A, Toh H, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314(5797):267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 131.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16(8):840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 133.Raghavan R, Sage A, Ochman H. Genome-wide identification of transcription start sites yields a novel thermosensing RNA and new cyclic AMP receptor protein-regulated genes in Escherichia coli. J. Bacteriol. 2011;193(11):2871–2874. doi: 10.1128/JB.00398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Felden B, Vandenesch F, Bouloc P, Romby P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 2011;7(3):e1002006. doi: 10.1371/journal.ppat.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Woodard A, Wood DO. Analysis of convergent gene transcripts in the obligate intracellular bacterium Rickettsia prowazekii. PLoS ONE. 2011;6(1):e16537. doi: 10.1371/journal.pone.0016537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Toledo-Arana A, Dussurget O, Nikitas G, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459(7249):950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 137.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38(3):868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 139.Merhej V, Notredame C, Royer-Carenzi M, Pontarotti P, Raoult D. The rhizome of life: the sympatric Rickettsia felis paradigm demonstrates the random transfer of DNA sequences. Mol. Biol. Evol. 2011;28(11):3213–3223. doi: 10.1093/molbev/msr239. [DOI] [PubMed] [Google Scholar]
- 140.Fenn K, Blaxter M. Wolbachia genomes: revealing the biology of parasitism and mutualism. Trends Parasitol. 2006;22(2):60–65. doi: 10.1016/j.pt.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 141.Park J, Kim KJ, Choi KS, Grab DJ, Dumler JS. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 2004;6(8):743–751. doi: 10.1111/j.1462-5822.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- 142.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 143■■.Audoly G, Vincentelli R, Edouard S, et al. Effect of rickettsial toxin VapC on its eukaryotic host. PLoS ONE. 2011;6(10):e26528. doi: 10.1371/journal.pone.0026528. [First evidence for the presence of toxin–antitoxin module(s) in a few Rickettsia species, and identification of VapC as a rickettsial toxin detrimental to mammalian host cells through RNase activity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Georgiades K. Genomics of epidemic pathogens. Clin. Microbiol. Infect. 2012;18(3):213–217. doi: 10.1111/j.1469-0691.2012.03781.x. [DOI] [PubMed] [Google Scholar]
- 145.Renesto P, Rovery C, Schrenzel J, et al. Rickettsia conorii transcriptional response within inoculation eschar. PLoS ONE. 2008;3(11):e3681. doi: 10.1371/journal.pone.0003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Hackstadt T. Limited transcriptional responses of Rickettsia rickettsii exposed to environmental stimuli. PLoS ONE. 2009;4(5):e5612. doi: 10.1371/journal.pone.0005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Audia JP, Patton MC, Winkler HH. DNA microarray analysis of the heat shock transcriptome of the obligate intracytoplasmic pathogen Rickettsia prowazekii. Appl. Environ. Microbiol. 2008;74(24):7809–7812. doi: 10.1128/AEM.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vitorino L, Ze-Ze L, Sousa A, Bacellar F, Tenreiro R. rRNA intergenic spacer regions for phylogenetic analysis of Rickettsia species. Ann. NY Acad. Sci. 2003;990:726–733. doi: 10.1111/j.1749-6632.2003.tb07451.x. [DOI] [PubMed] [Google Scholar]
- 149.Vitorino L, De Sousa R, Bacellar F, Ze-Ze L. Characterization of a tandem repeat polymorphism in Rickettsia strains. J. Med. Microbiol. 2005;54(Pt 9):833–841. doi: 10.1099/jmm.0.45956-0. [DOI] [PubMed] [Google Scholar]
- 150.Vitorino L, De Sousa R, Bacellar F, Ze-Ze L. Automated method based in VNTR analysis for rickettsiae genotyping. Ann. NY Acad. Sci. 2006;1078:582–586. doi: 10.1196/annals.1374.116. [DOI] [PubMed] [Google Scholar]
- 151.Eremeeva ME, Bosserman E, Zambrano M, Demma L, Dasch GA. Molecular typing of novel Rickettsia rickettsii isolates from Arizona. Ann. NY Acad. Sci. 2006;1078:573–577. doi: 10.1196/annals.1374.114. [DOI] [PubMed] [Google Scholar]
- 152.Fournier PE, Raoult D. Identification of rickettsial isolates at the species level using multi-spacer typing. BMC Microbiol. 2007;7:72. doi: 10.1186/1471-2180-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009;7:6. doi: 10.1186/1741-7007-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baldridge GD, Burkhardt N, Herron MJ, Kurtti TJ, Munderloh UG. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl. Environ. Microbiol. 2005;71(4):2095–2105. doi: 10.1128/AEM.71.4.2095-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baldridge GD, Burkhardt NY, Labruna MB, et al. Wide dispersal and possible multiple origins of low-copy-number plasmids in Rickettsia species associated with blood-feeding arthropods. Appl. Environ. Microbiol. 2010;76(6):1718–1731. doi: 10.1128/AEM.02988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fournier PE, El Karkouri K, Leroy Q, et al. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics. 2009;10:166. doi: 10.1186/1471-2164-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dong X, El Karkouri K, Robert C, Raoult D, Fournier PE. Genome sequence of Rickettsia australis, the agent of Queensland tick typhus. J. Bacteriol. 2012;194(18):5129. doi: 10.1128/JB.01117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Munderloh UG, Felsheim RF, Burkhardt NY, et al. The way forward: improving genetic systems. In: Palmer GH, Azad AF, editors. Intracellular Pathogens II: Rickettsiales. ASM Press; Washington, DC, USA: 2012. pp. 416–432. [Google Scholar]
- 159.Driskell LO, Tucker AM, Winkler HH, Wood DO. Rickettsial metK-encoded methionine adenosyltransferase expression in an Escherichia coli metK deletion strain. J. Bacteriol. 2005;187(16):5719–5722. doi: 10.1128/JB.187.16.5719-5722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tucker AM, Winkler HH, Driskell LO, Wood DO. S-adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 2003;185(10):3031–3035. doi: 10.1128/JB.185.10.3031-3035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sahni SK, Rydkina E. Progress in the functional analysis of rickettsial genes through directed mutagenesis of Rickettsia prowazekii phospholipase D. Future Microbiol. 2009;4(10):1249–1253. doi: 10.2217/fmb.09.99. [DOI] [PubMed] [Google Scholar]
- 162.Clark TR, Lackey AM, Kleba B, et al. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J. Bacteriol. 2011;193(18):4993–4995. doi: 10.1128/JB.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Clark TR, Ellison DW, Kleba B, Hackstadt T. Complementation of Rickettsia rickettsii RelA/SpoT restores a nonlytic plaque phenotype. Infect. Immun. 2011;79(4):1631–1637. doi: 10.1128/IAI.00048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164■.Burkhardt NY, Baldridge GD, Williamson PC, et al. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS ONE. 2011;6(12):e29511. doi: 10.1371/journal.pone.0029511. [Development of vectors based on Rickettsia amyblyommii plasmids; also offers a proof-of-principle that Rickettsia can be stably transformed with extrachromosomal elements.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165■■.Wood DO, Hines A, Tucker AM, et al. Establishment of a replicating plasmid in Rickettsia prowazekii. PLoS ONE. 2012;7(4):e34715. doi: 10.1371/journal.pone.0034715. [The first study demonstrating replication of a plasmid in R. prowazekii, thereby providing an important genetic tool for elucidating the function(s) of key proteins involved in pathogenesis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu ZM, Tucker AM, Driskell LO, Wood DO. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 2007;73(20):6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Welch MD, Reed SC, Lamason RL, Serio AW. Expression of an epitope-tagged virulence protein in Rickettsia parkeri using transposon insertion. PLoS ONE. 2012;7(5):e37310. doi: 10.1371/journal.pone.0037310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pornwiroon W, Bourchookarn A, Paddock CD, Macaluso KR. Proteomic analysis of Rickettsia parkeri strain Portsmouth. Infect. Immun. 2009;77(12):5262–5271. doi: 10.1128/IAI.00911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tucker AM, Driskell LO, Pannell LK, Wood DO. Differential proteomic analysis of Rickettsia prowazekii propagated in diverse host backgrounds. Appl. Environ. Microbiol. 2011;77(14):4712–4718. doi: 10.1128/AEM.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kobir A, Shi L, Boskovic A, Grangeasse C, Franjevic D, Mijakovic I. Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta. 2011;1810(10):989–994. doi: 10.1016/j.bbagen.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 171.Abeykoon AH, Chao CC, Wang G, Gucek M, Yang DC, Ching WM. Two protein lysine methyltransferases methylate outer membrane protein B from Rickettsia. J. Bacteriol. 2012;194(23):6410–6418. doi: 10.1128/JB.01379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Subramanian S, Abendroth J, Phan IQ, et al. Structure of 3-ketoacyl-(acyl-carrier-protein) reductase from Rickettsia prowazekii at 2.25 A resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67(Pt 9):1118–1122. doi: 10.1107/S1744309111030673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Phan I, Subramanian S, Olsen C, et al. Structure of fumarate hydratase from Rickettsia prowazekii, the agent of typhus and suspected relative of the mitochondria. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67(Pt 9):1123–1128. doi: 10.1107/S174430911102690X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gerarden KP, Fuchs AM, Koch JM, et al. Solution structure of the cold-shock-like protein from Rickettsia rickettsii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68(Pt 11):1284–1288. doi: 10.1107/S174430911203881X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Myers WF, Provost PJ, Wisseman CL., Jr Permeability properties of Rickettsia mooseri. J. Bacteriol. 1967;93(3):950–960. doi: 10.1128/jb.93.3.950-960.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Audia JP, Winkler HH. Study of the five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transports ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J. Bacteriol. 2006;188(17):6261–6268. doi: 10.1128/JB.00371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Frohlich KM, Roberts RA, Housley NA, Audia JP. Rickettsia prowazekii uses an snglycerol-3-phosphate dehydrogenase and a novel dihydroxyacetone phosphate transport system to supply triose phosphate for phospholipid biosynthesis. J. Bacteriol. 2010;192(17):4281–4288. doi: 10.1128/JB.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rovery C, Renesto P, Crapoulet N, et al. Transcriptional response of Rickettsia conorii exposed to temperature variation and stress starvation. Res. Microbiol. 2005;156(2):211–218. doi: 10.1016/j.resmic.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 179.Gomes MT, Campos PC, Oliveira FS, et al. Critical role of ASC inflammasomes and bacterial type IV secretion system in caspase-1 activation and host innate resistance to Brucella abortus infection. J. Immunol. 2013;190(7):3629–3638. doi: 10.4049/jimmunol.1202817. [DOI] [PubMed] [Google Scholar]
- 180.Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14(6):1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Weinert LA, Welch JJ, Jiggins FM. Conjugation genes are common throughout the genus Rickettsia and are transmitted horizontally. Proc. Biol. Sci. 2009;276(1673):3619–3627. doi: 10.1098/rspb.2009.0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kelkar YD, Ochman H. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics. 2013;193(1):303–307. doi: 10.1534/genetics.112.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gillespie JJ, Wattam AR, Cammer SA, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect. Immun. 2011;79(11):4286–4298. doi: 10.1128/IAI.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Seattle Structural Genomics Center for Infectious Disease www.ssgcid.org.